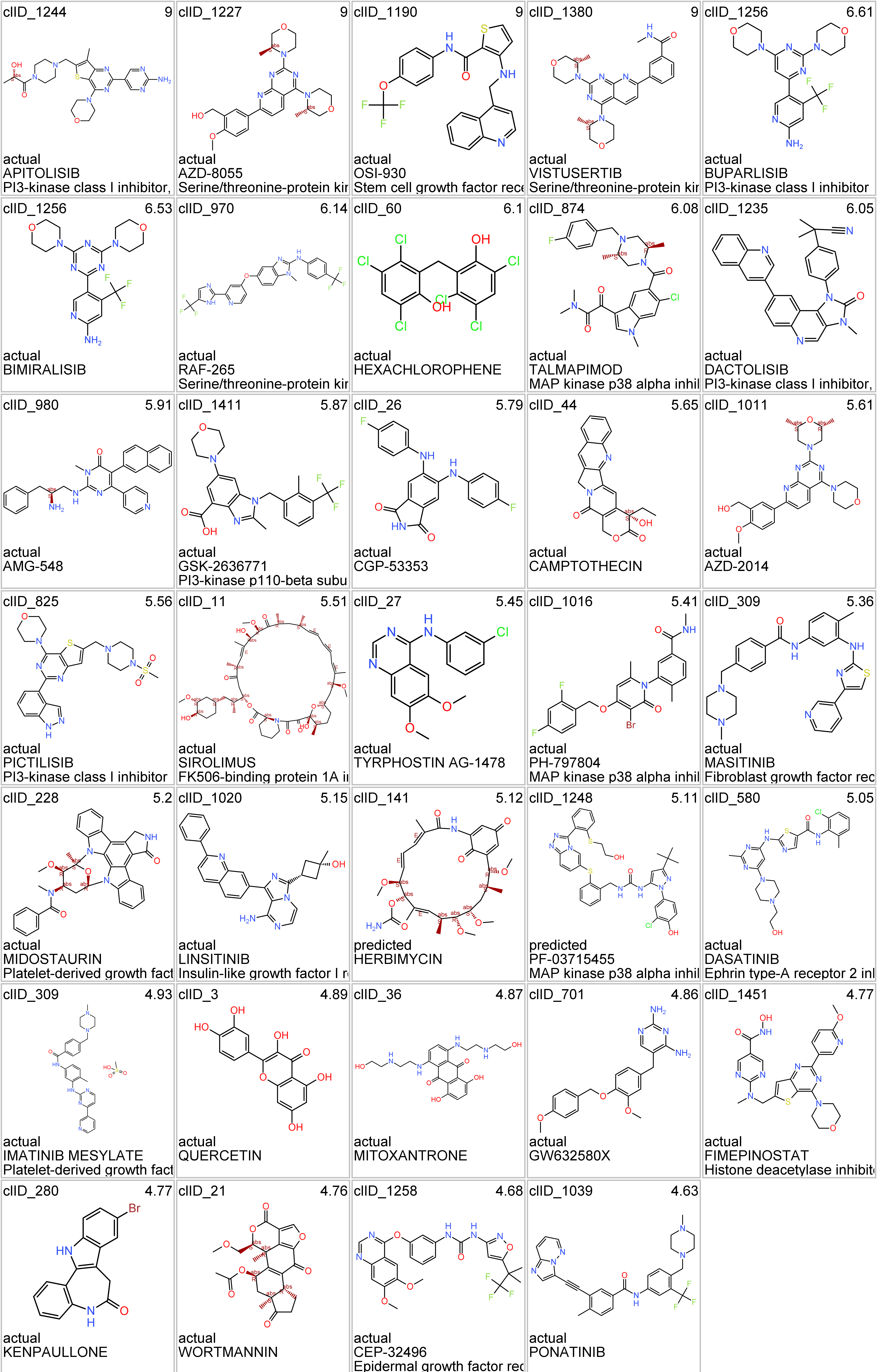
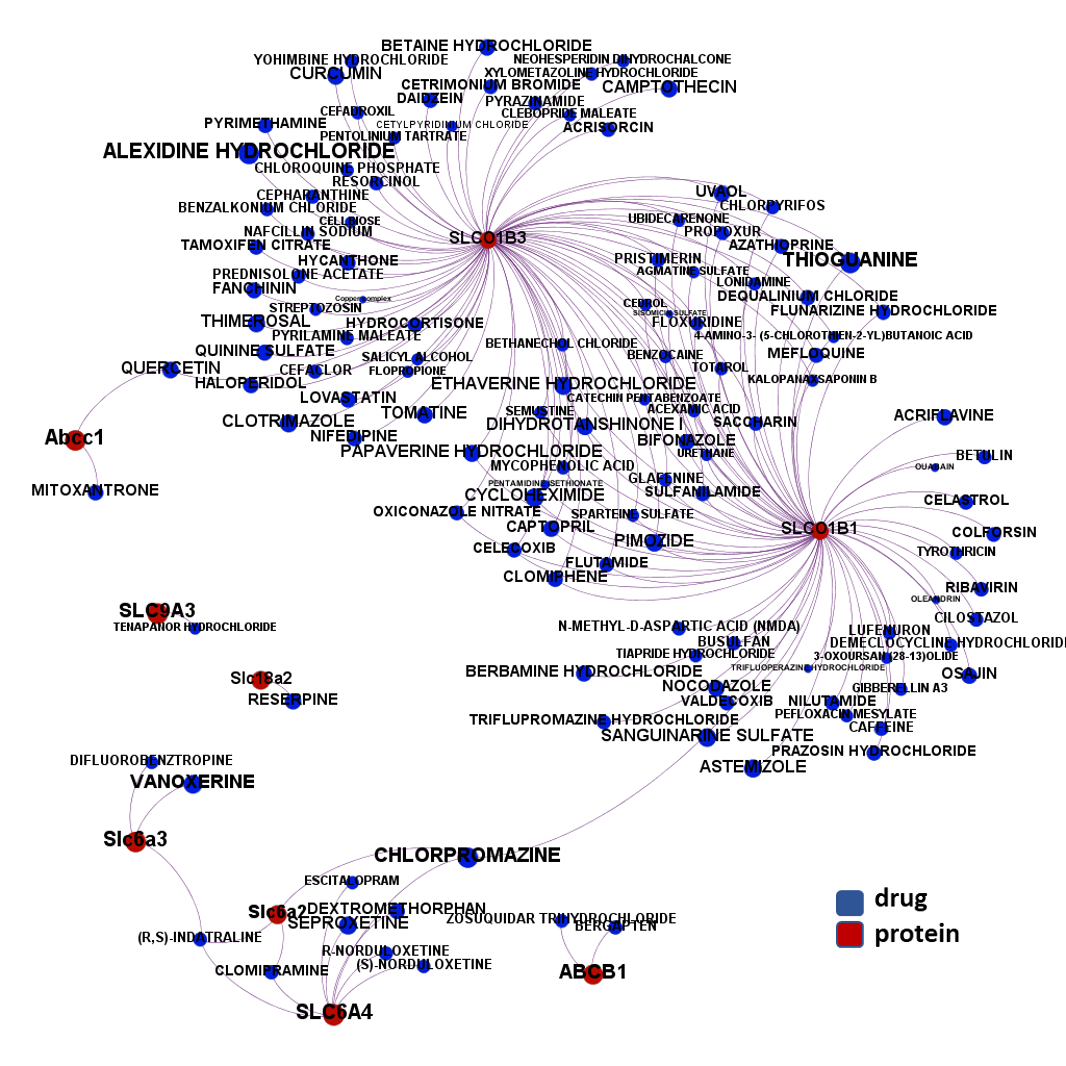
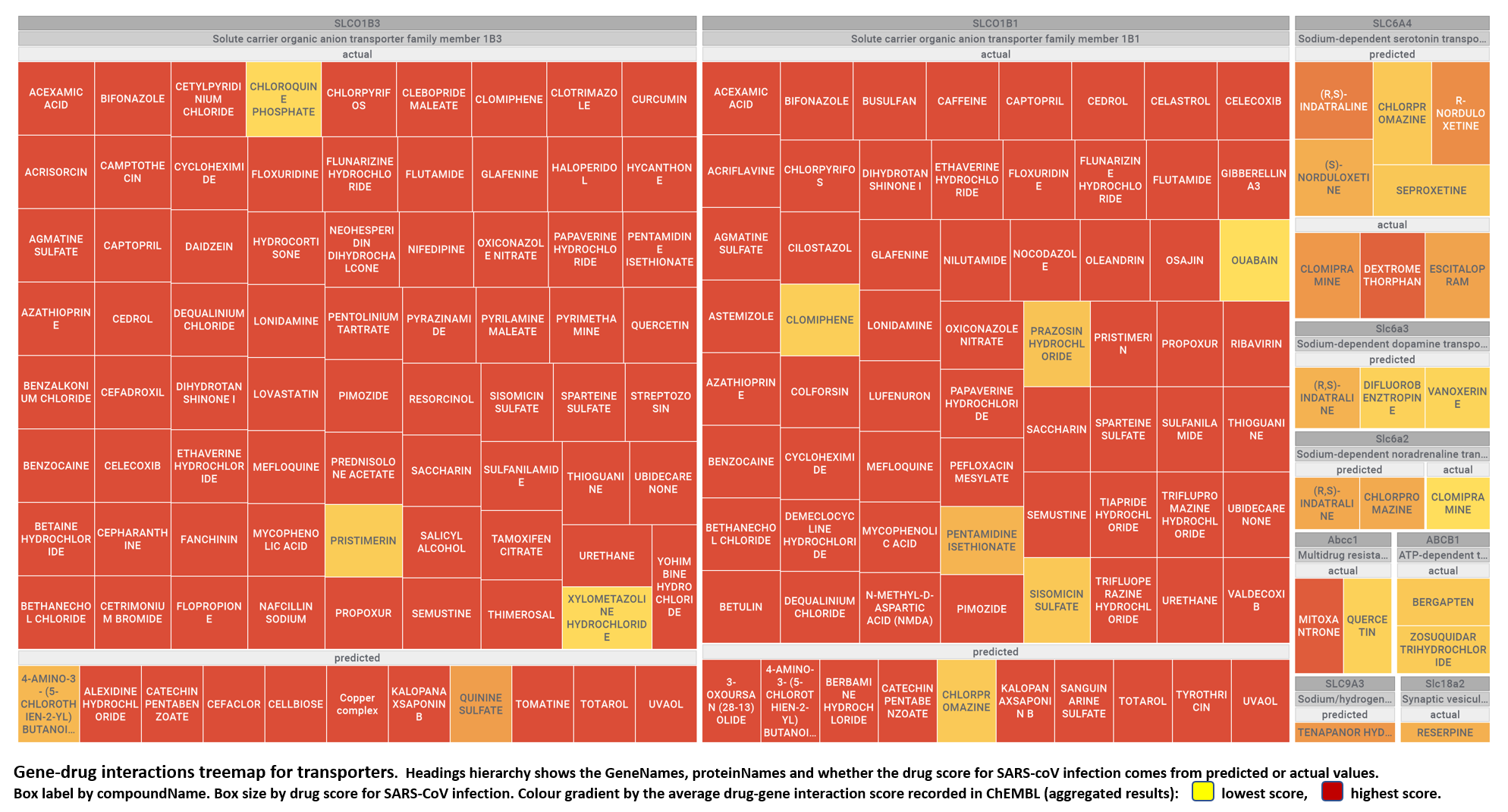
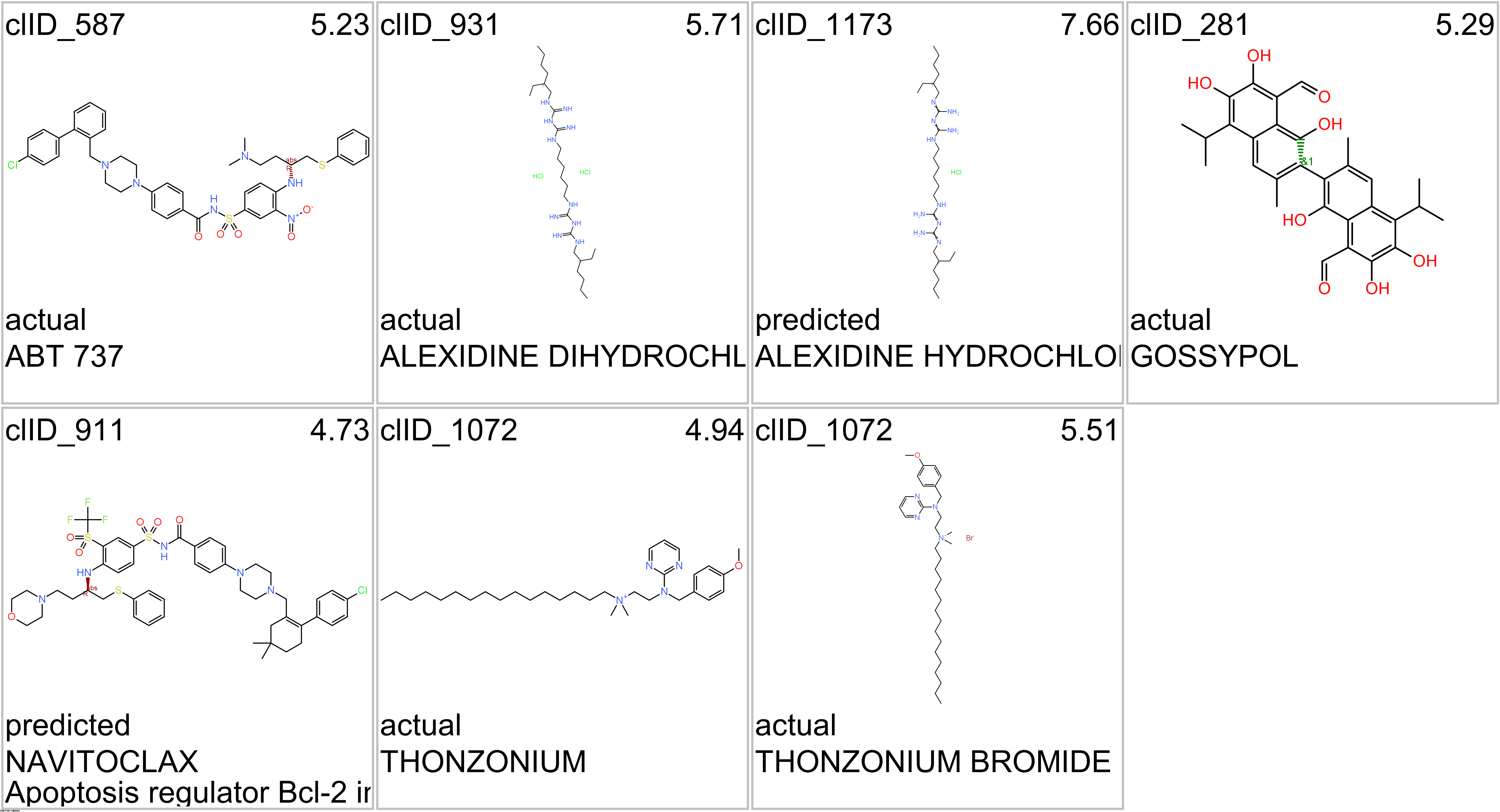
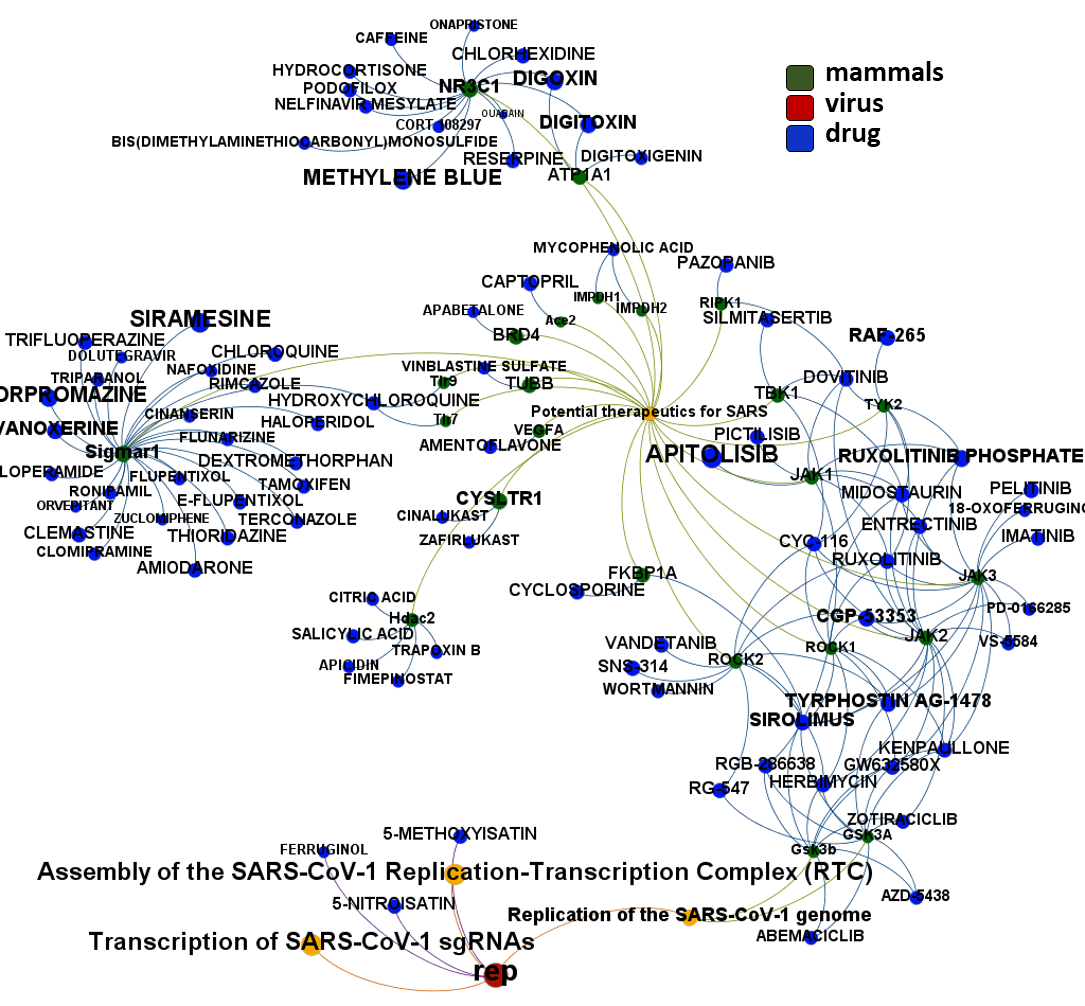
