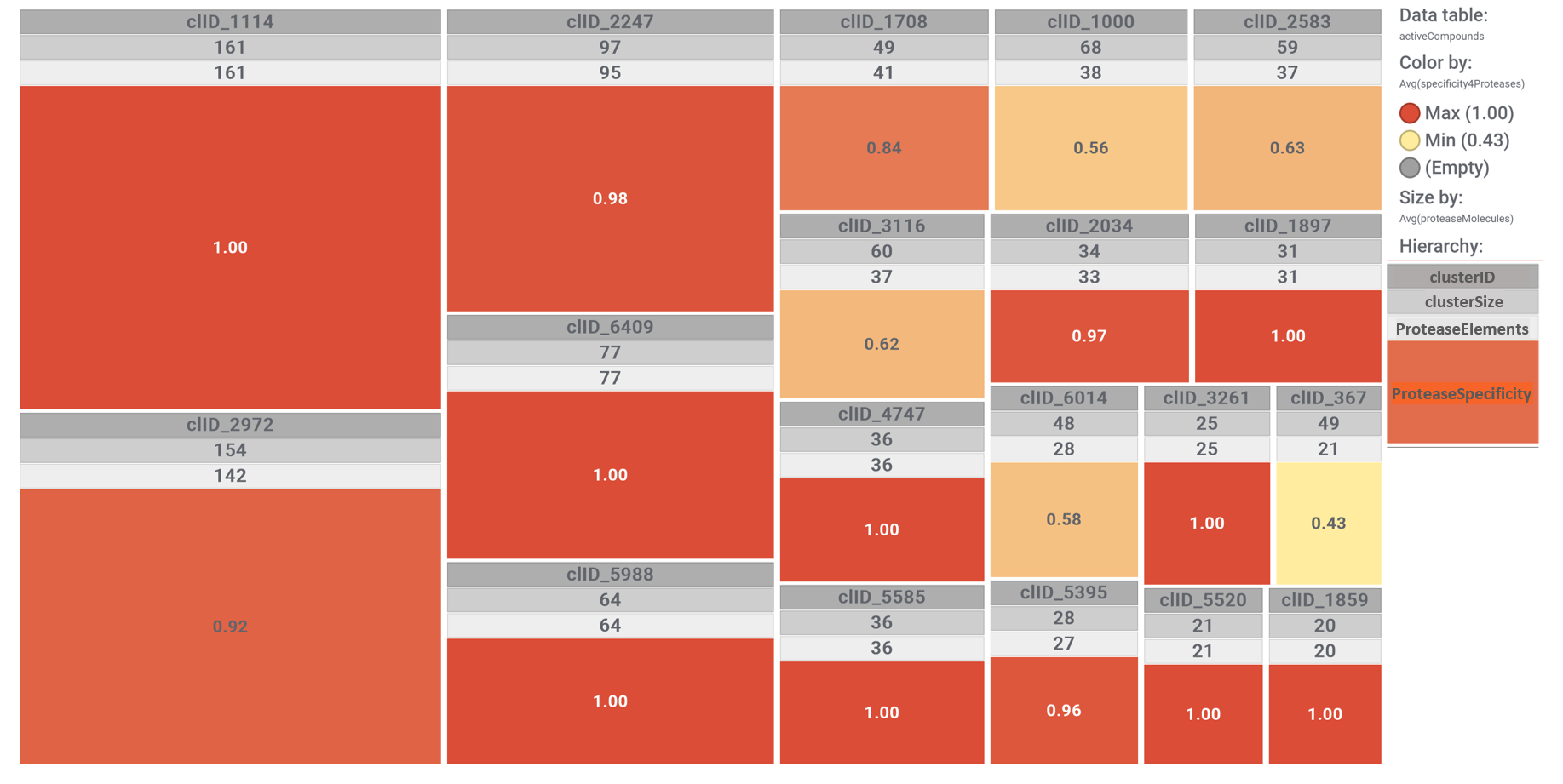
Now, we can select the compounds belonging to clusters that hit protease targets with higher specificity.
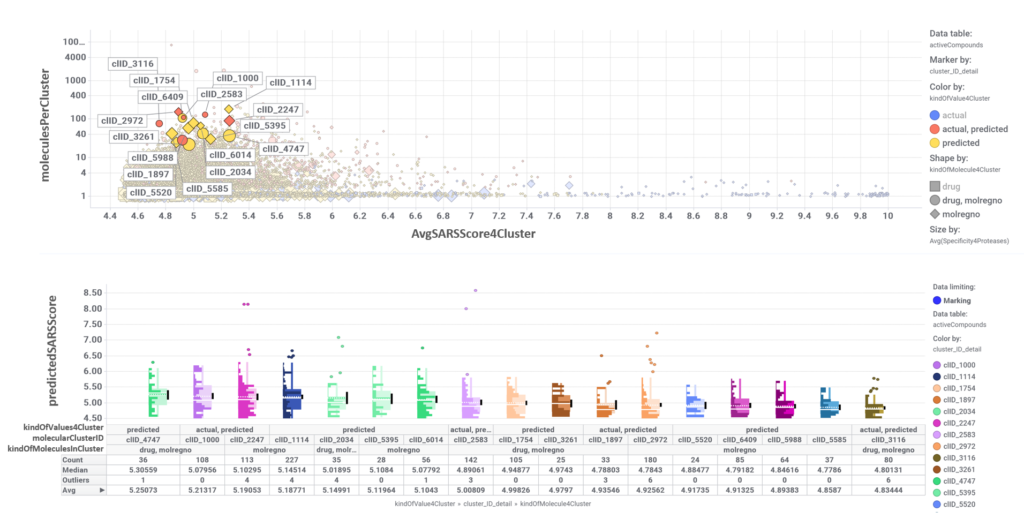
The tree map shows most specific molecular clusters, based on how many compounds hit on proteases vs the total number of compounds that have molecular interaction records in ChEMBL. On the right, these protease specific lusters are highlighted in the scatter plot representing SARS predicted or actual potency vs total number of molecules per cluster. The box plot represents the distribution of SARS score for each cluster.
Just click on each plot to get an expanded view.
