Among the 64k molecules predicted to be active against SARS viruses, there are 2700 hitting on 87 different ion channel types
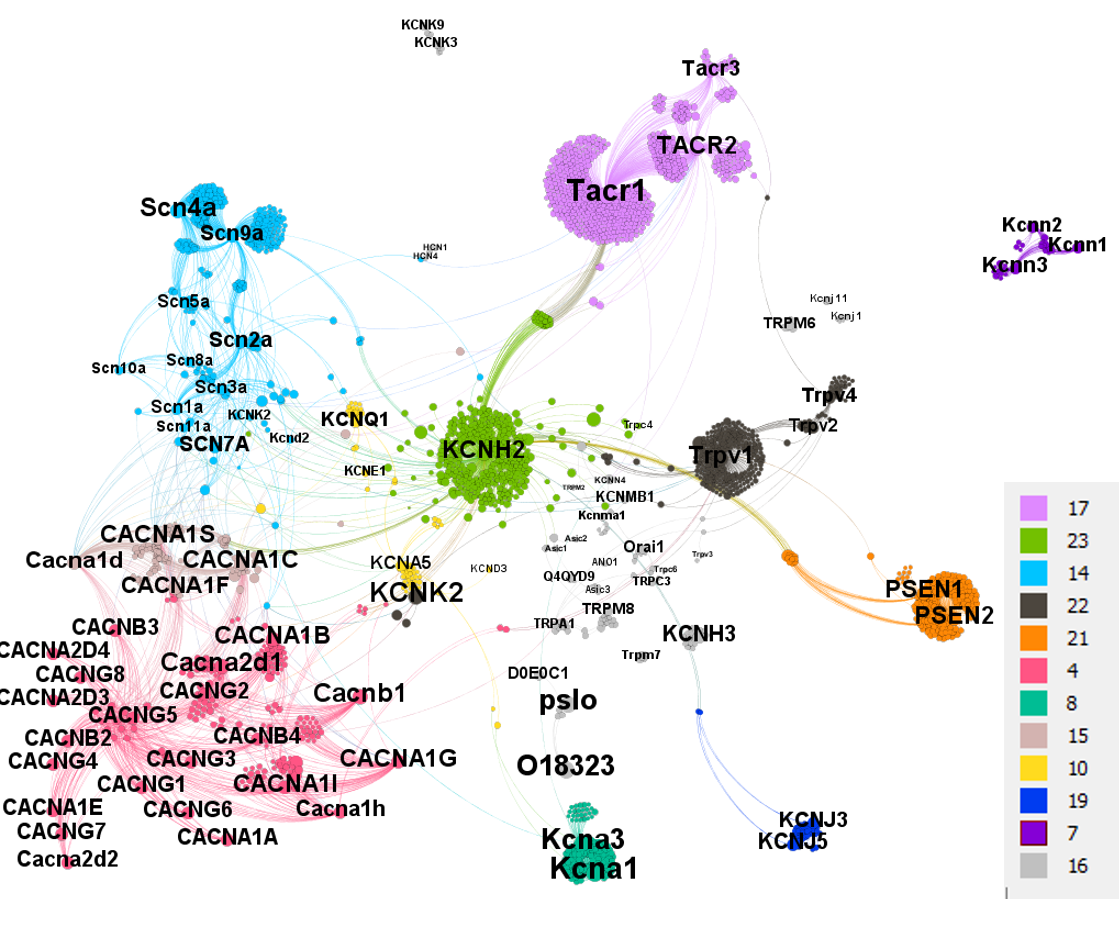
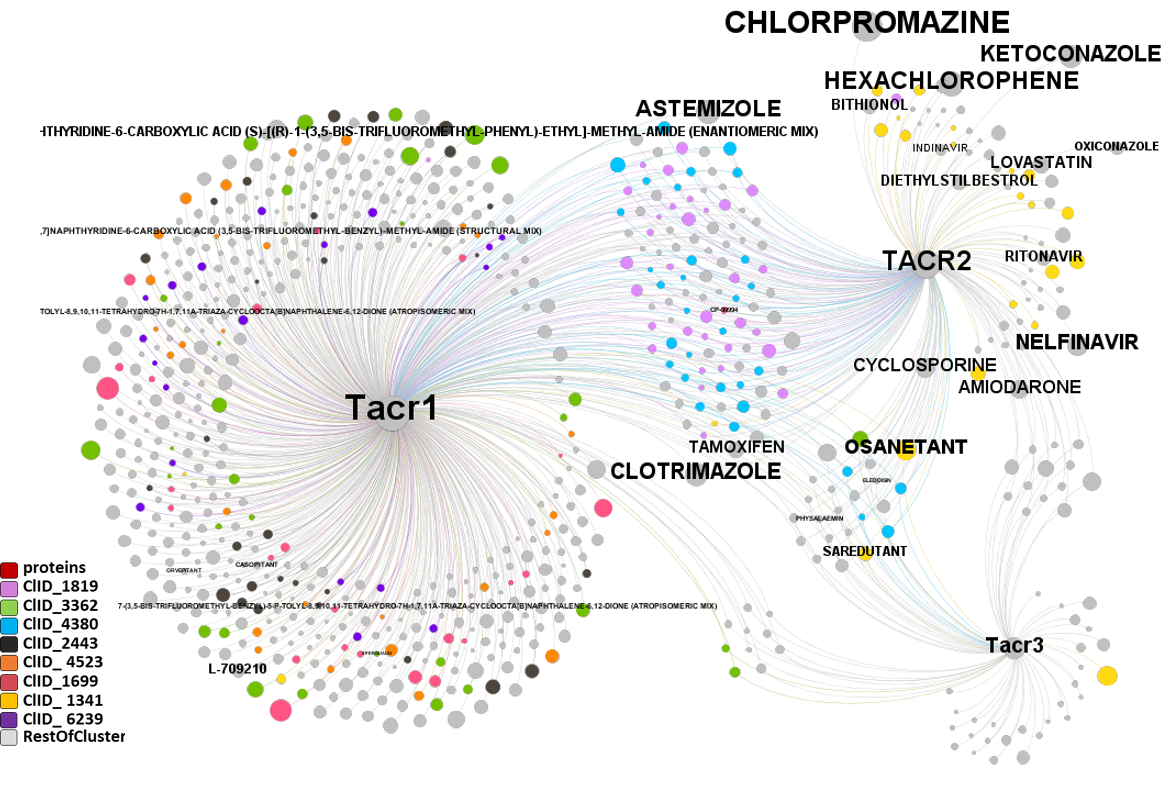
The modularity analysis identifies two additional non ion channel protein