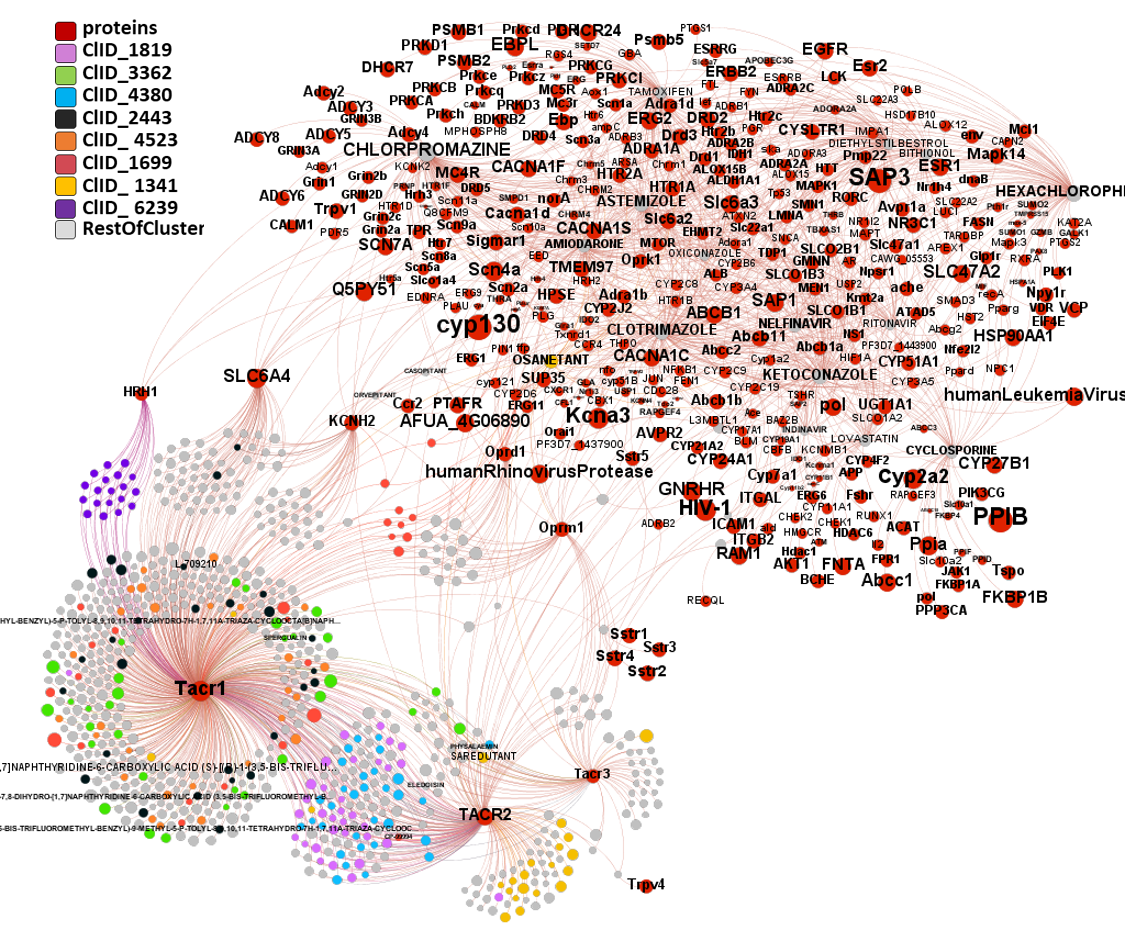
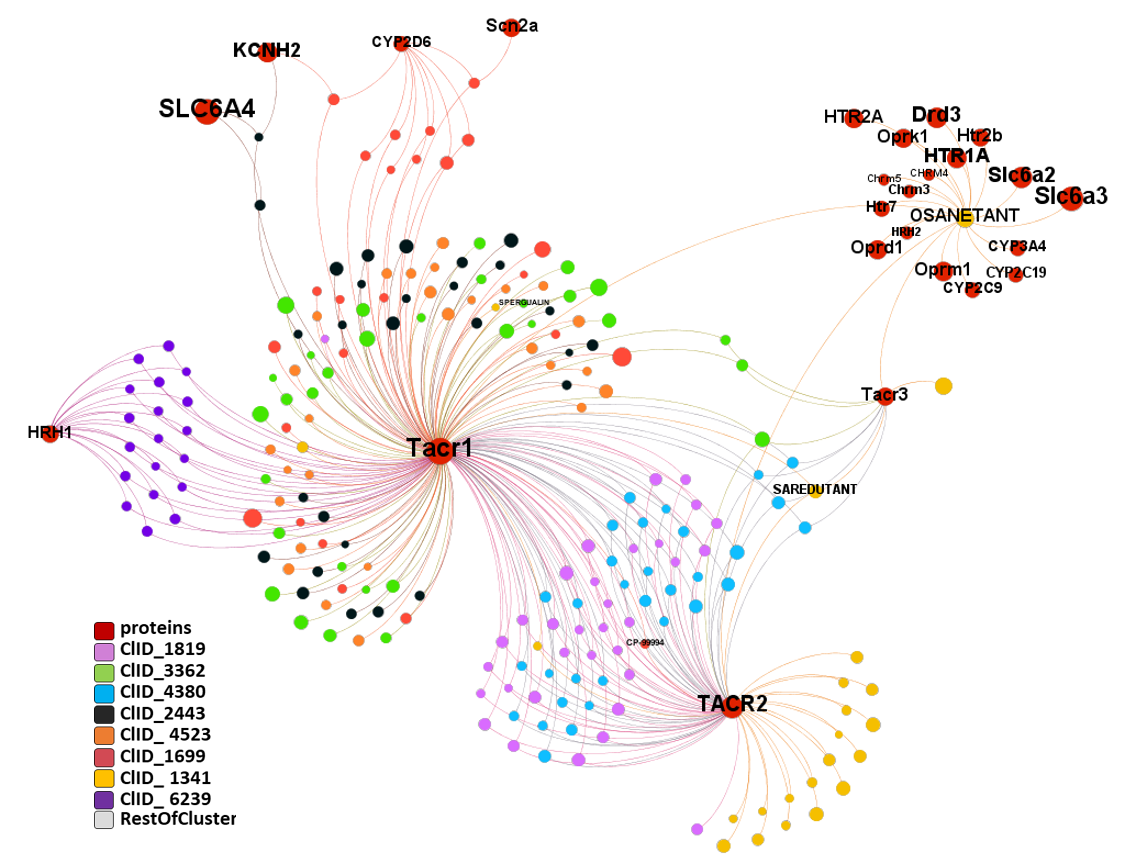
217 molecules predicted as SARS active interact with the three known tachykinin receptors TACR1, TACR2 and TACR3 (aka NK1R, NK2R and NK3R, respectively). All SARS active molecules have been classified in clusters attending to their structural similarity. Graphs summarize all molecule-protein interactions for the TACRs module, obtained by interactions-based segmentation, plus any other interaction with other proteins recorded in th ChEMBL DB in order to discriminate most TACRs -specific molecular clusters. Although TACRs are not ion channels but GPCRs are enclosed in then ion channels module because their common interactions with ion channels molecules.
ChEMBL molecule registry number (molregnos) labels have been removed in the networks to facilitate visualization of interactions. Only known drugs and protein names remain.
In this tab, we show interactions of most VGCC-specific clusters.
ClID_3362, ClID_4523 and ClID_2443 andClID_1699 show clear preference for TACR1 with occasional records for cytochromes ion channels and the Sodium-dependent serotonin transporter SCL6A4. ClID_6239, shows also bias for TACR1 but in this case shared with the histamine receptor H1 (HRH1). ClID_1341, that includes the known TACR antagonists saredutant and osanetant, hits preferentially TACR2, although a few members interact with TACR1&3 and osanetant keeps ChEMBL records for GPCRs, cytochromes and amine transporters. There is a significantly smaller number of records for TACR3 interactions with ClID_4380, ClID_3362 and ClID_1341. ClID_1819 and ClID_4380 show les selectivity for the different TACRs subtypes.
Additional molecular clusters hit on TACRs with lower specificity.