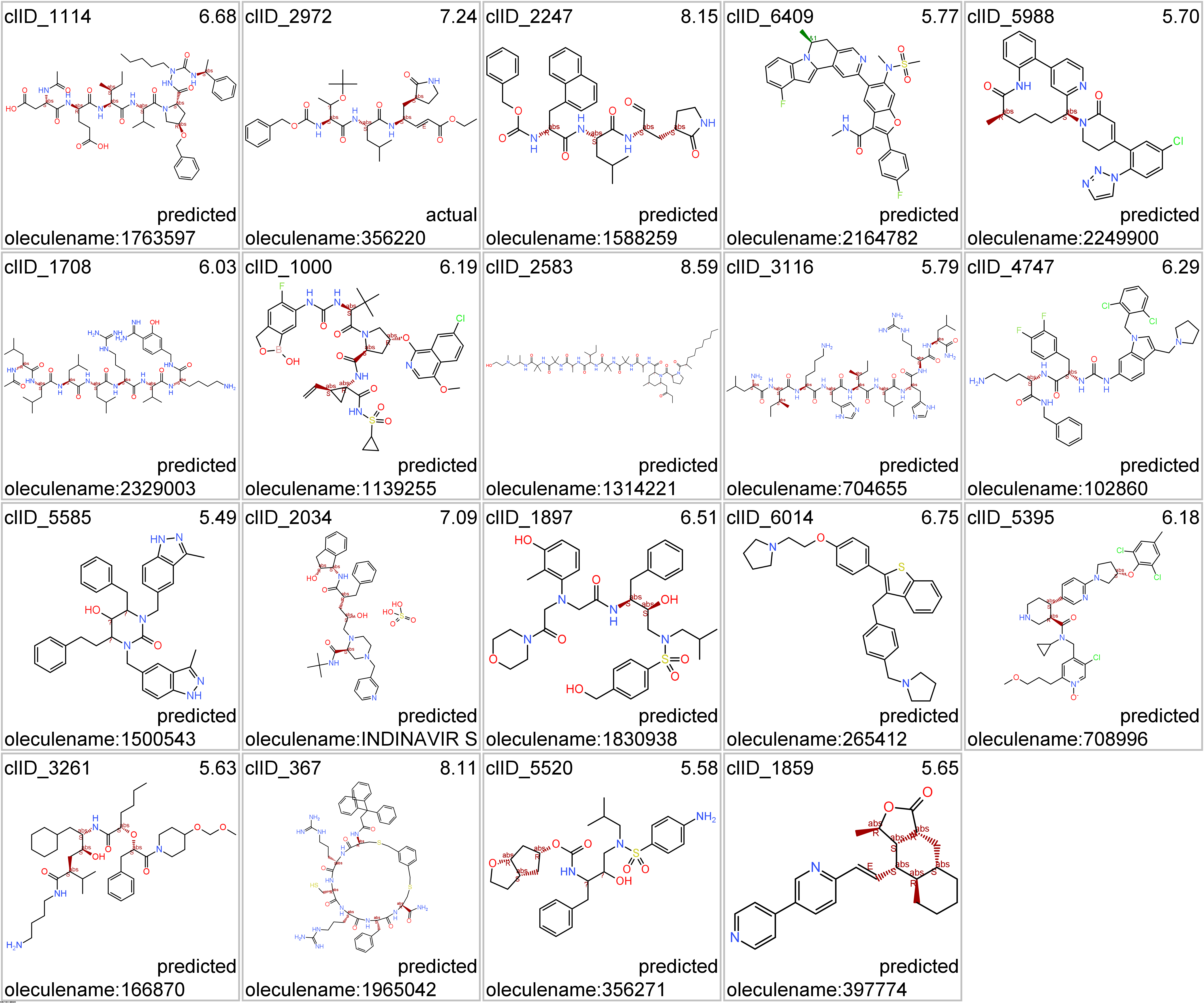
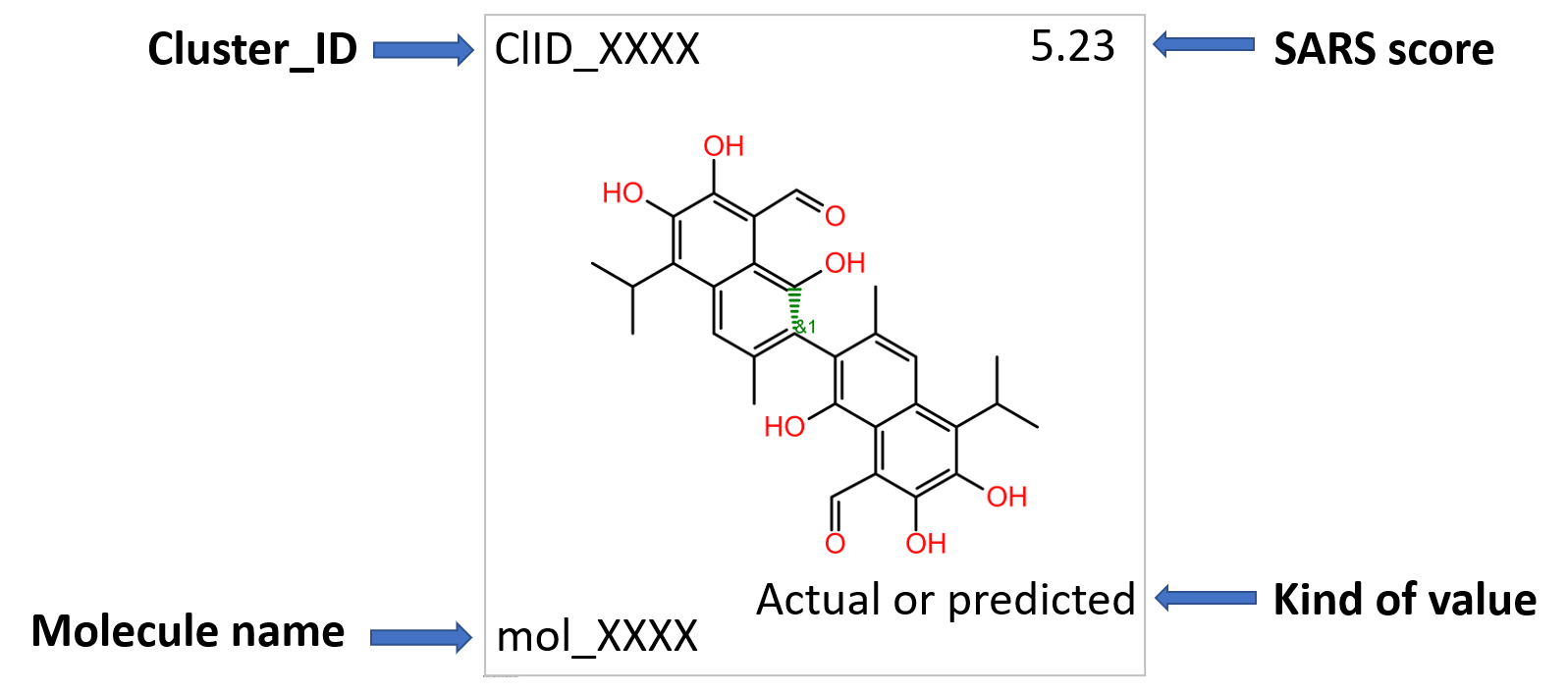
Representative structures of protease-specific clusters.
Click on the tabs below to visualize interactions and structures of molecules classified in most representative clusters.