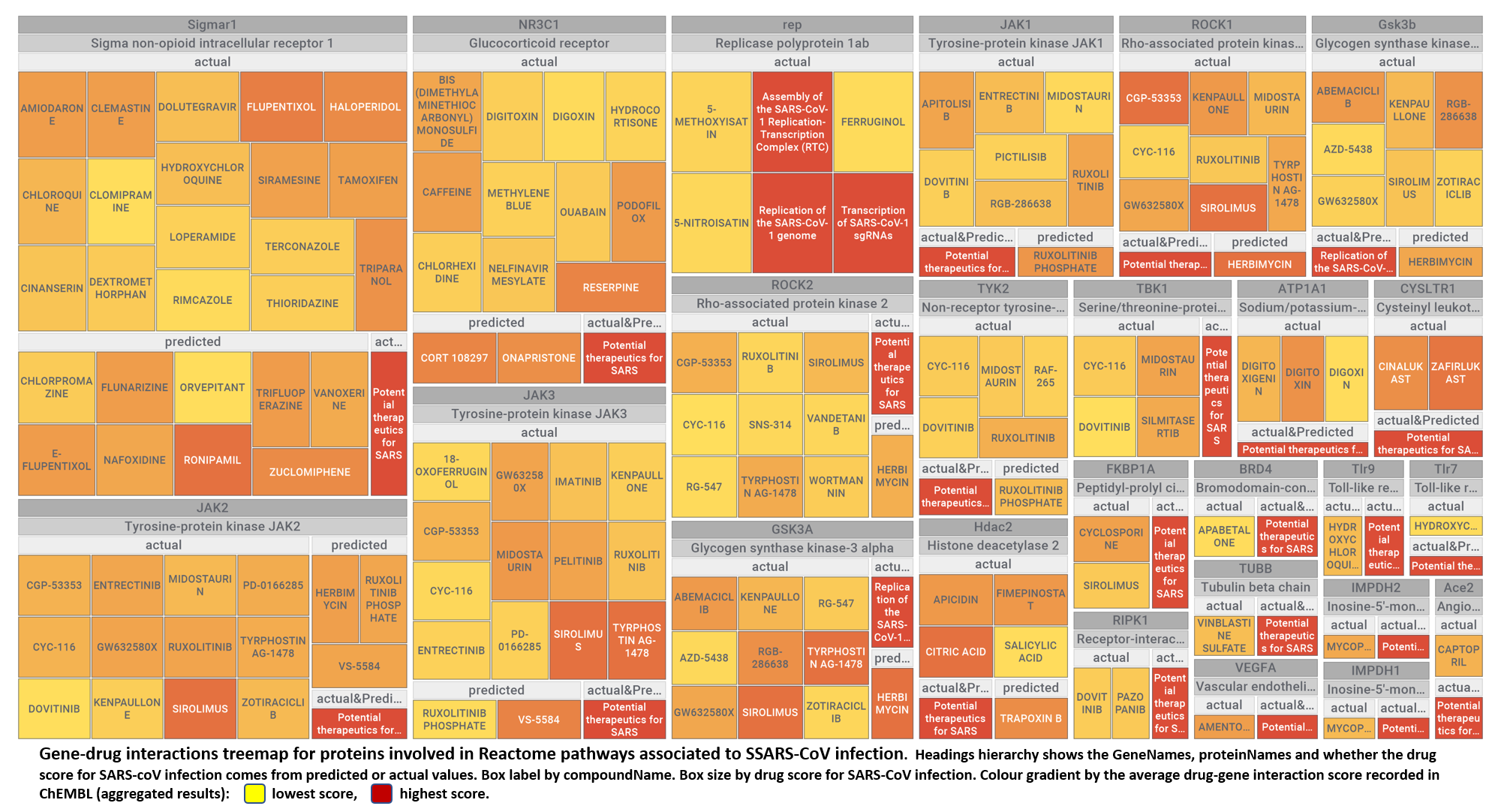
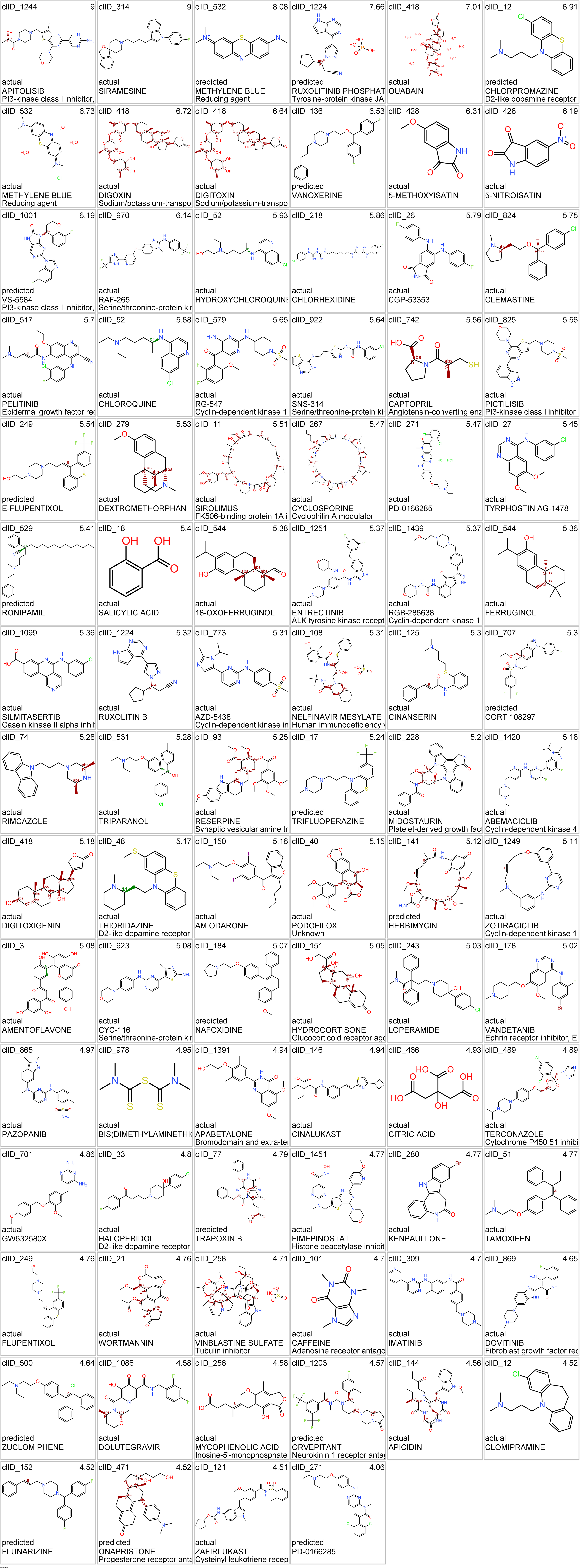
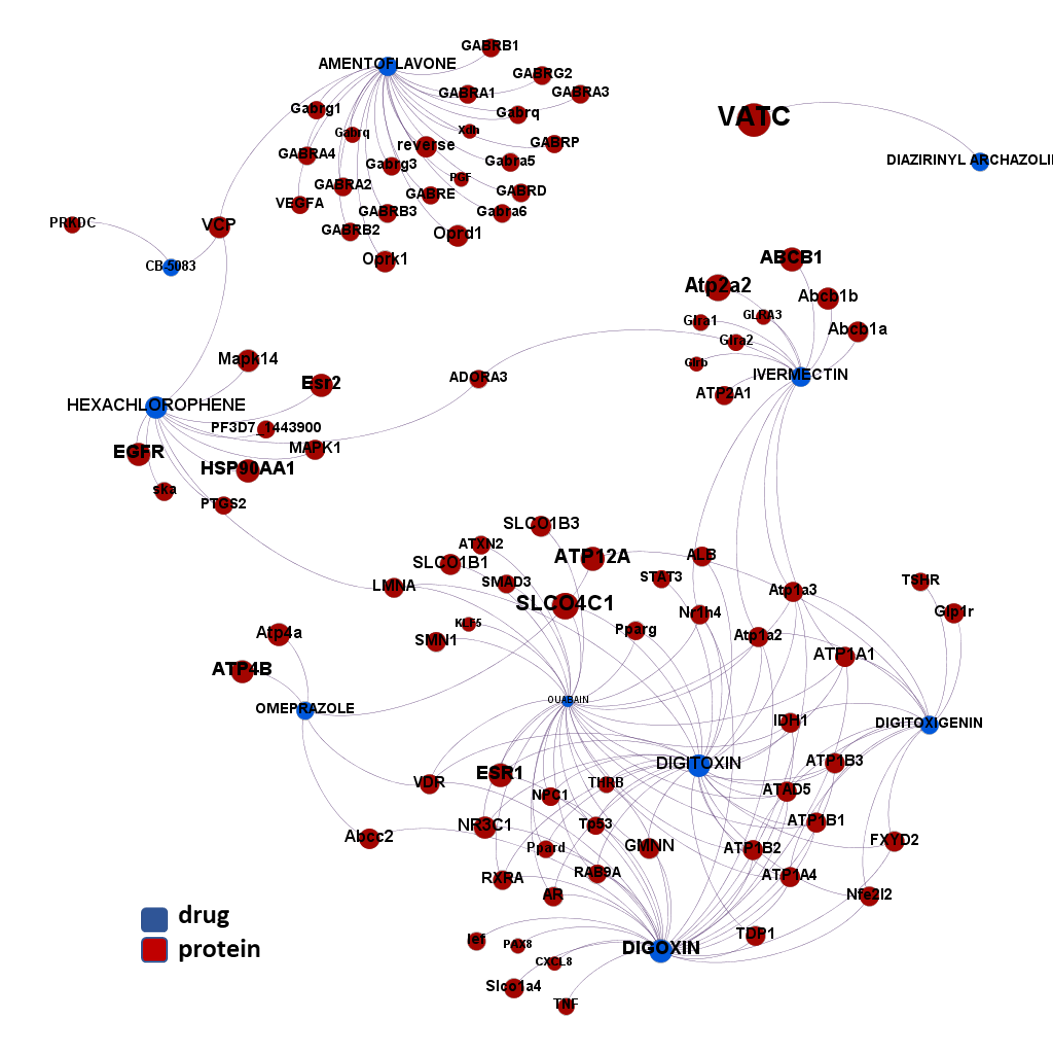
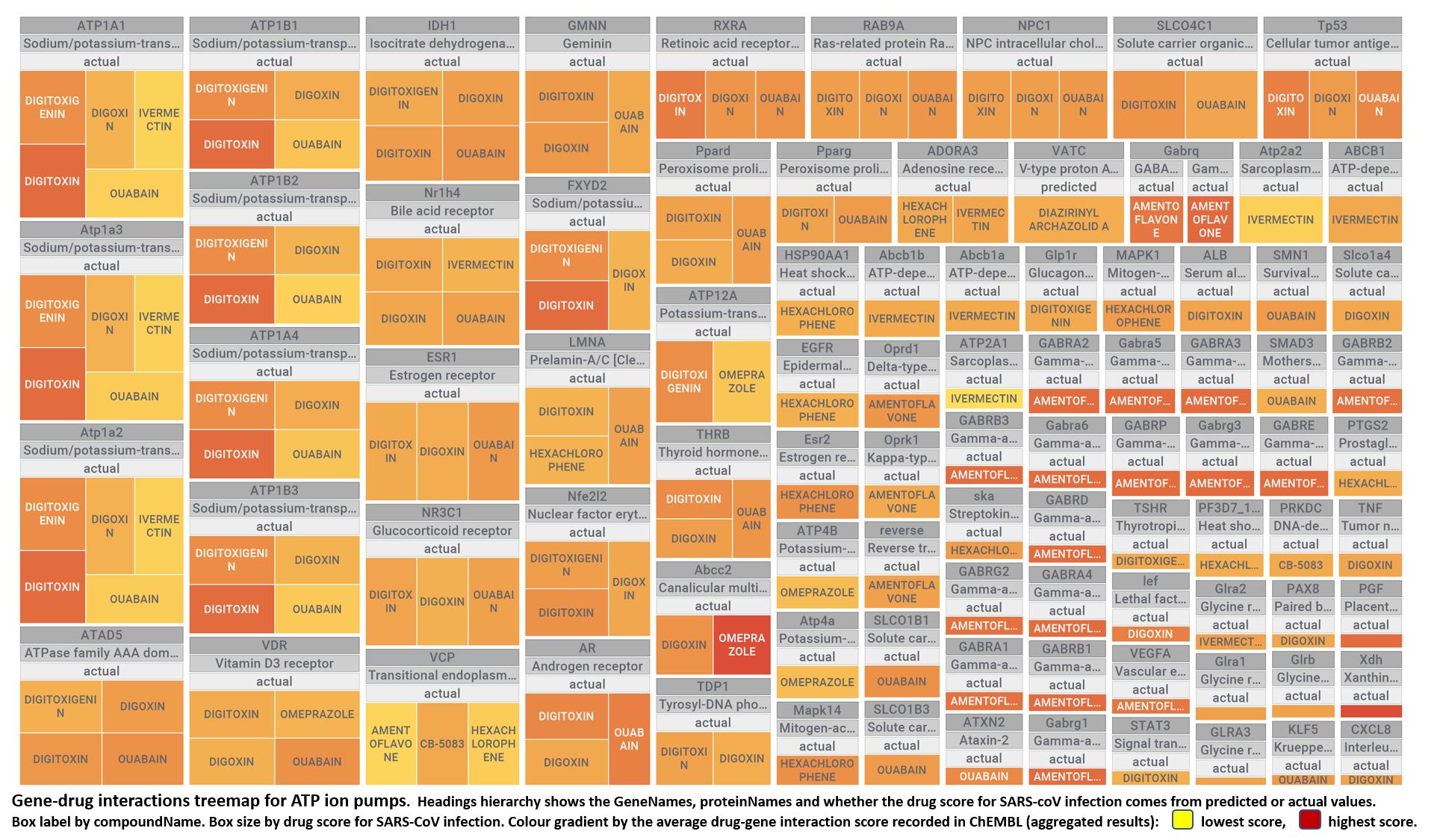
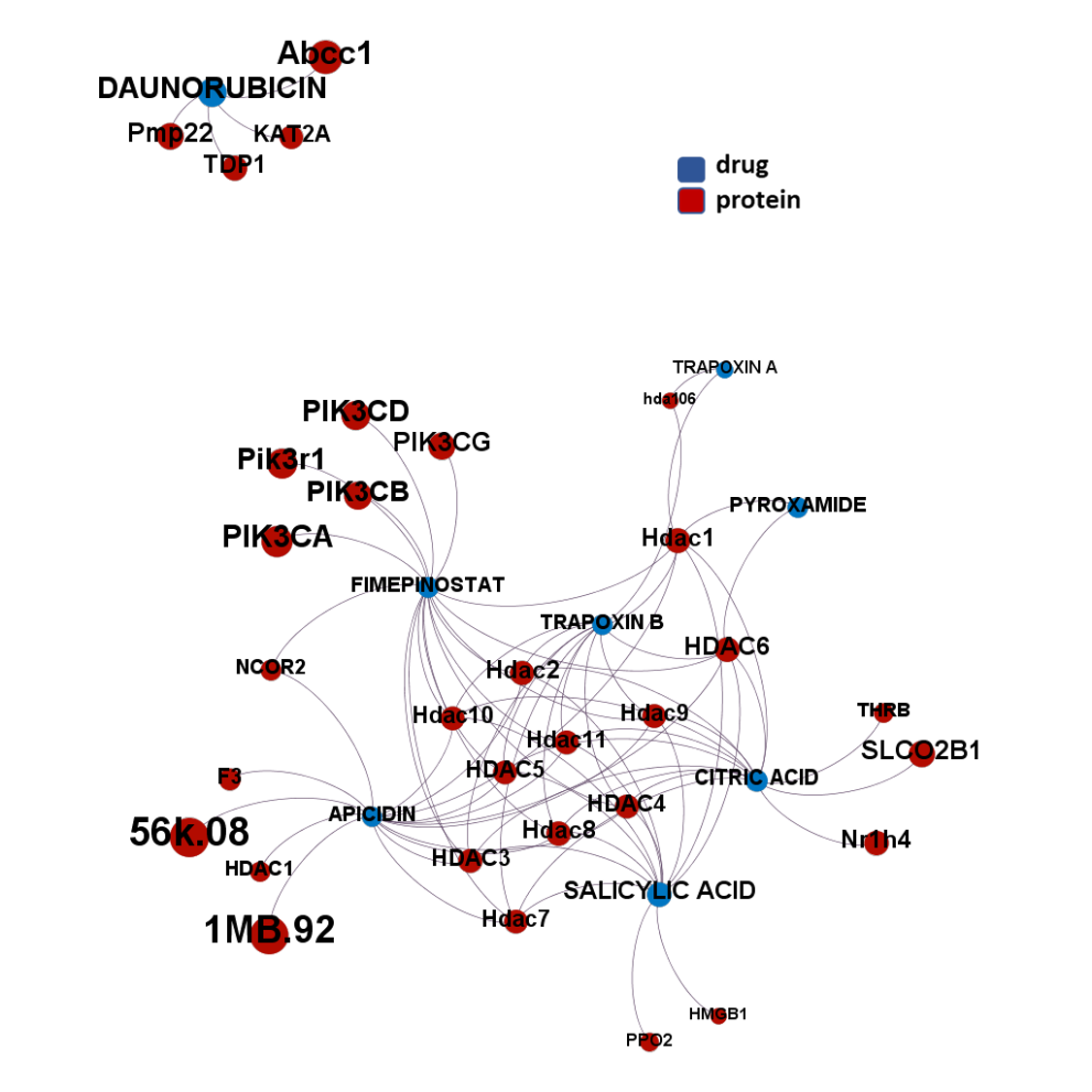