groups, tachykinin receptors and presenilin units, connected to ion channels network by common interacting compounds
Presenilins highlighted in ClID_2972 compounds interactions in the proteases section
Find below the interaction networks between compounds and proteins.
Just click on each chart to get an expanded view.
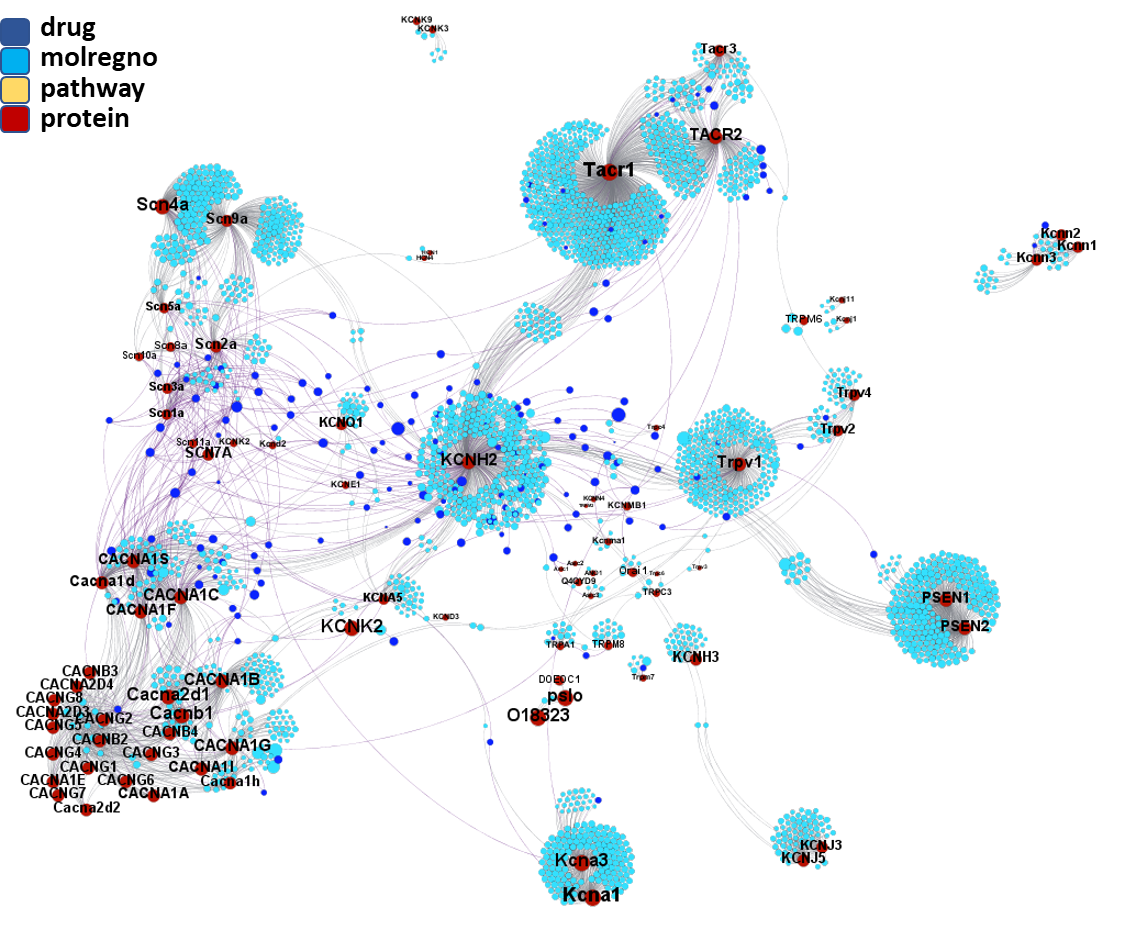
Compounds-ion channels interaction network. Compound nodes are colored in blue, dark blue when corresponds to a known drug. Drug names and molecule registry numbers (molregno) have been removed to facilitate the visualization of the graph.