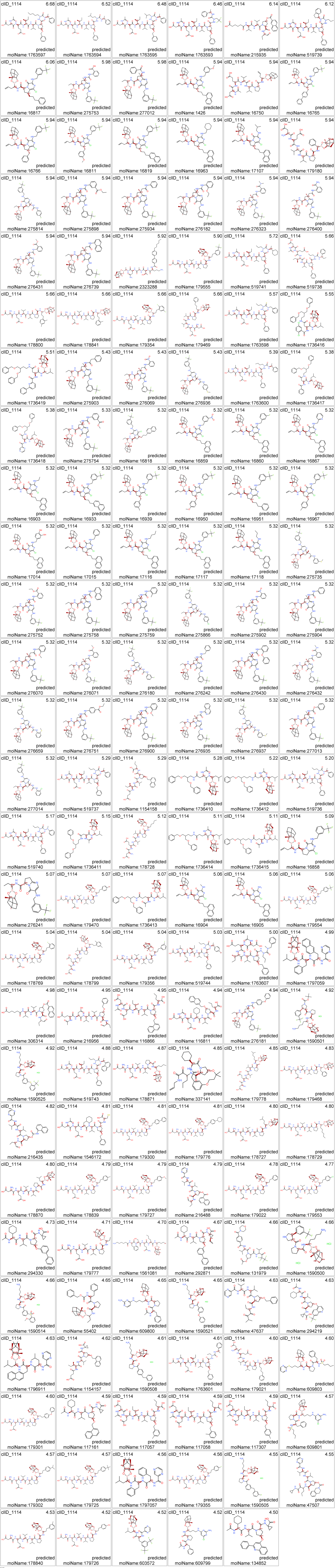
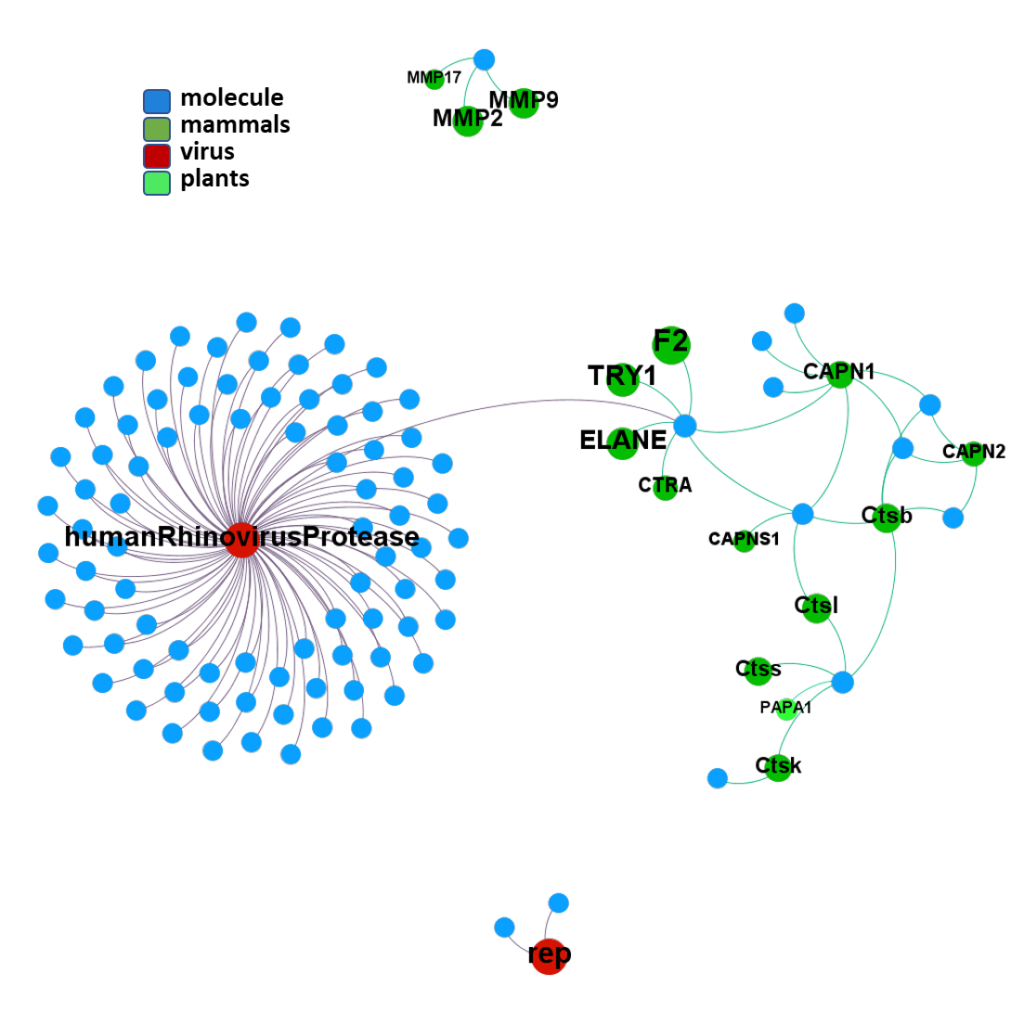
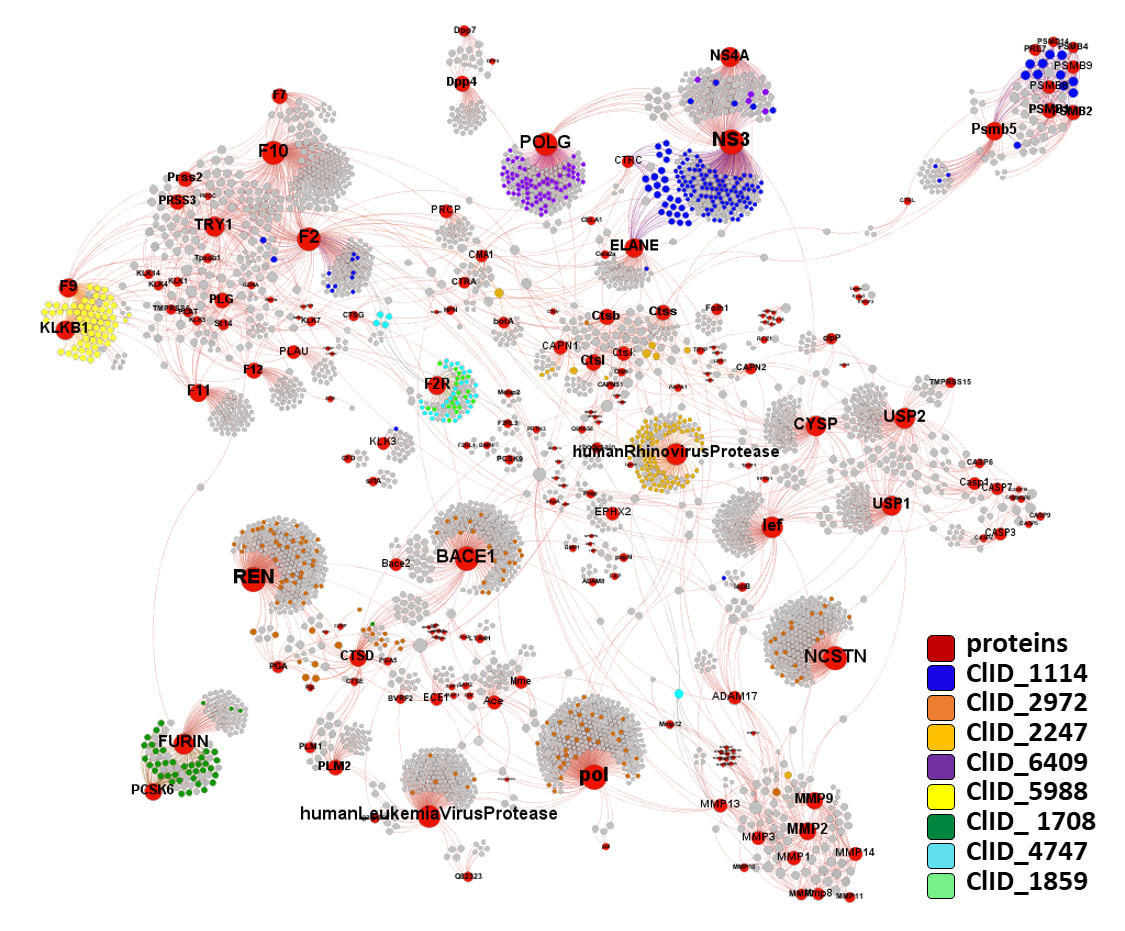
ClID_1114 compounds interact with viral NS3 and NS4 proteases, but show some cross reactivity with mammalian elastases,trypsin-like and proteasome subunits with protease activity.