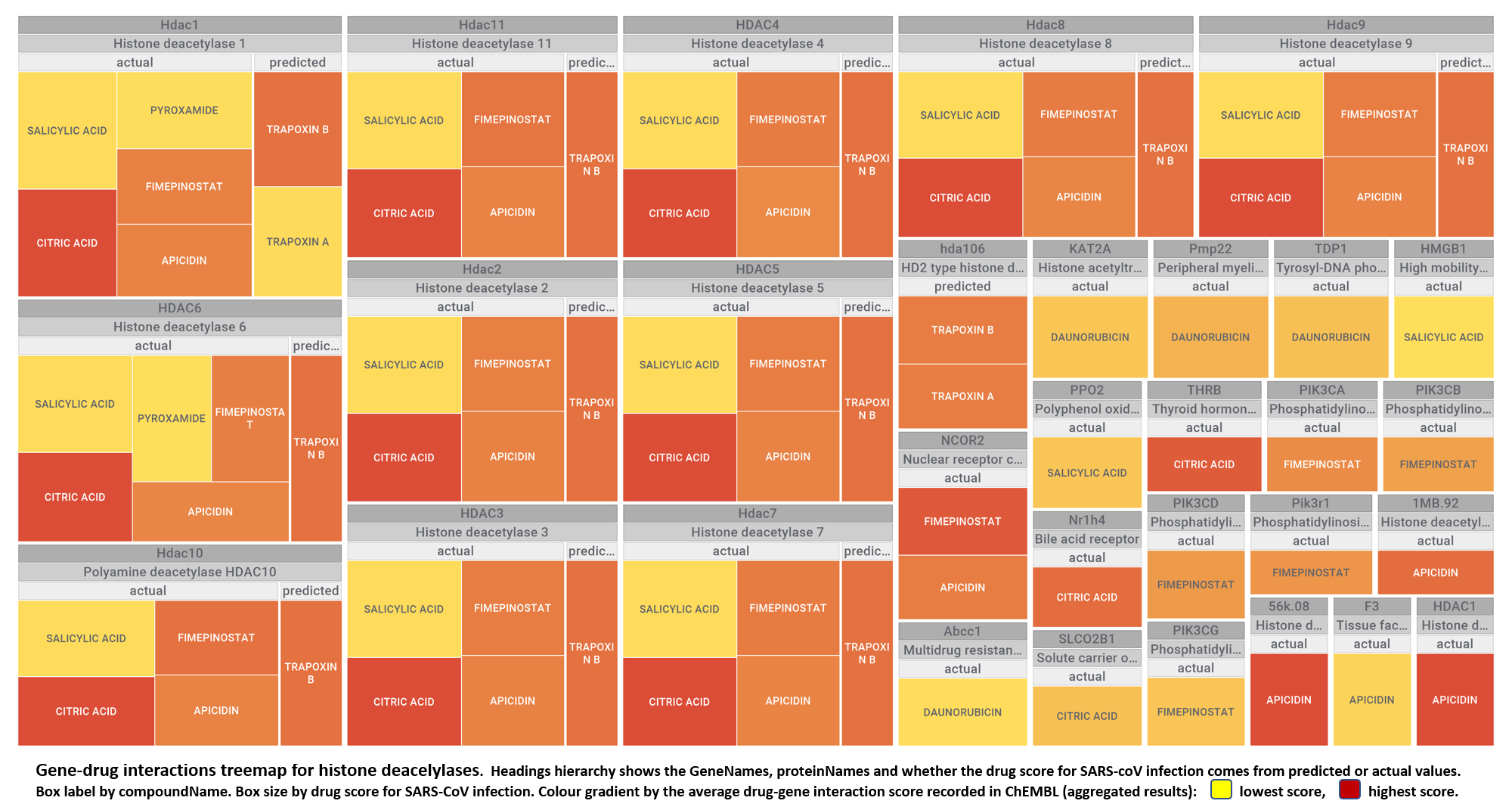
Here, we will examine some of the most relevant drugs identified as
antiviral and their molecular interactions recorded in ChEMBL DB.