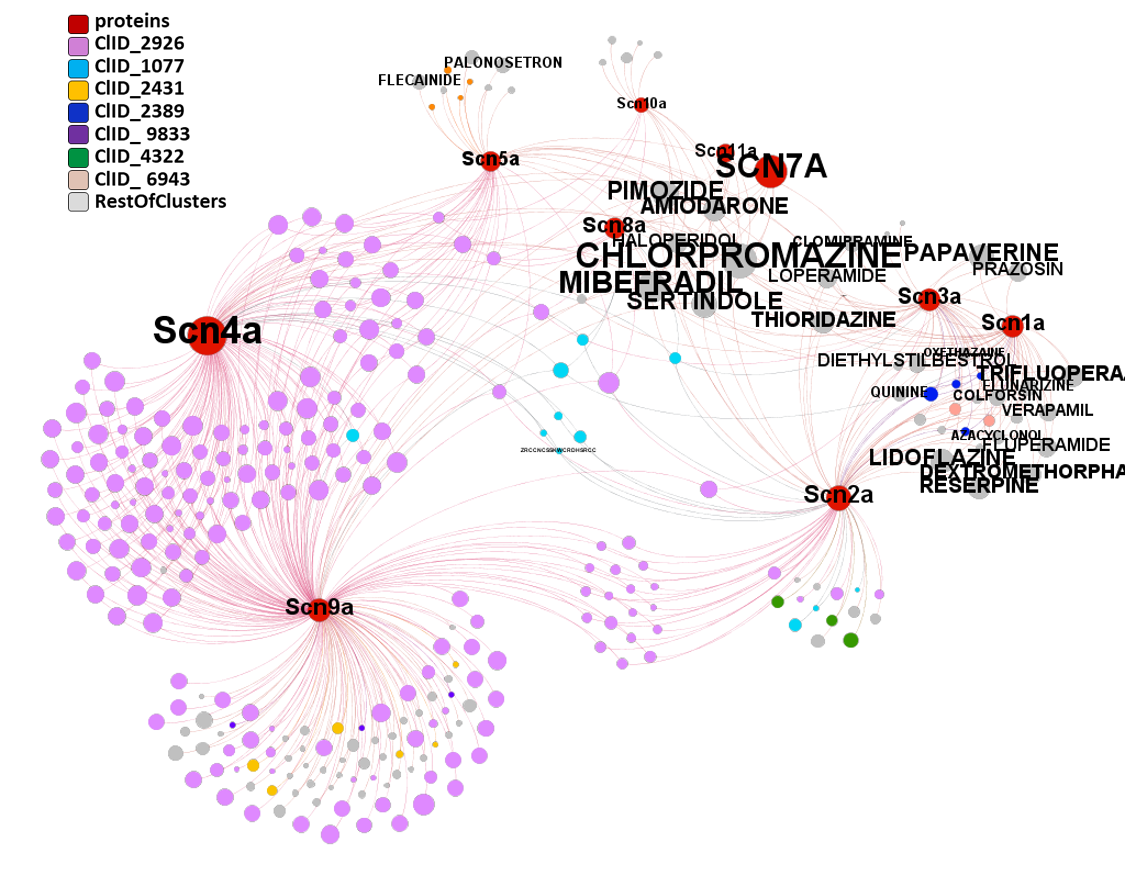
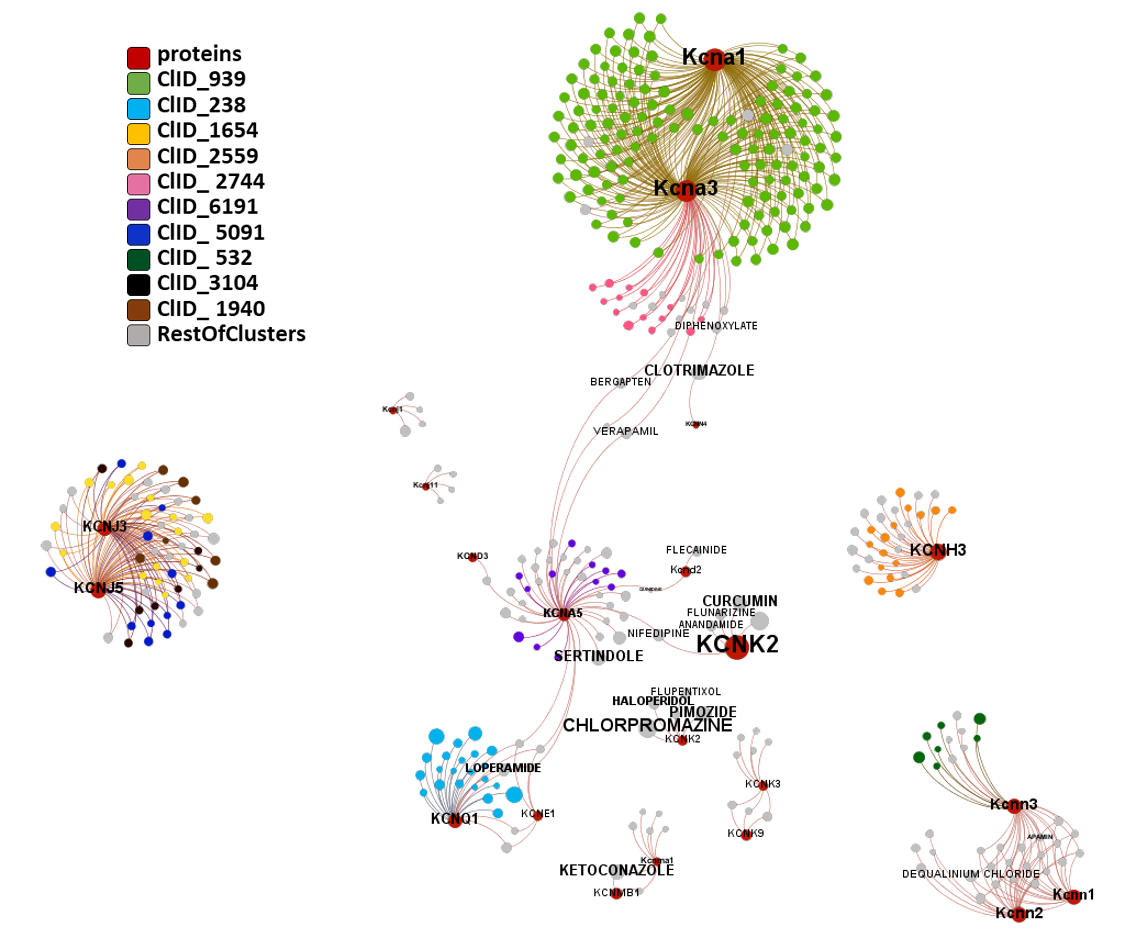
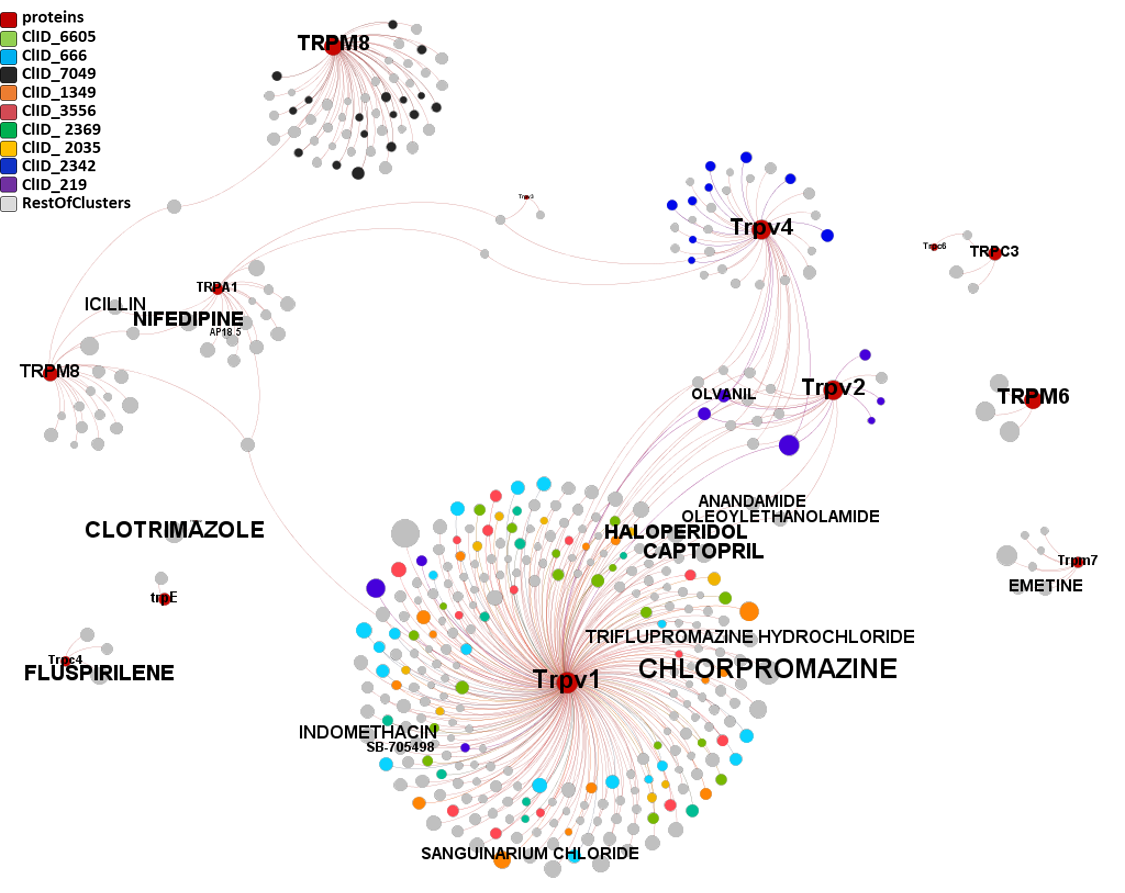
Segmentation based on interactions based on the graph on this left. The procedure discriminates specific separate groups for voltage-gated sodium channels, voltage-gated calcium channels and transient receptor potential channels. Voltage-gated potassium channels appear in different groups. Tachykinin receptors and presenilins appear as associated non ion channel groups by modularity analysis.