seeProteinNames
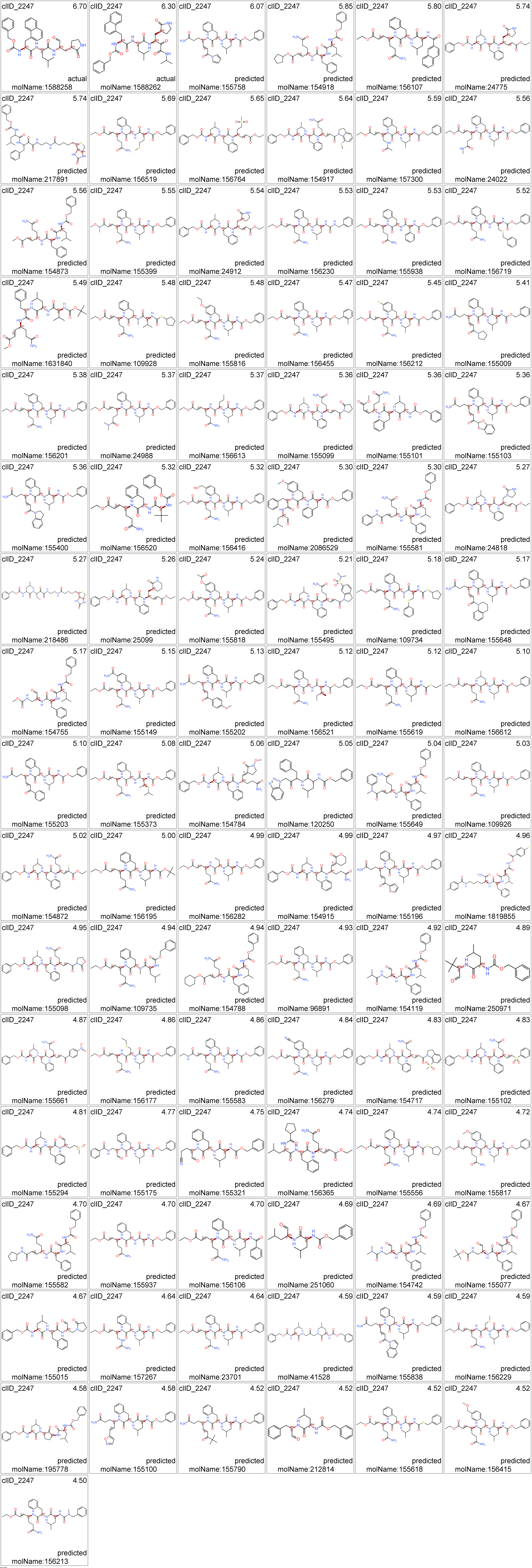
ClID_2247 compounds main target is rhinovirus protease, but show some cross reactivity with mammalian calpains, cathepsins, trypsin-like and elastases.
seeProteinNames
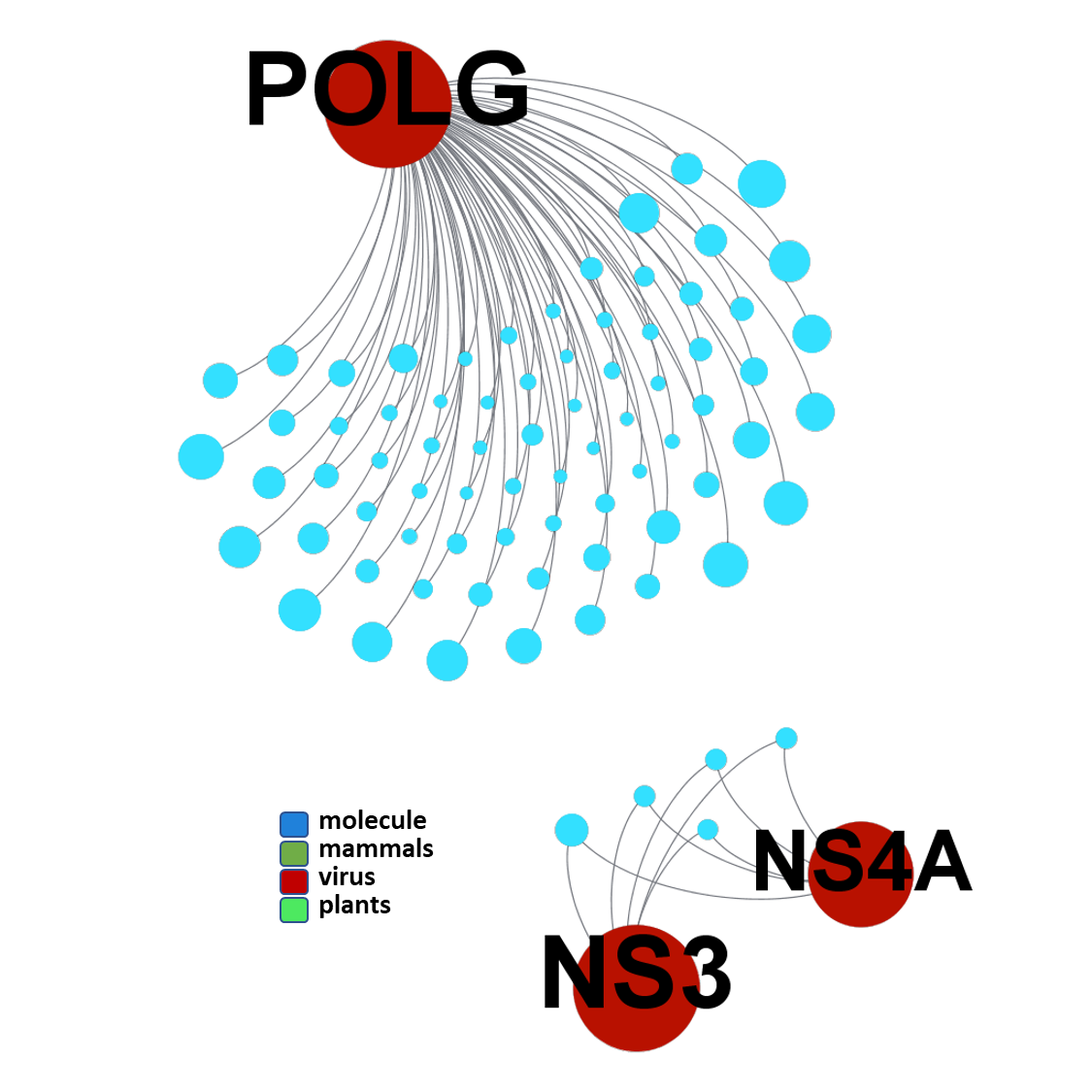
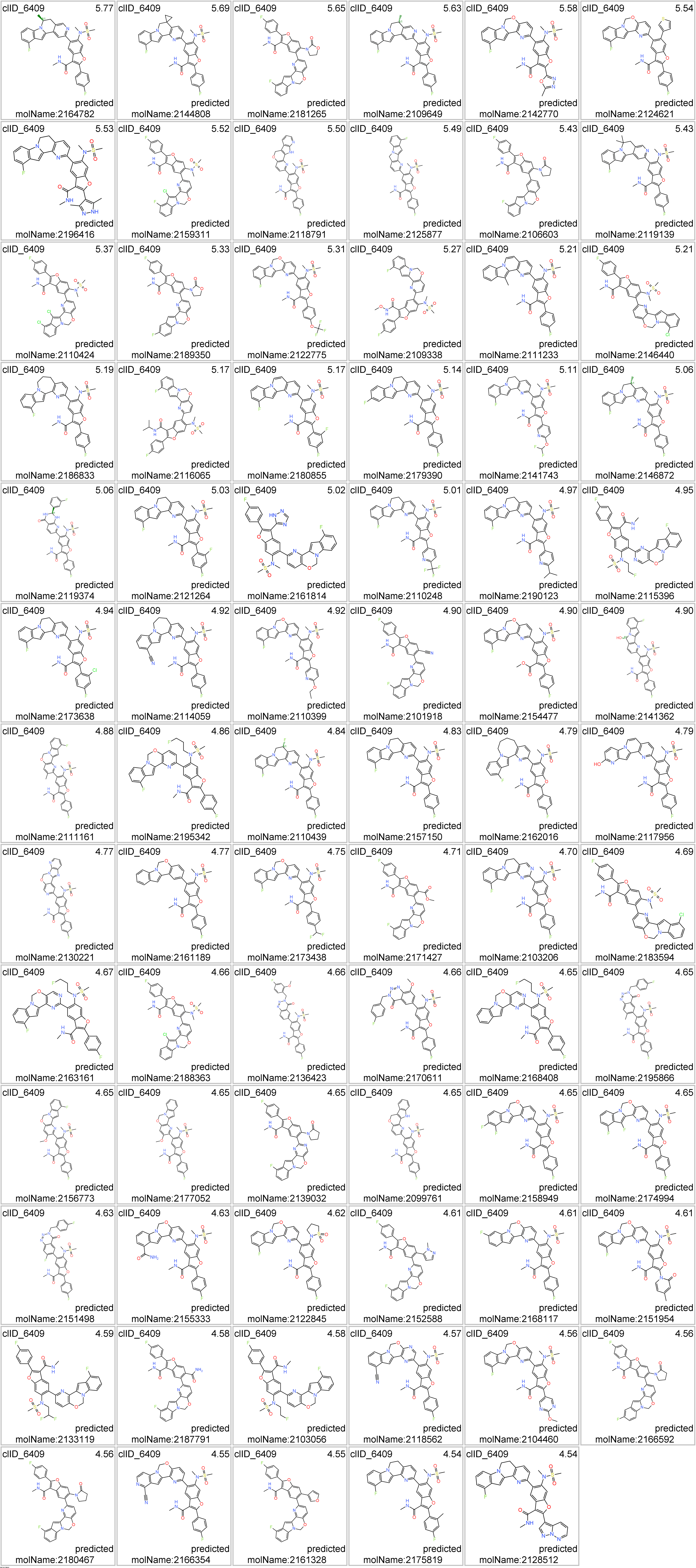
ClID_6409 compounds interact with NS3/4 viral proteases. POLG, although coding several structural, polymerases, proteases and others, is the gene ChEMBL annotation for the several protease assays aggregated as POLG in this graph.