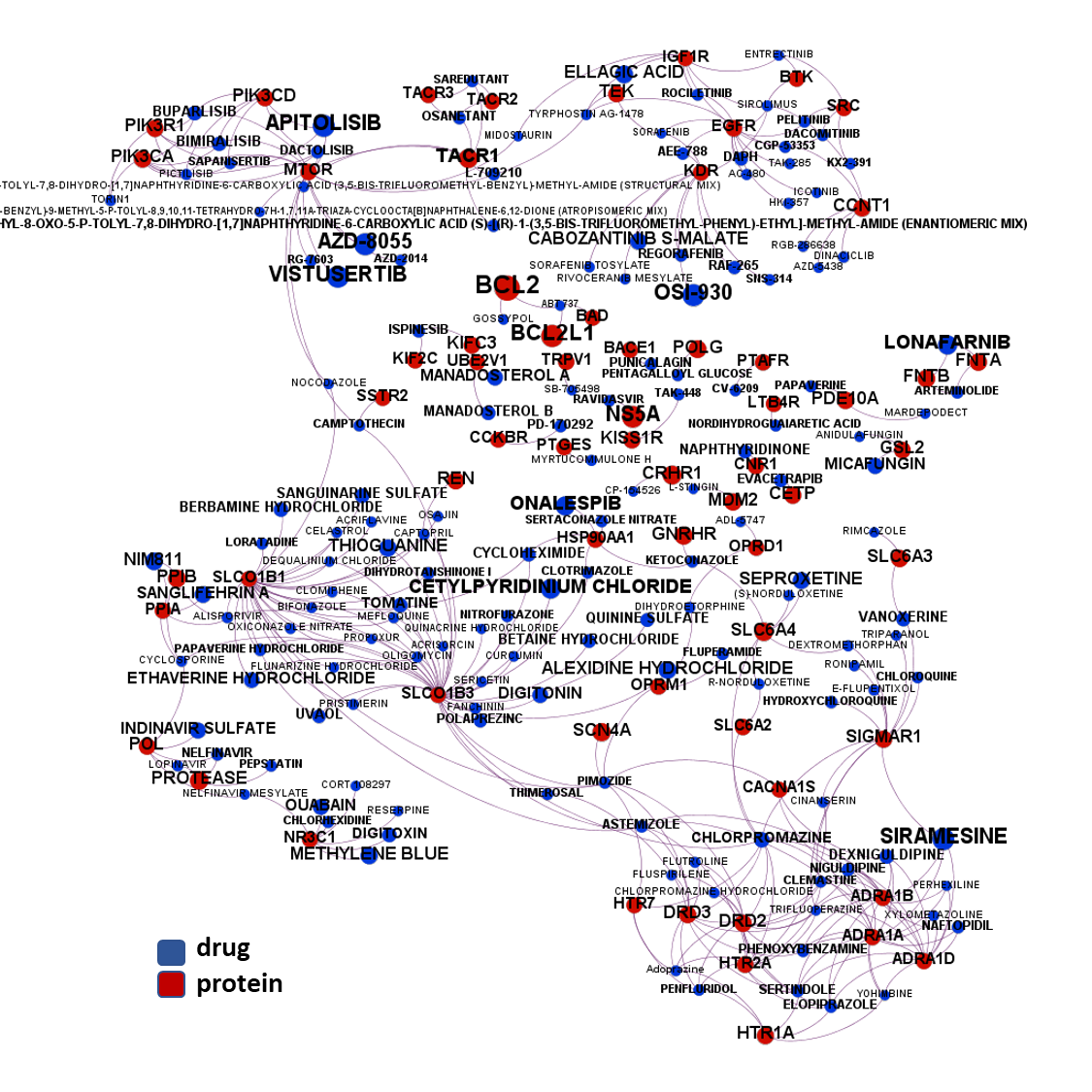
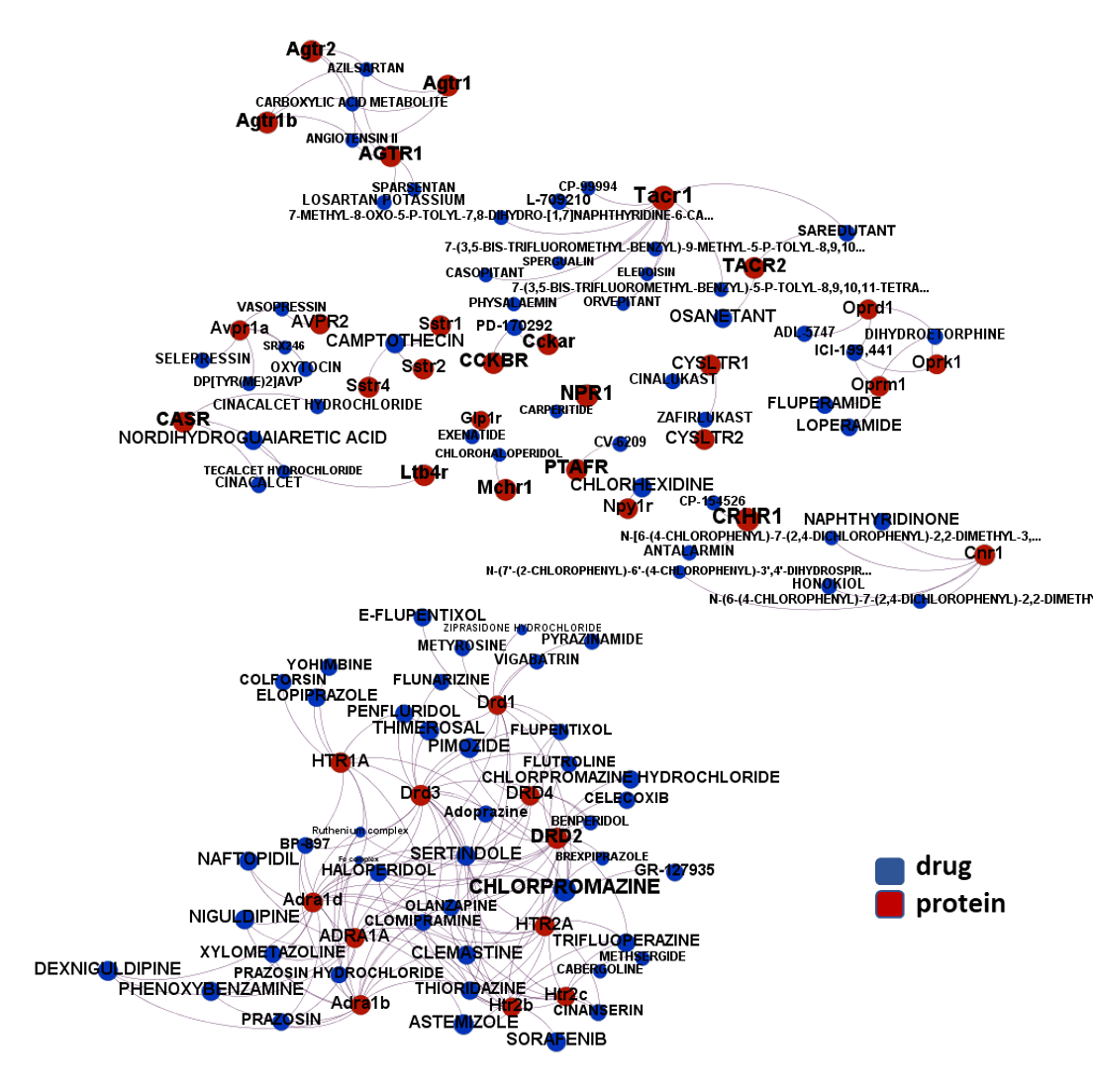
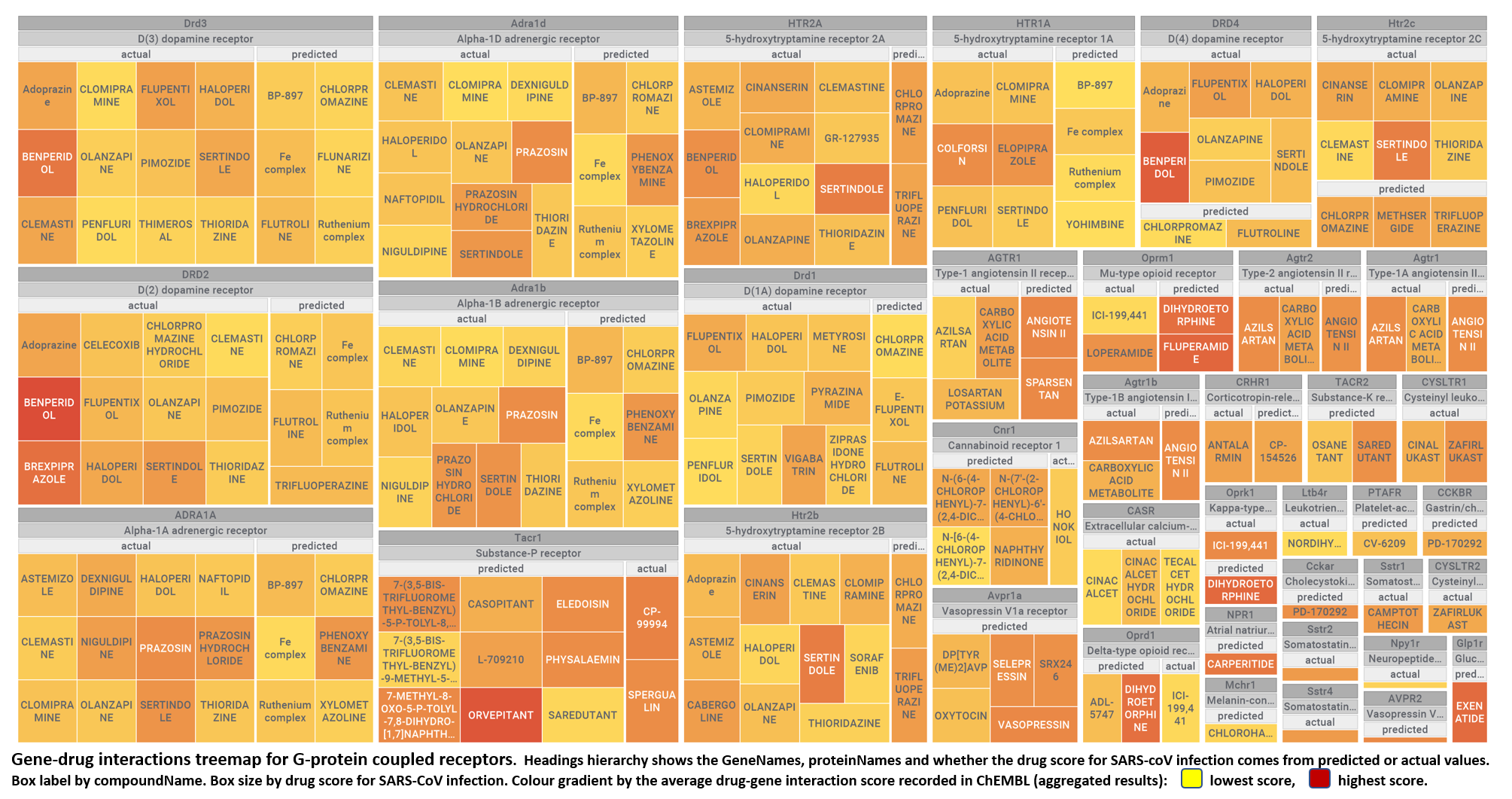
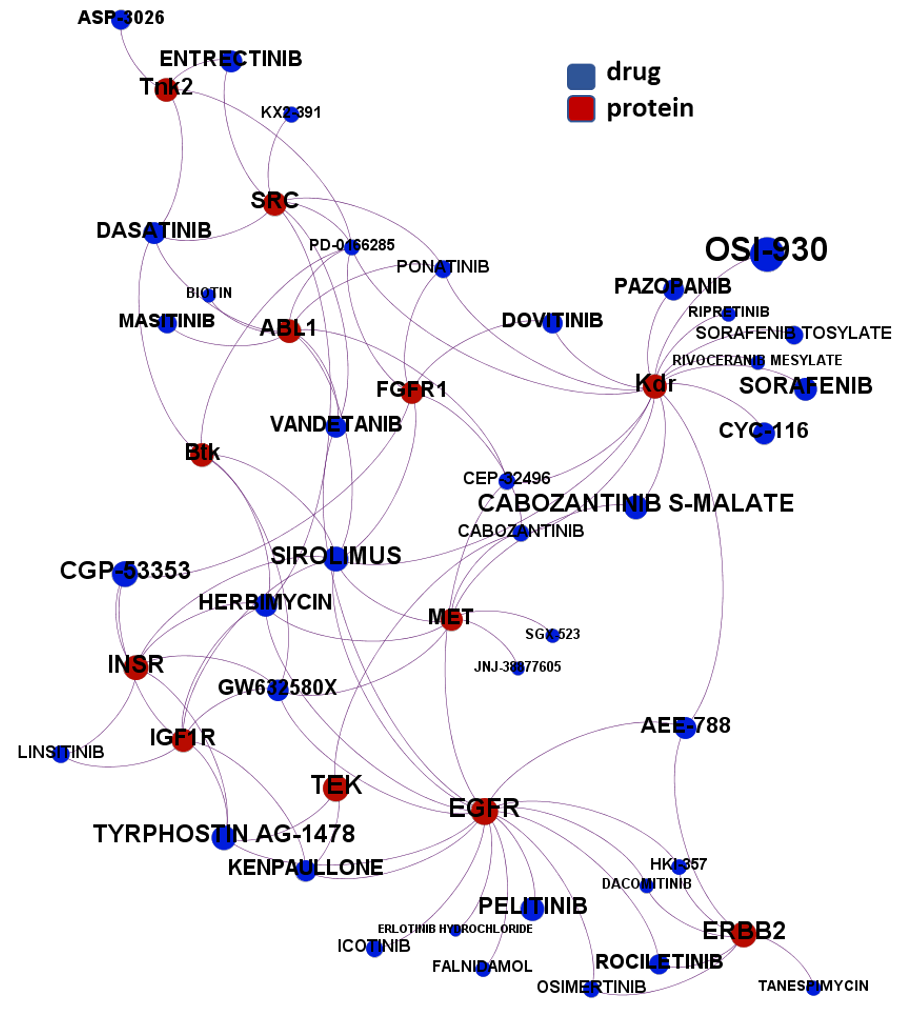
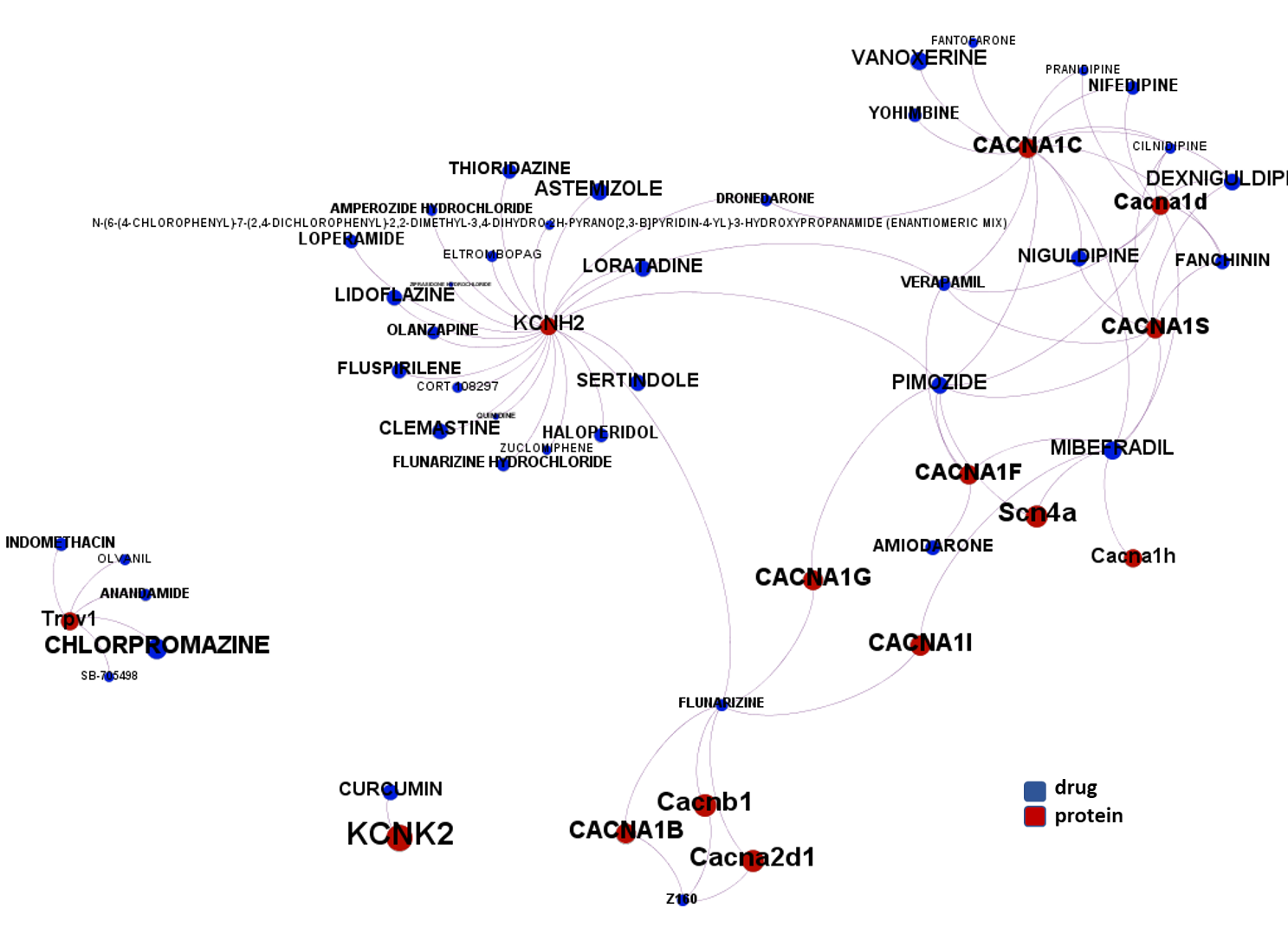
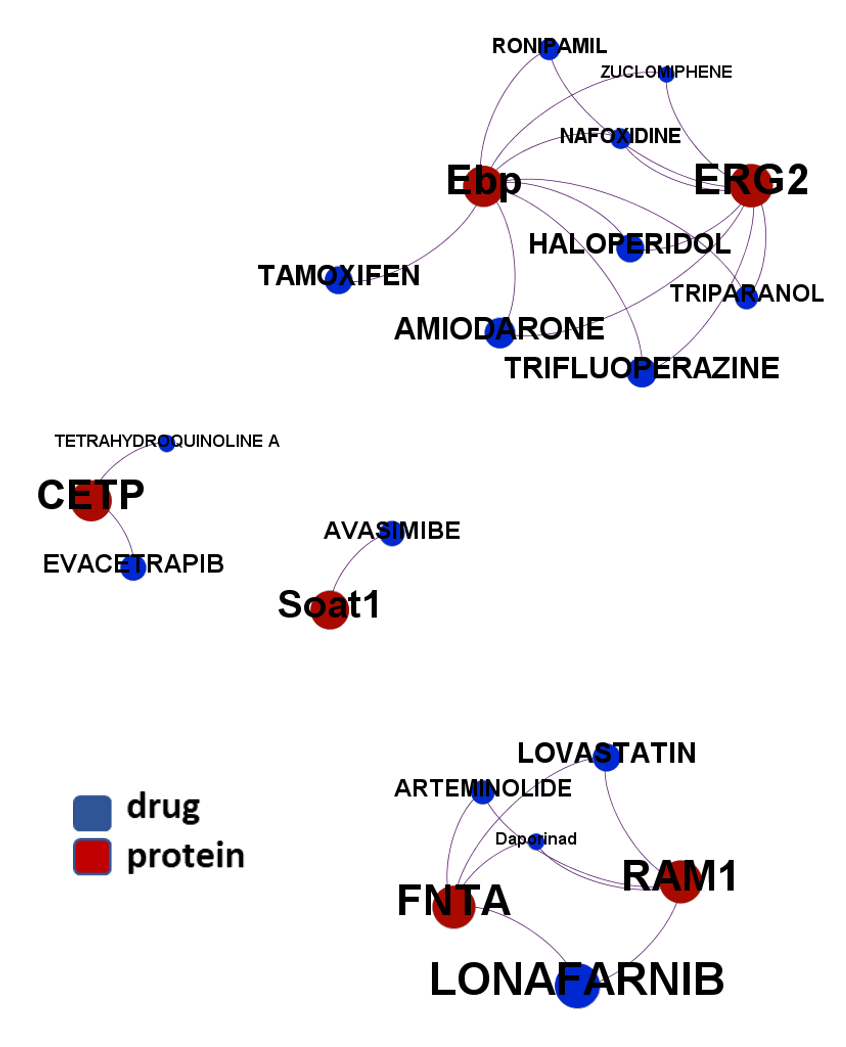