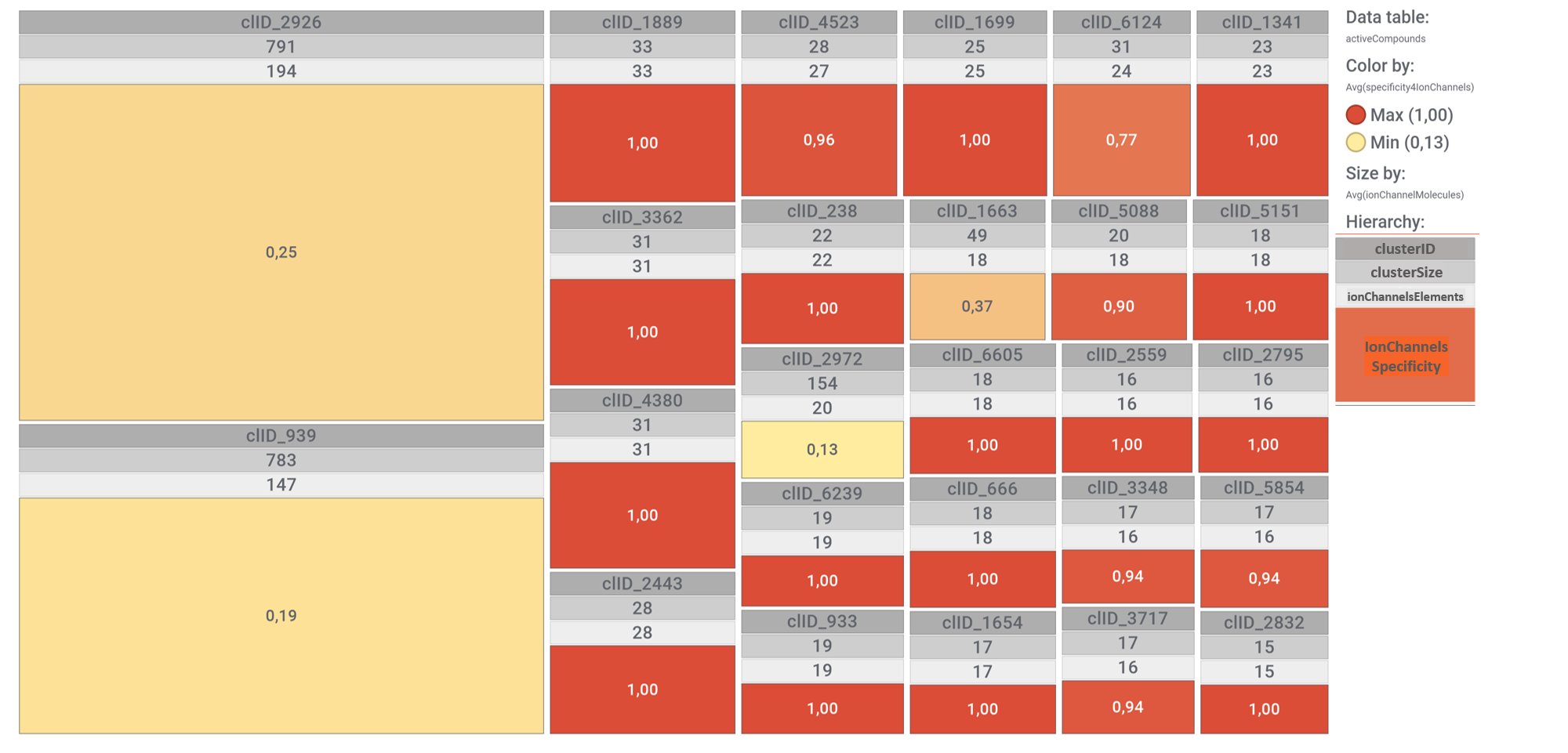
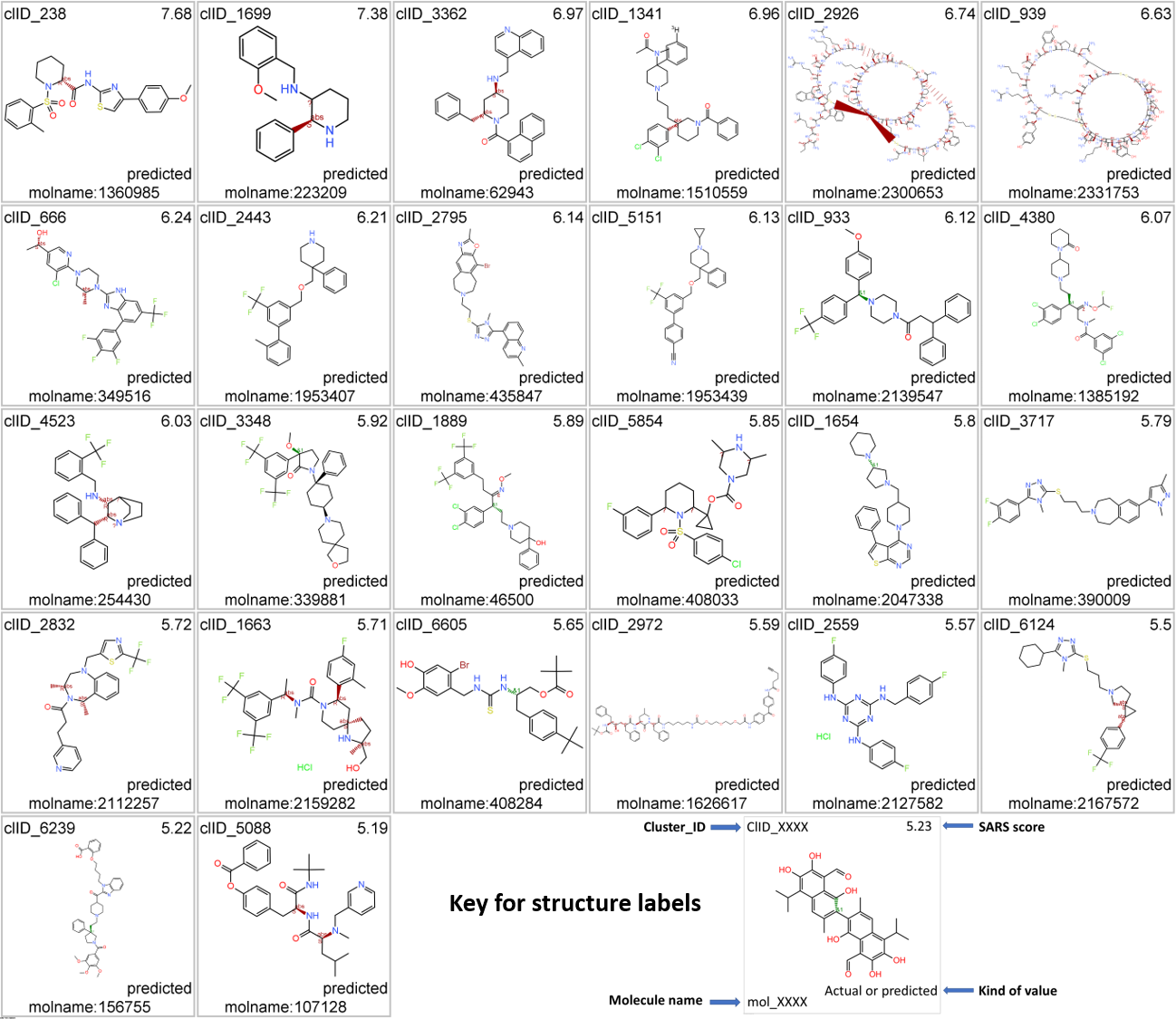
All predicted SARS active molecules have been clustered by their structural analogy. The tree map shows most representative ion channel molecular clusters attending to the number of molecules interacting with ion channels (ionChannelElements in the tree map 3rd level hierarchy) and the ionChannelsSpecificity, calculated as ionChannelElements /clusterSize.
Now, let’s have a more specific look to this interactions. There is one for each module obtained by segmentation based on closest interactions.