seeProteinNames
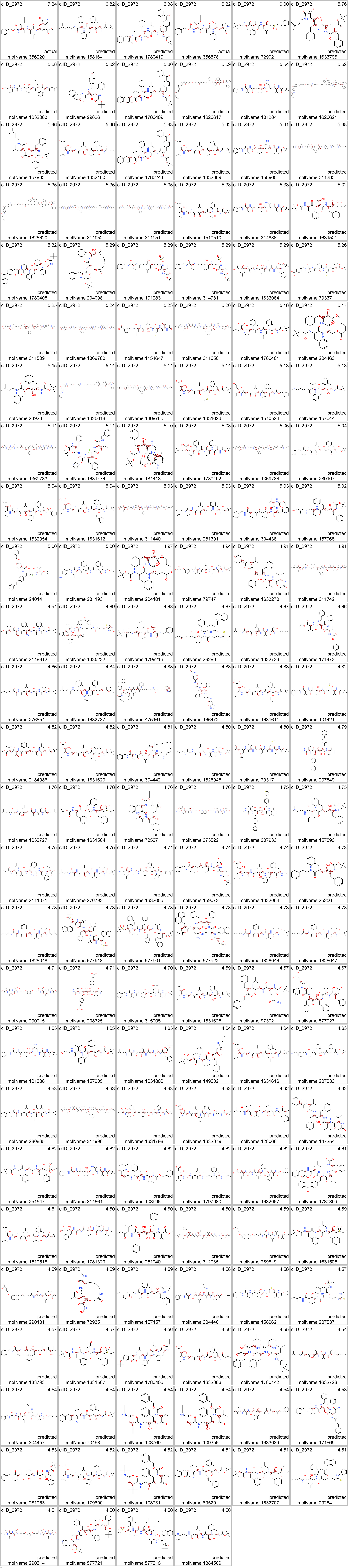
ClID_2972 compounds interact with NS3/4 viral proteases, 1a and pol polyproteins, renin, beta secretase, gamma secretase (including preselinins, nicastrin and APH1A/B as parts of the complex), pepsin, cathepsin, matrix metalloproteinases and some others. 1a and pol, actually, are the gene ChEMBL annotations for the several protease assays aggregated as POLG in this graph.