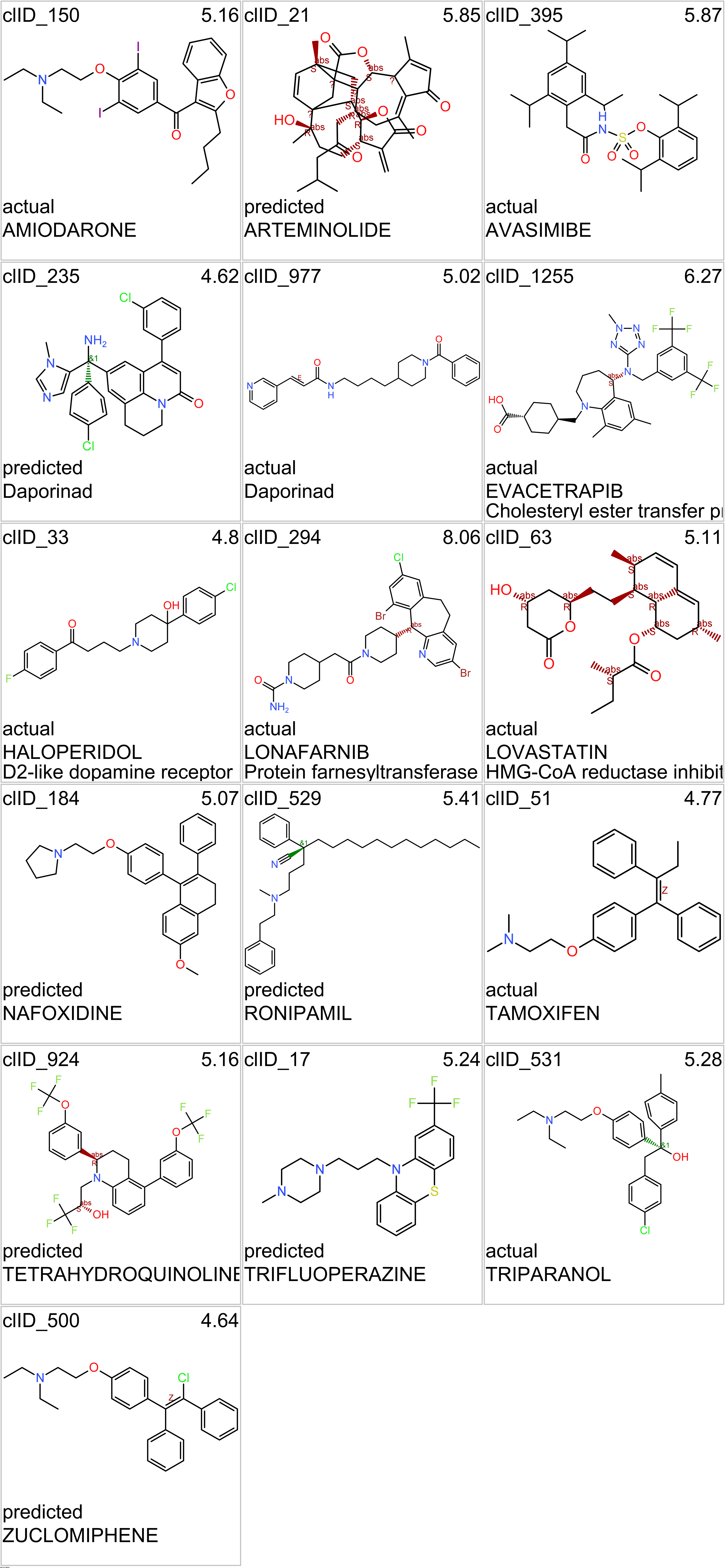
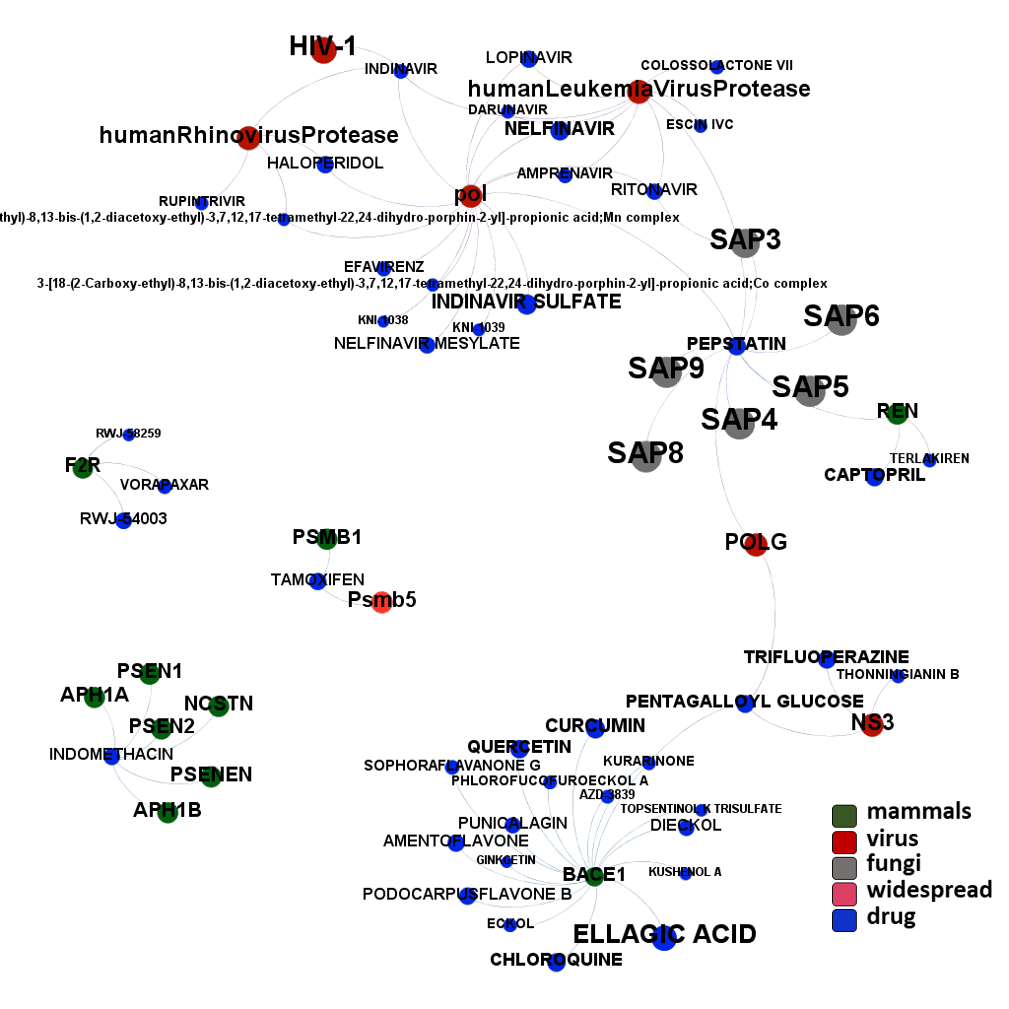
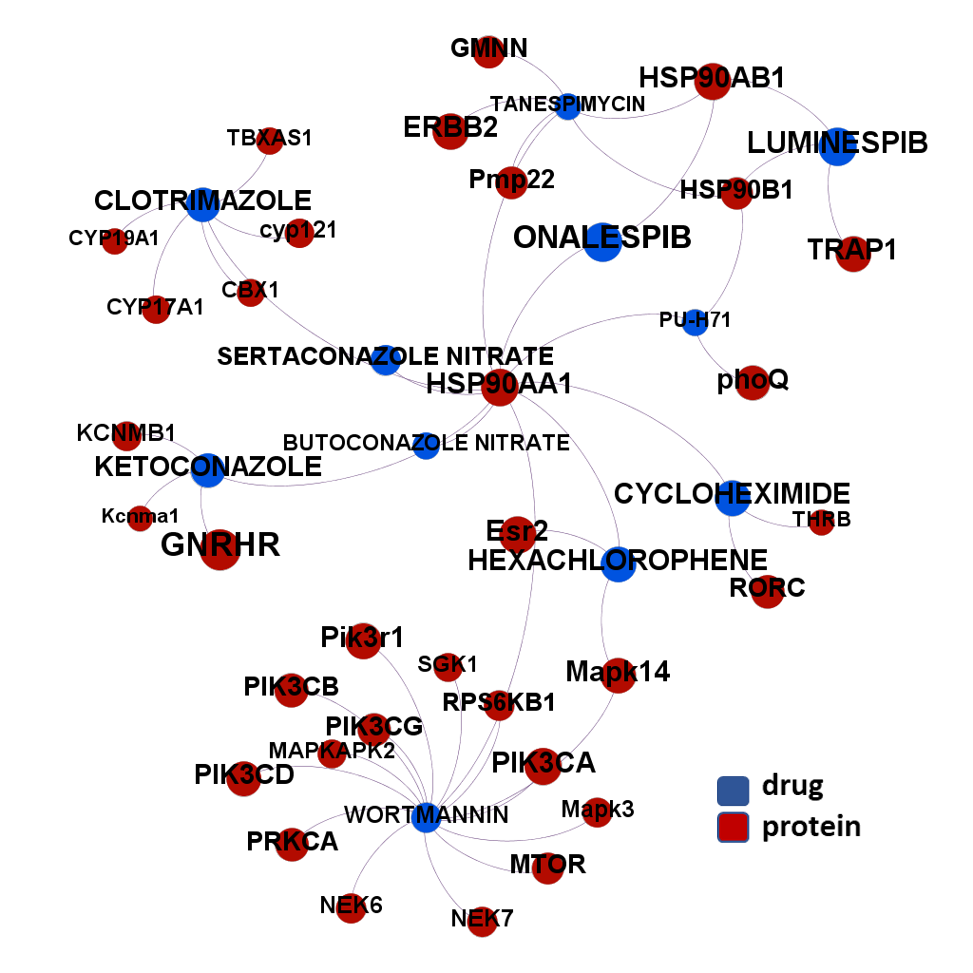
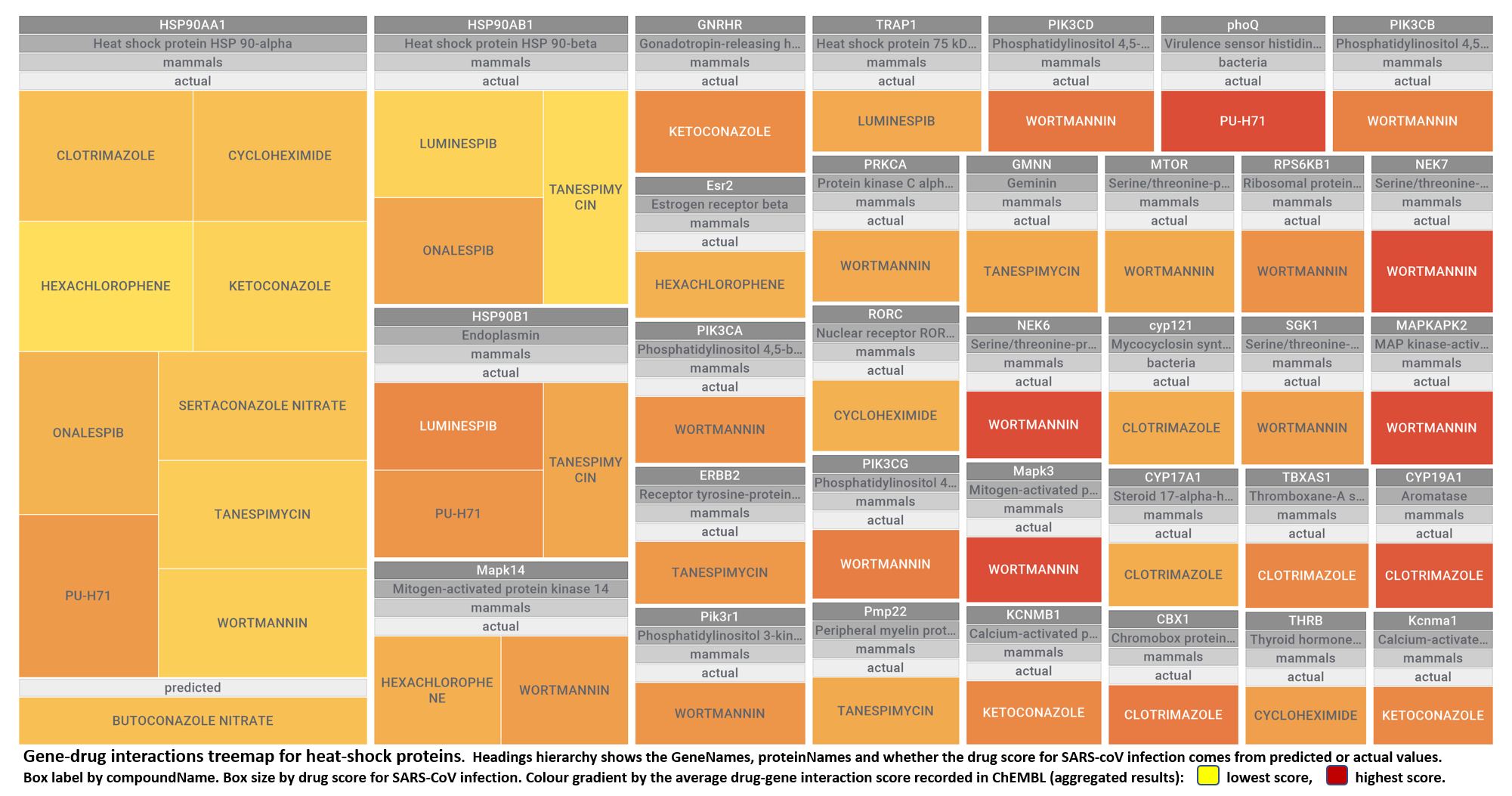
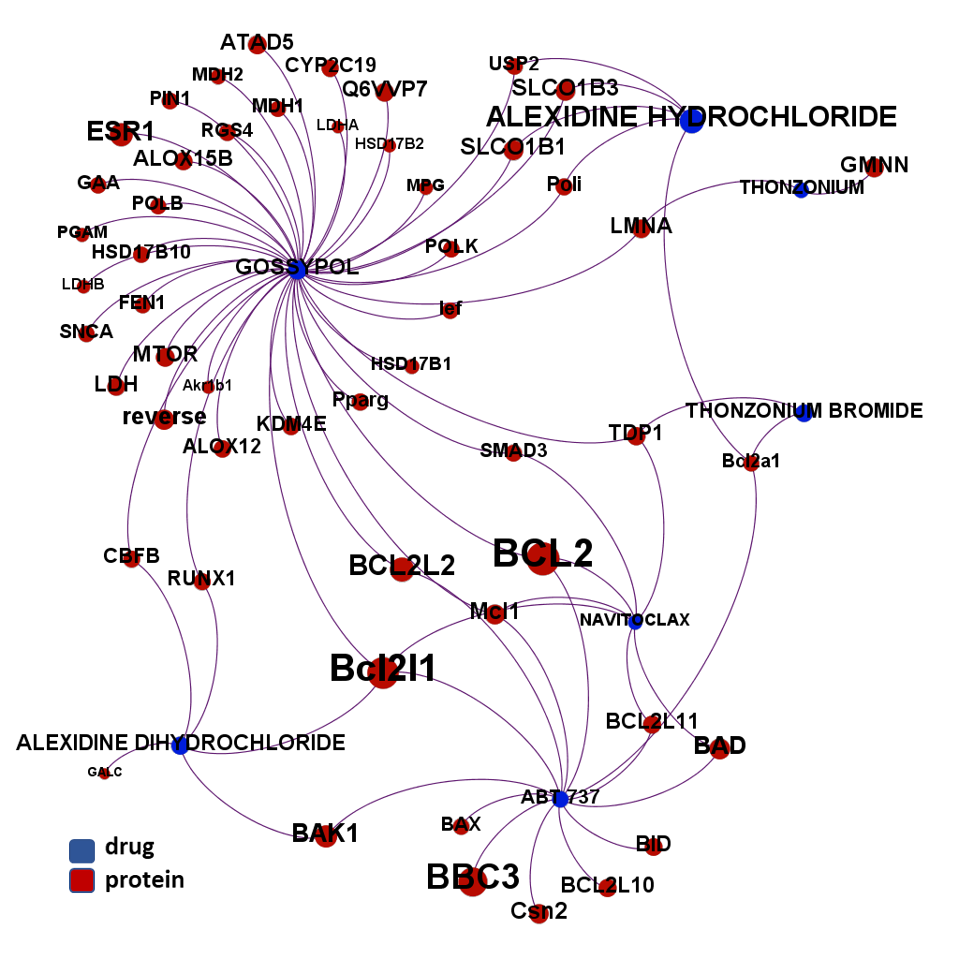
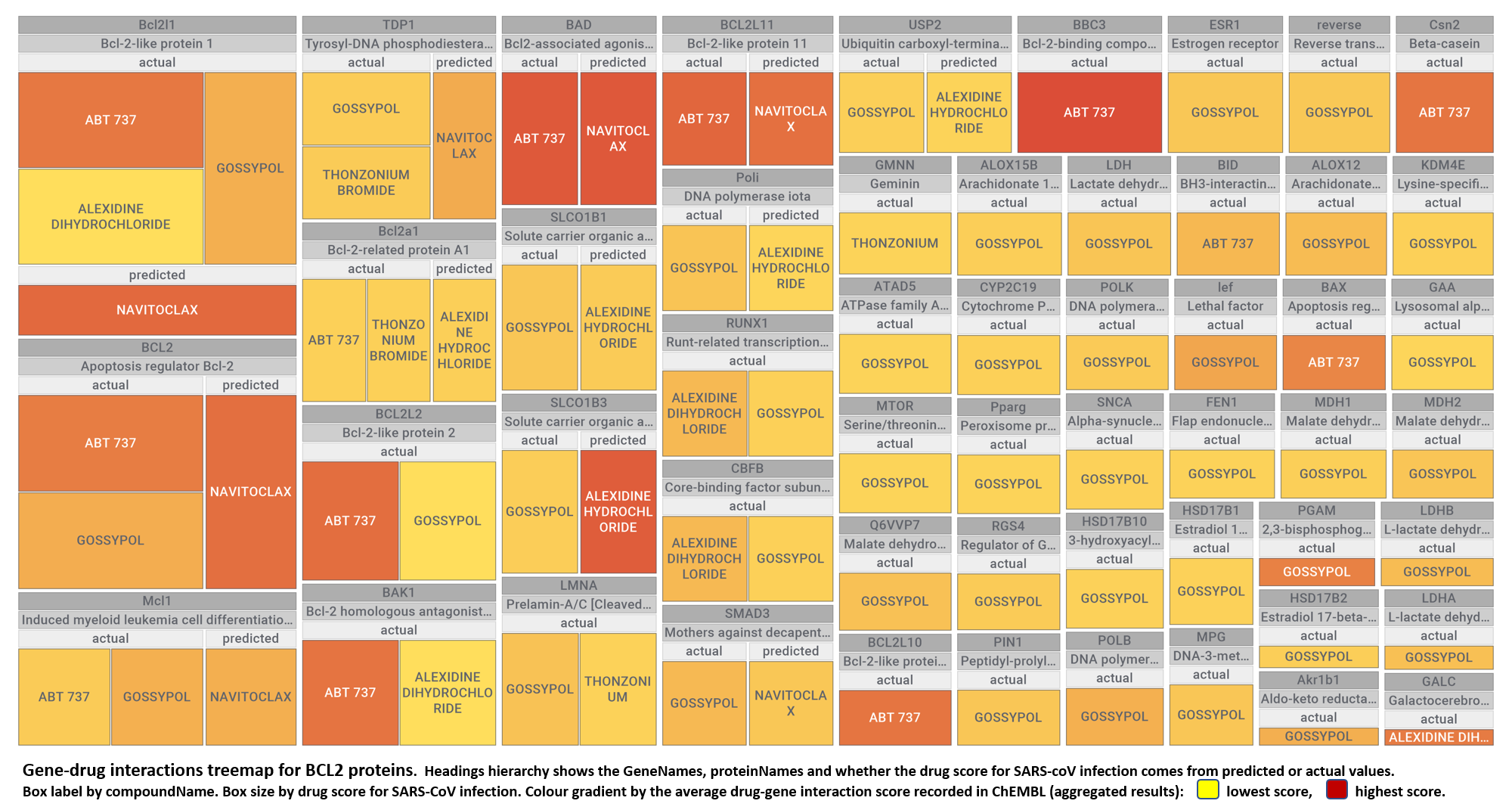
The 4k repurposed substances show a complex interaction map, as depicted in the down-left graph. It’s difficult to see anything, but we can apply filters to select best scored drugs and molecular targets with most potent recorded interactions to make them visible.
just click to expand charts.
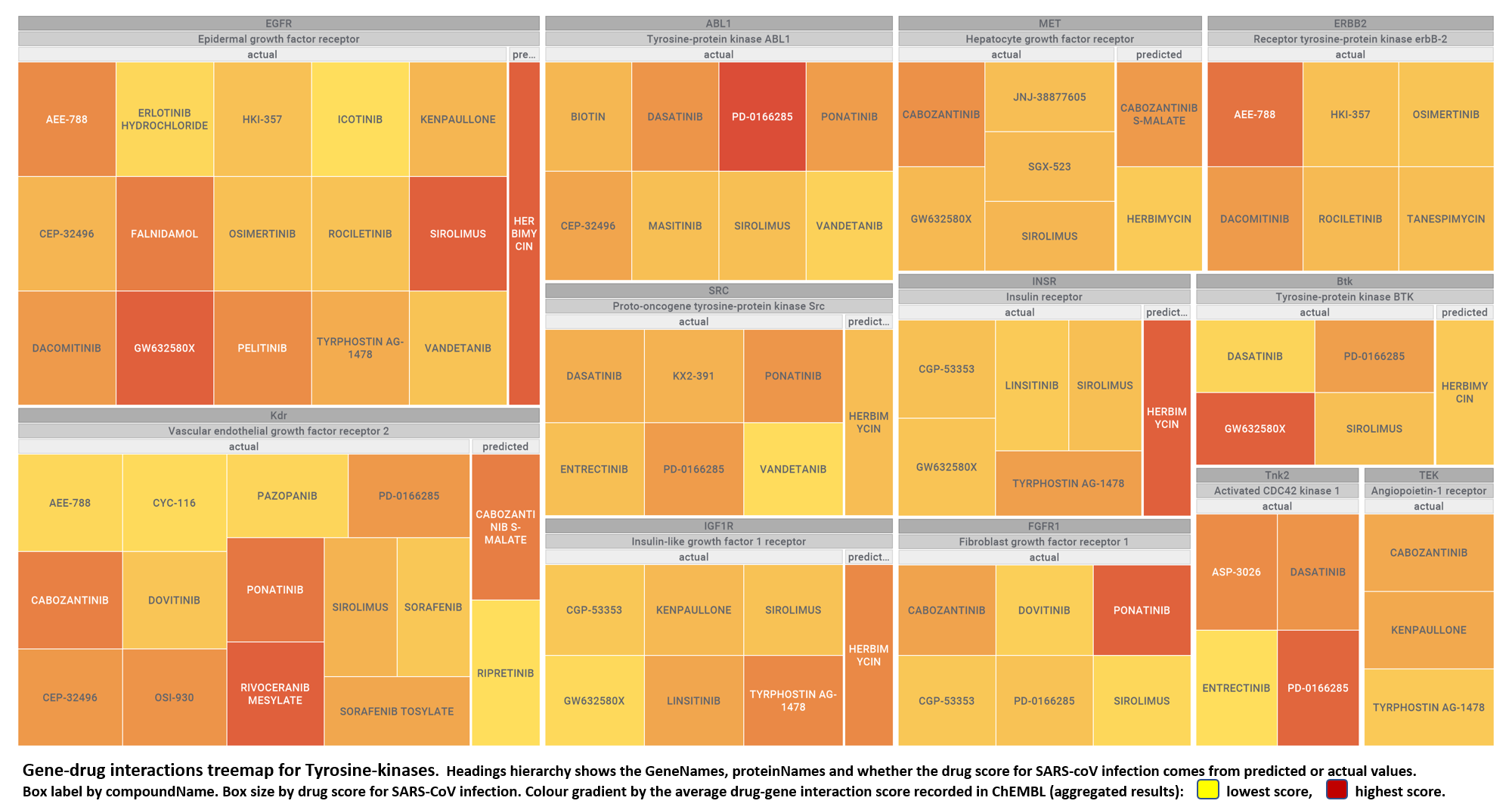
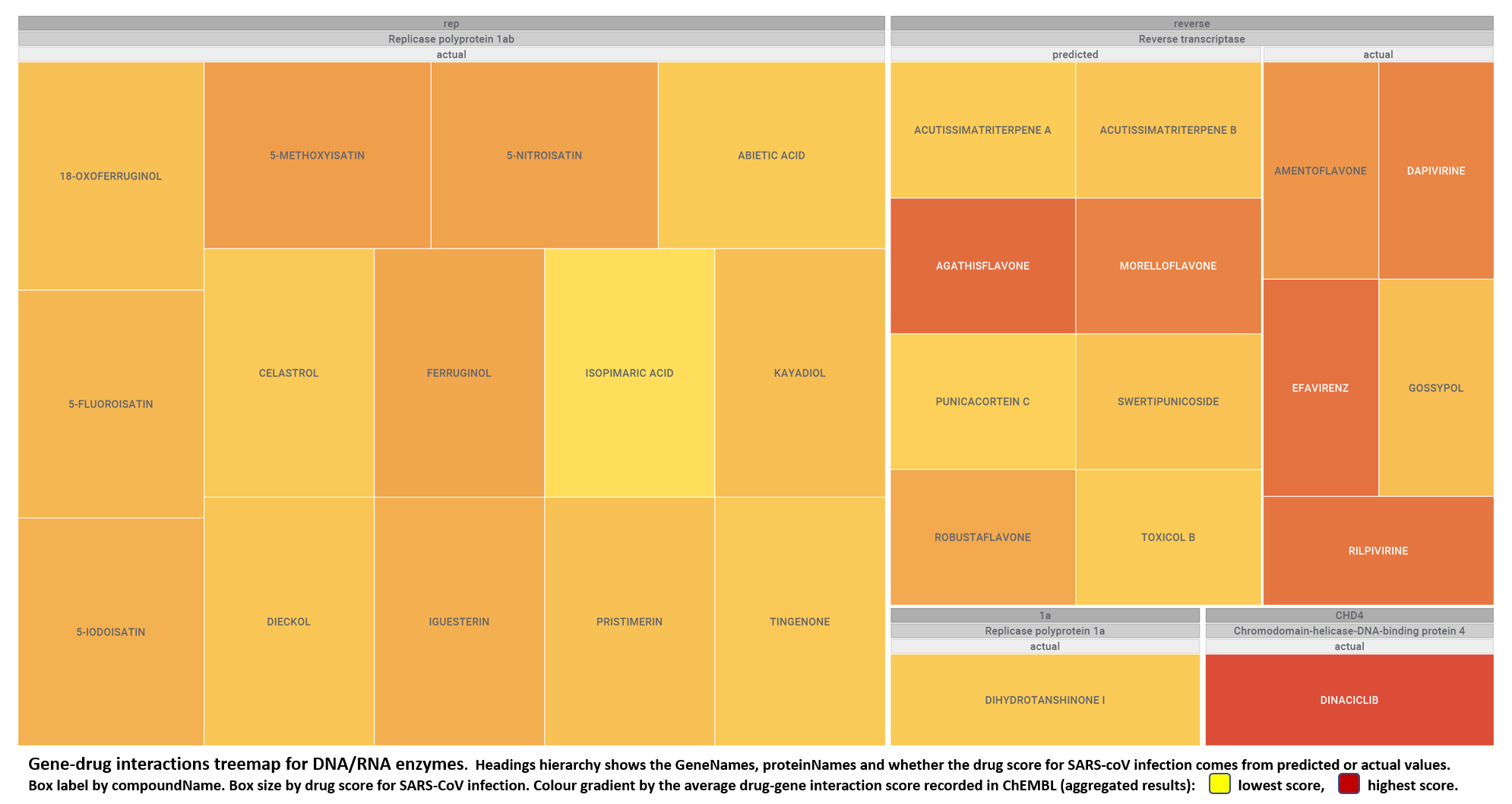
But, for more specific views, it may be better to go through specific mechanisms, just clicking on the tabs below.
Drugs acting through...