seeProteinNames
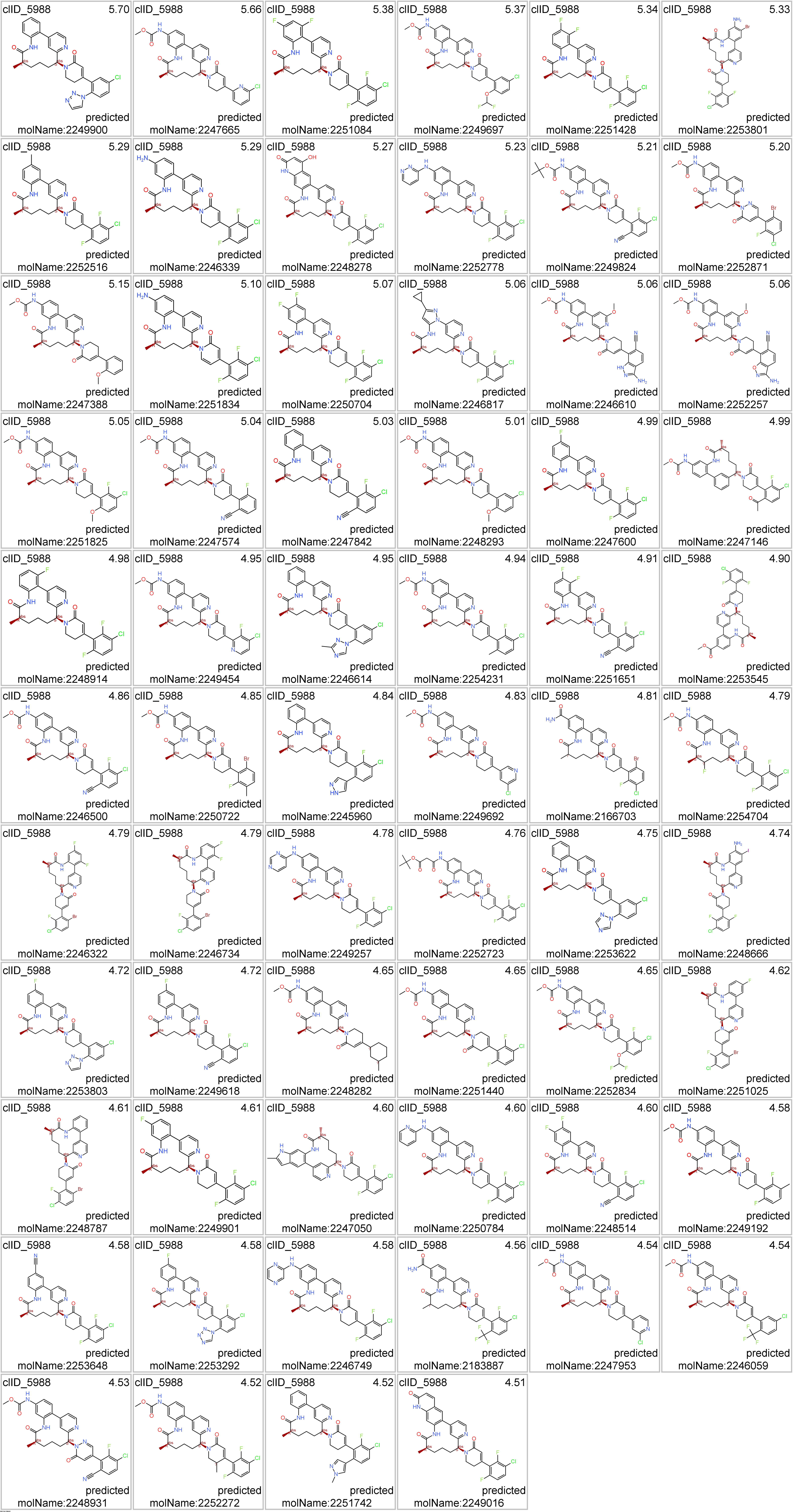
ClID_5988 compounds interact with kallikrein and coagulation factor IX.
seeProteinNames
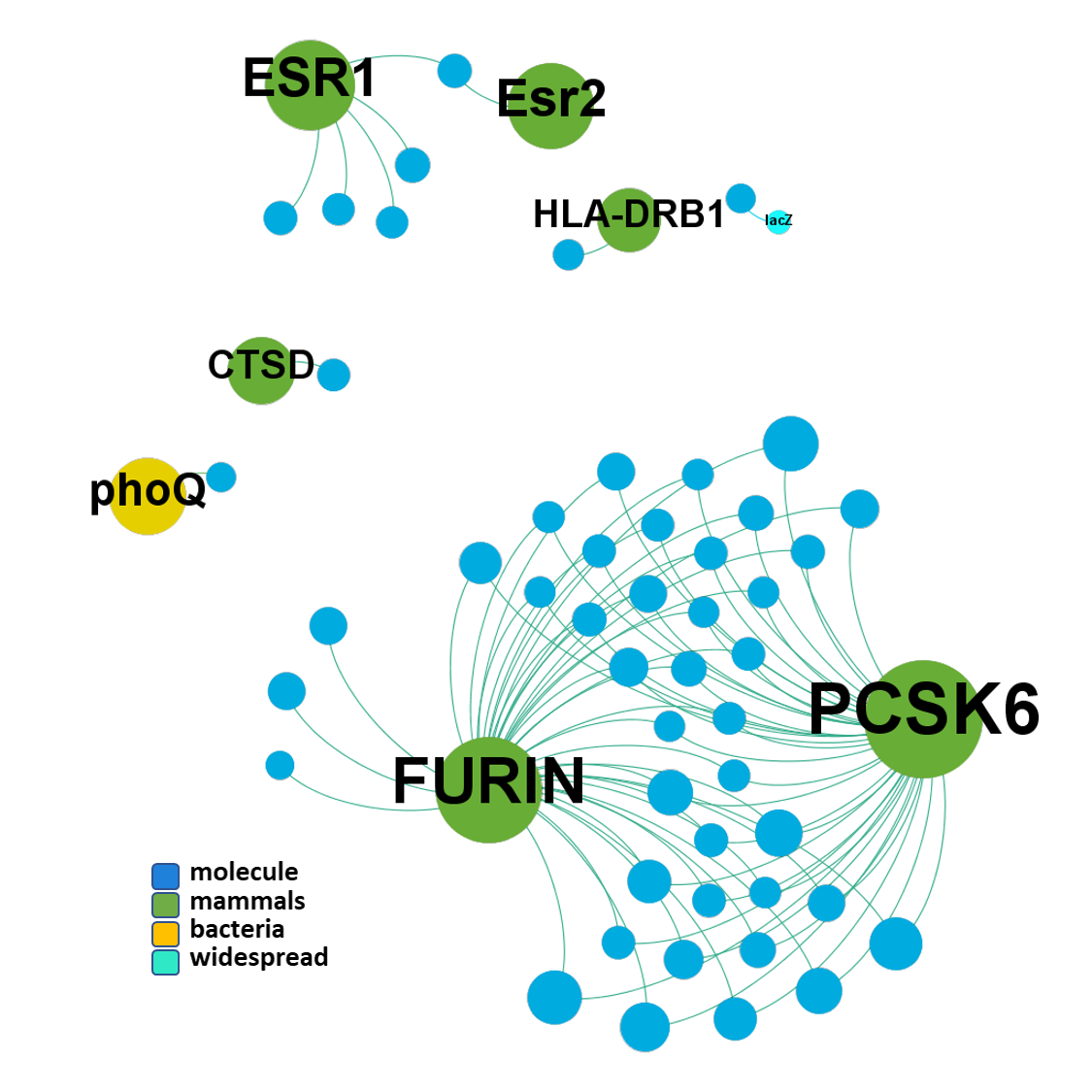
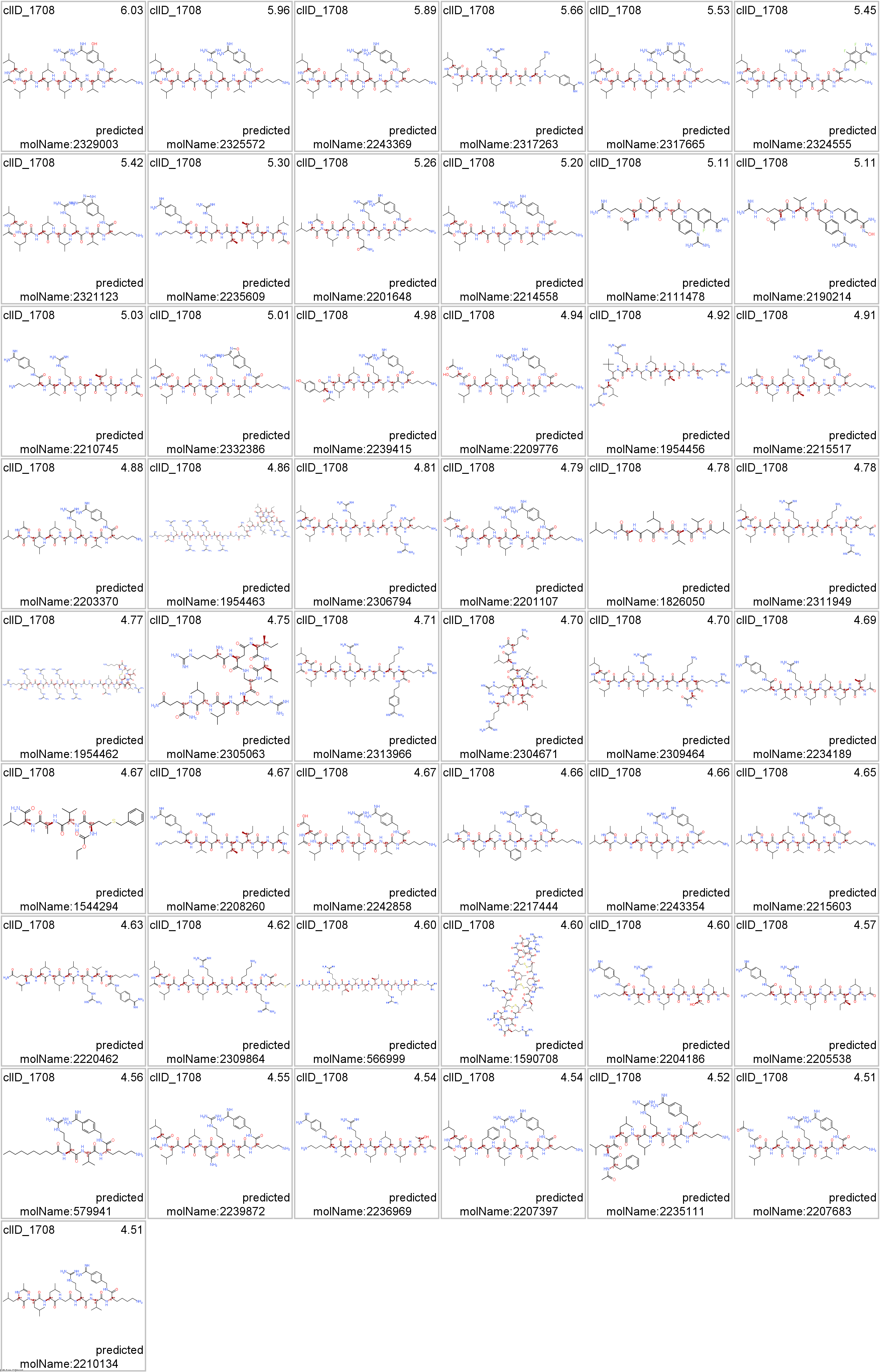
ClID_1708 compounds interact mainly with furin and PCSK6 proteases. Few compounds Also hit cathepsin, estrogen receptors and HLA complex.
seeProteinNames
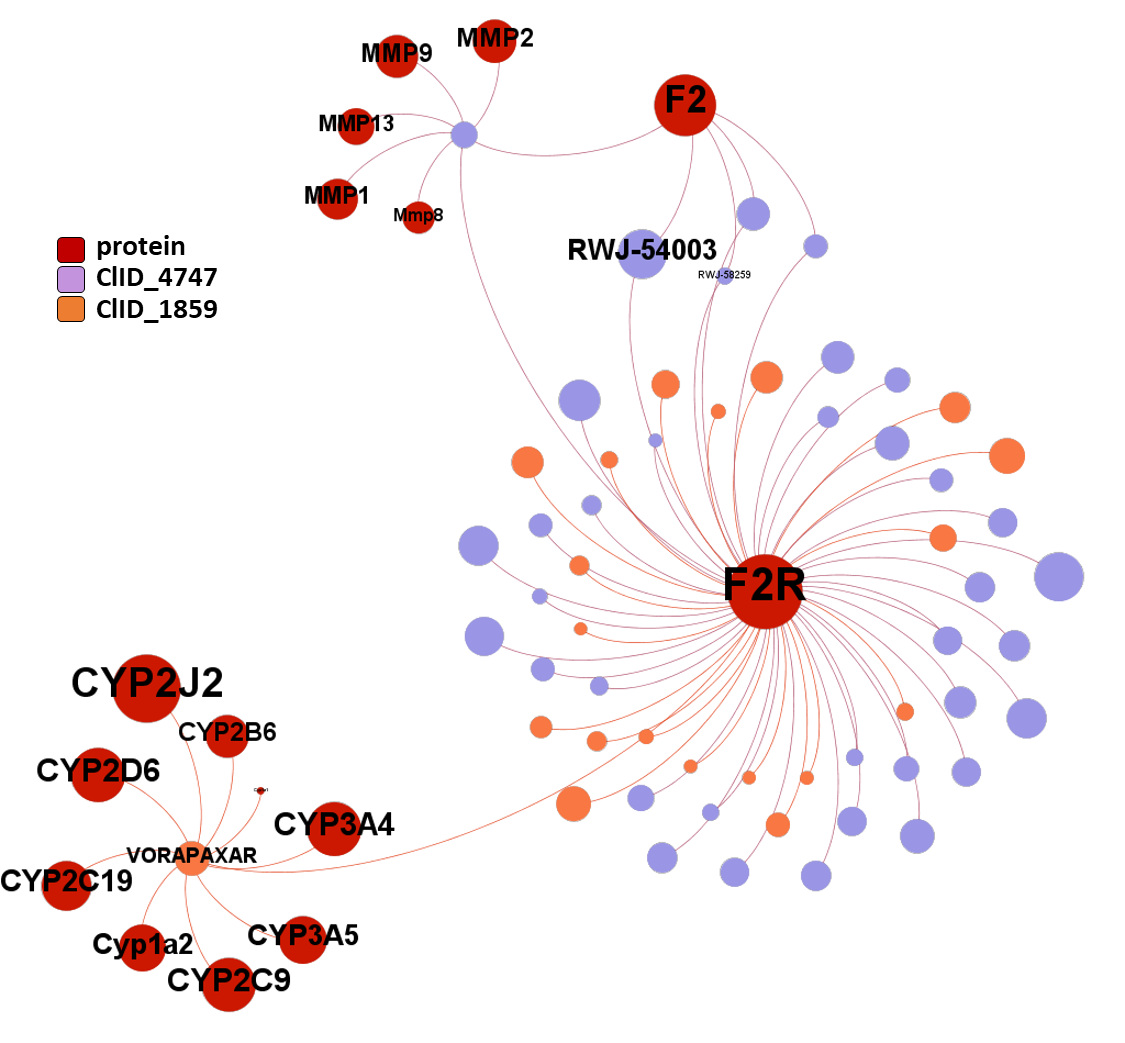
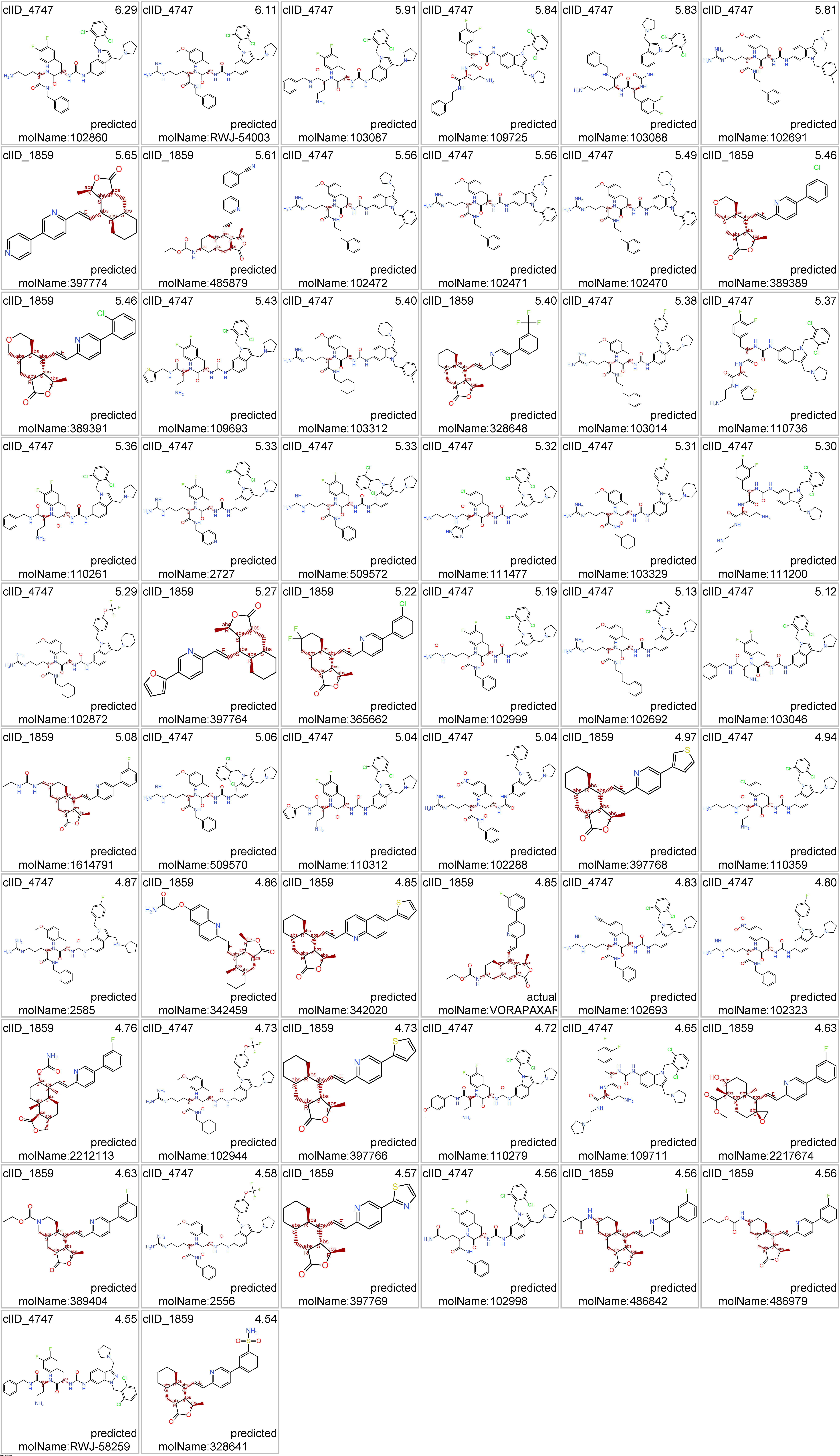
ClID_4747 and ClID_1859 compounds recorded interactions point to the proteinase activated receptor (F2R). A minor number of molecules show crossed interactions with prothrombin and metalloproteinases. Vorapaxar present additional records of interactions with cytochrome proteins, probably a consequence of its development as drug.
seeProteinNames
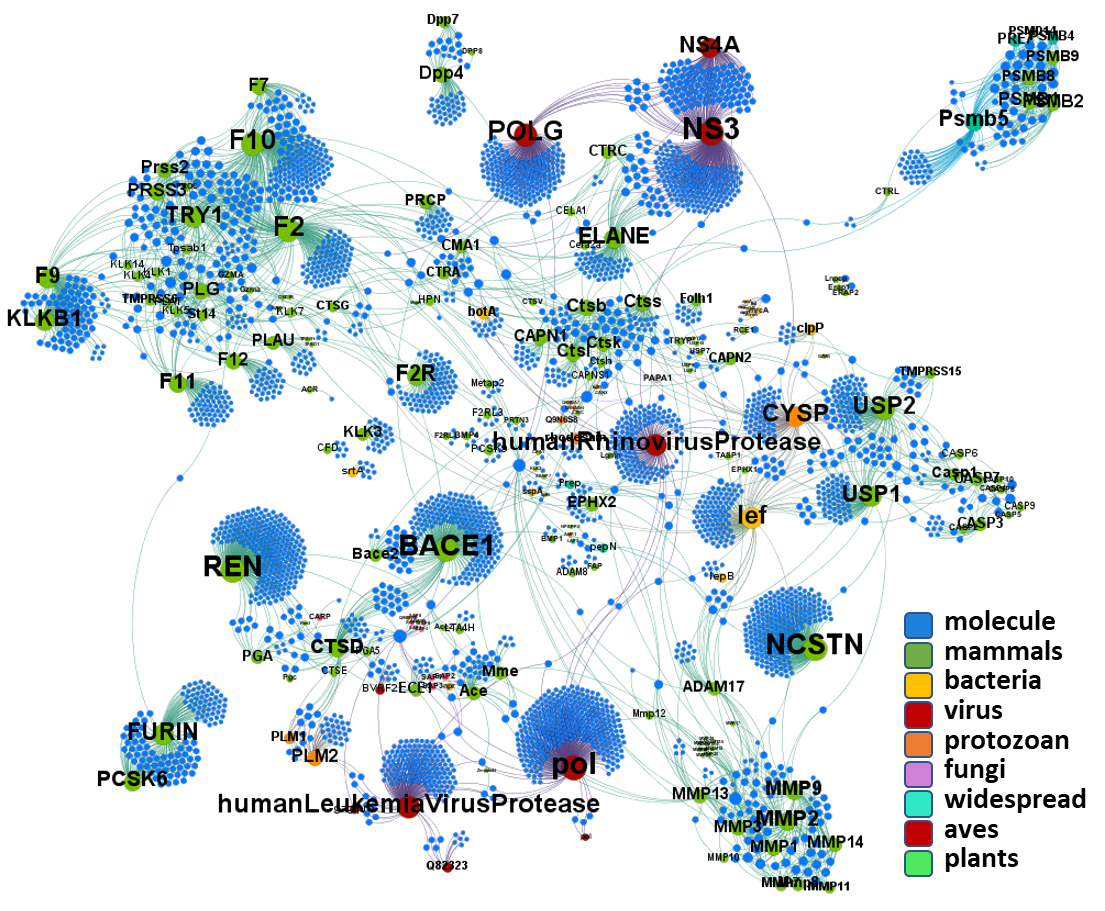
And finally, take a look on crossed reactions between molecules and different protease types. Either looking at organism type or clusters selected in this page.
Just click on the graphs to expand the views.