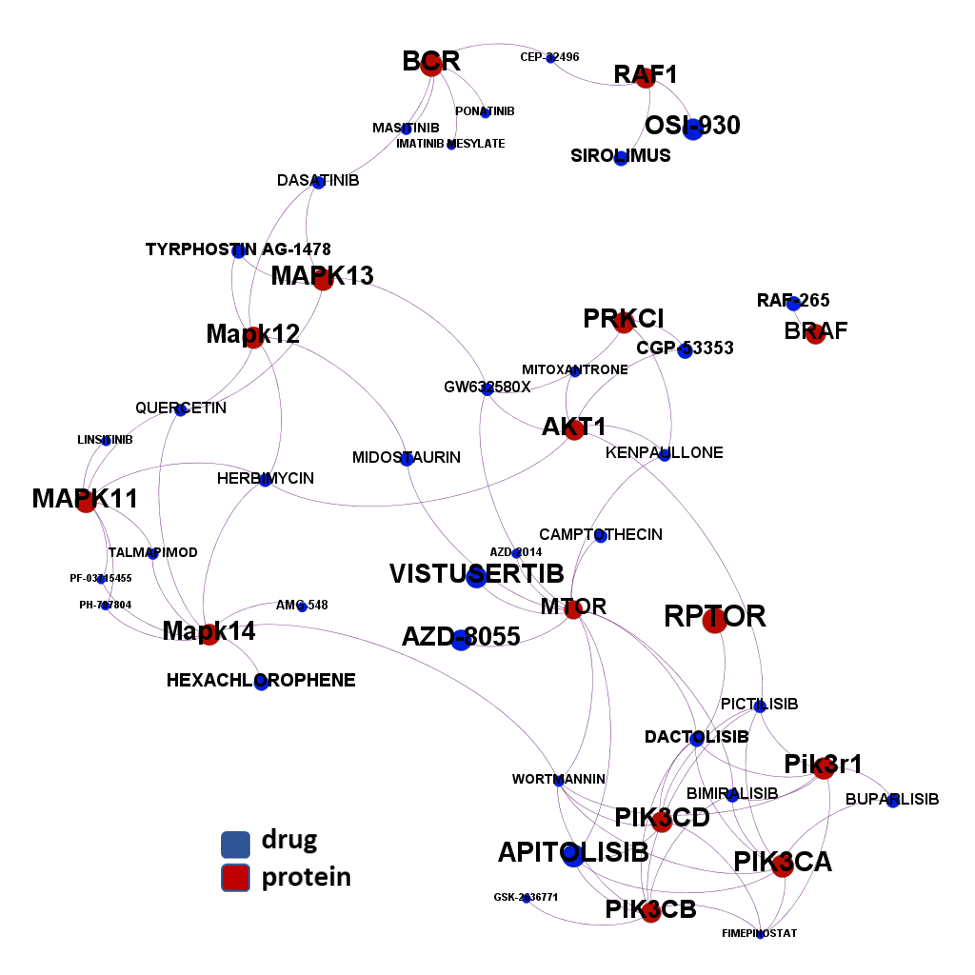
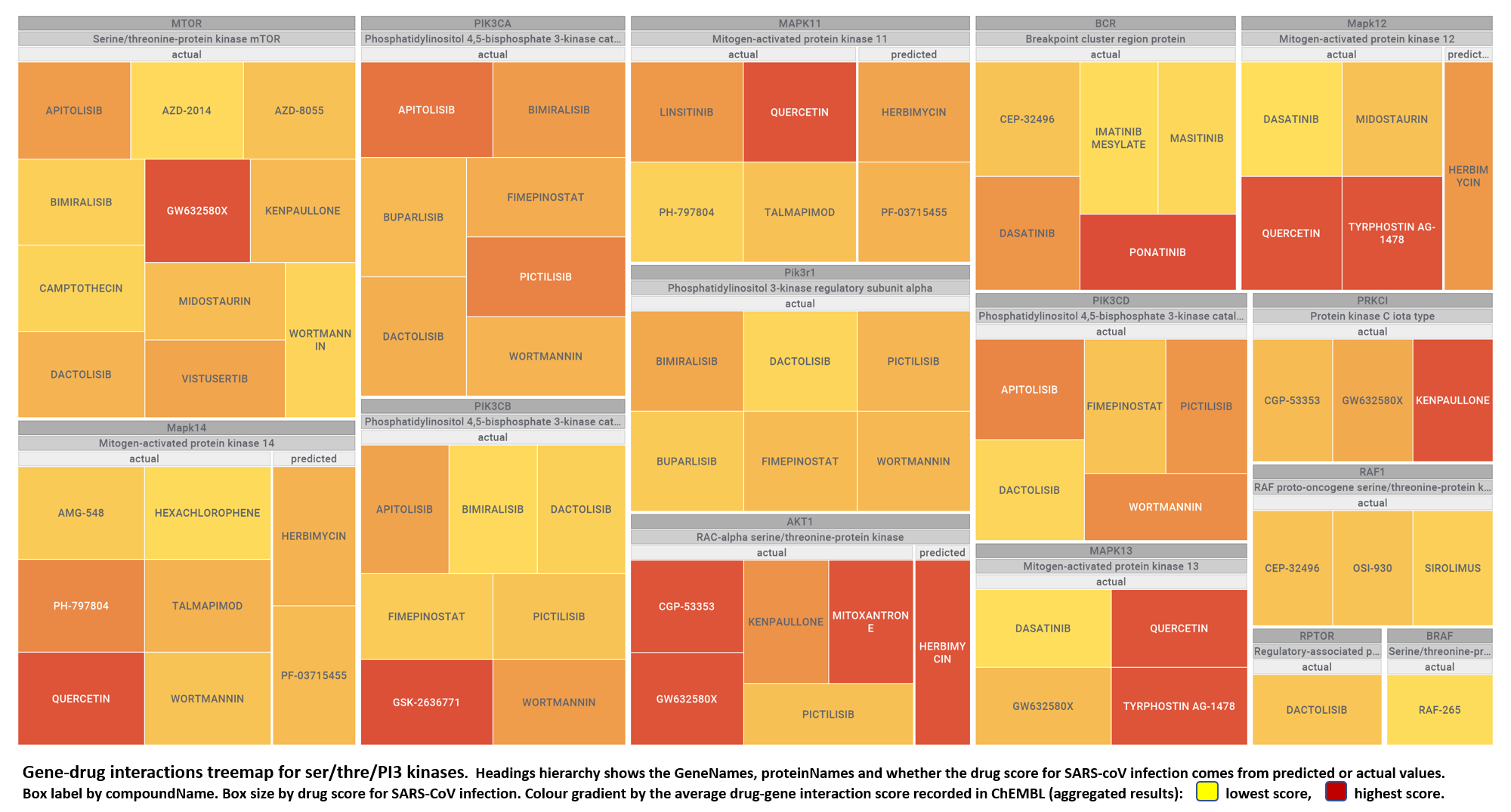
- Ser/Thre/PI3Kinases
- Transporters
- G-protein coupled receptors
- Tyrosine-kinases
- Nucleic acids interacting proteins
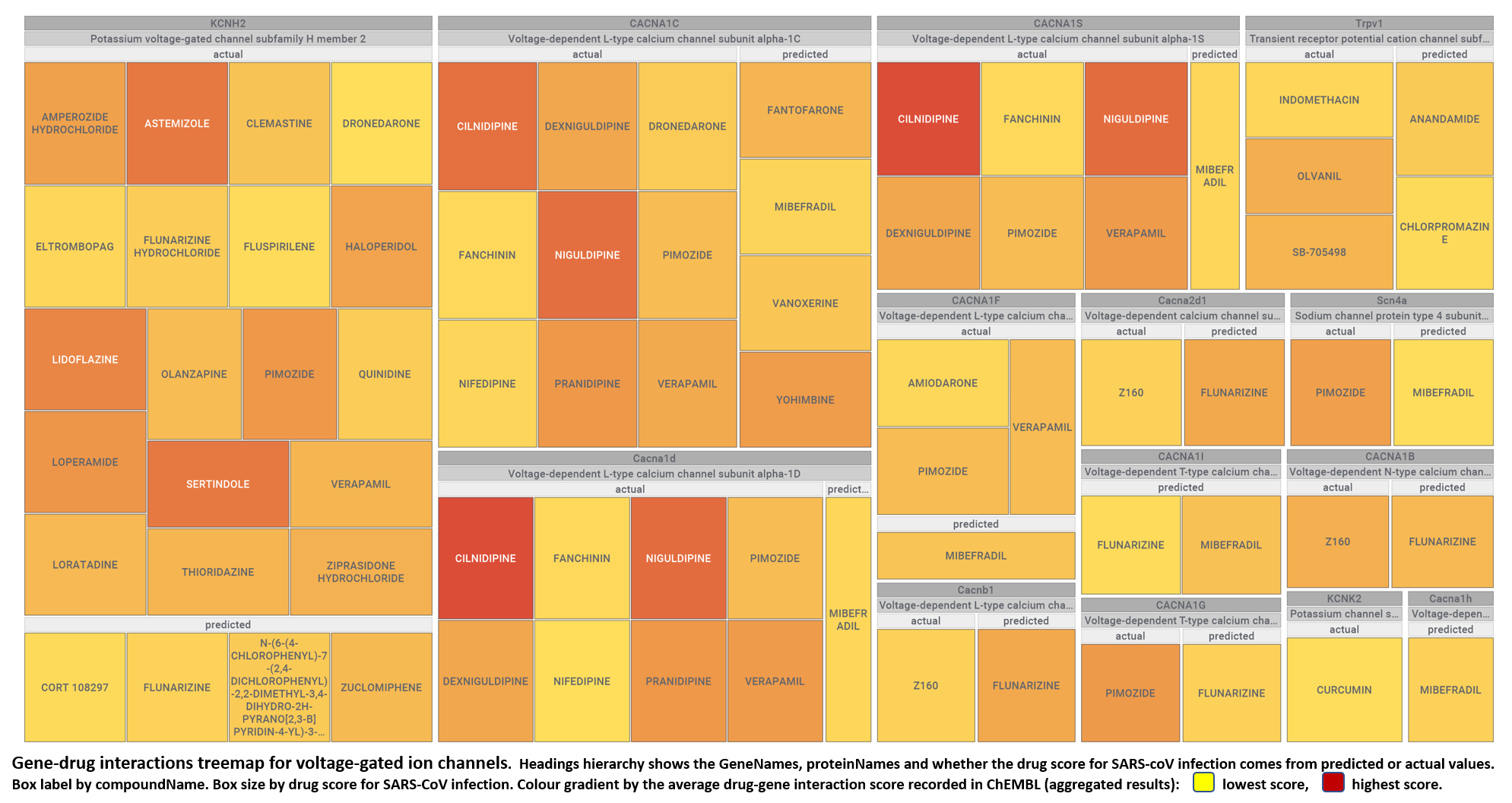
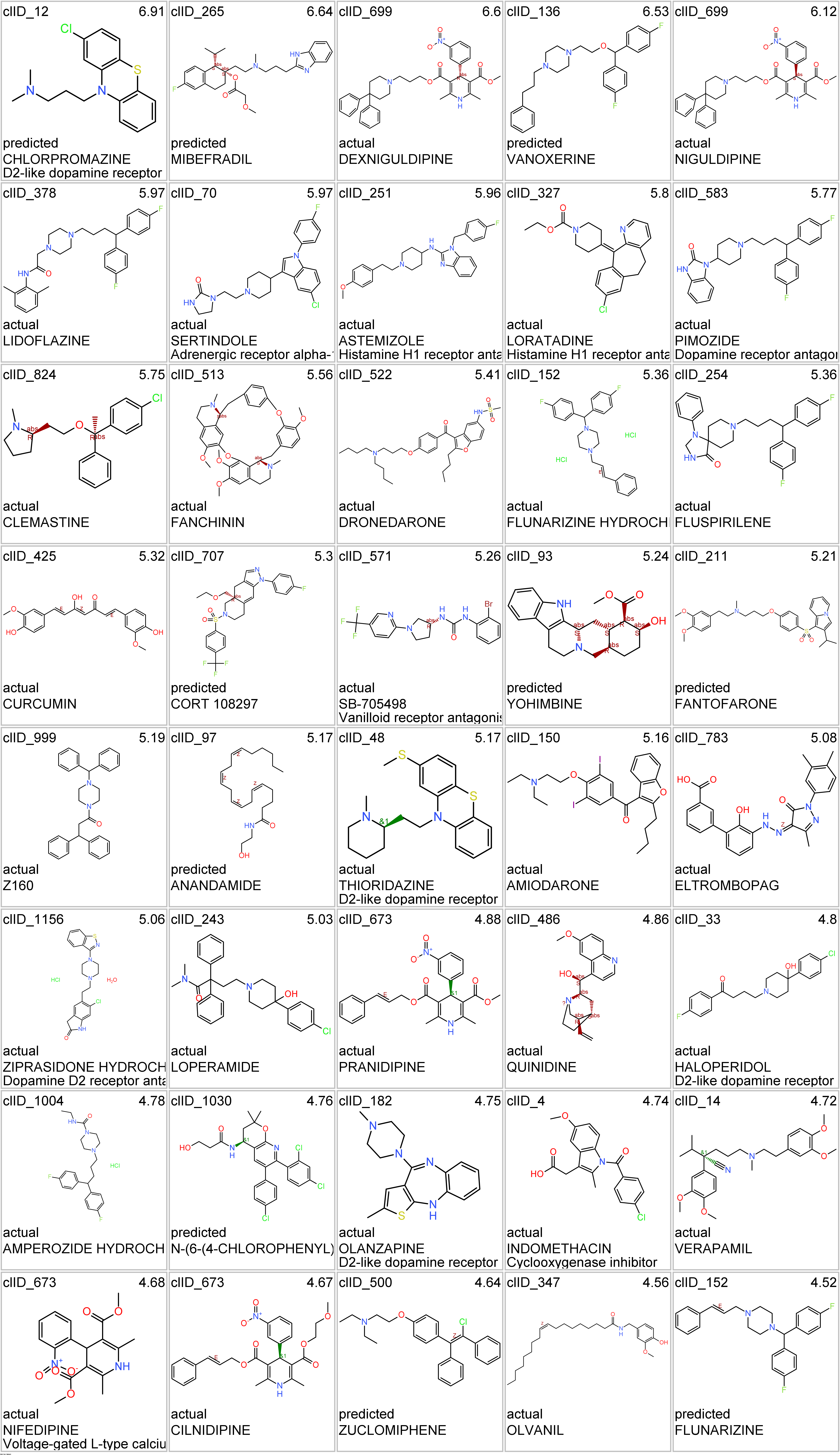
- Voltage-gated ion channels
- Steroid synthesis enzymes
- Ub-ligases & bromodomains
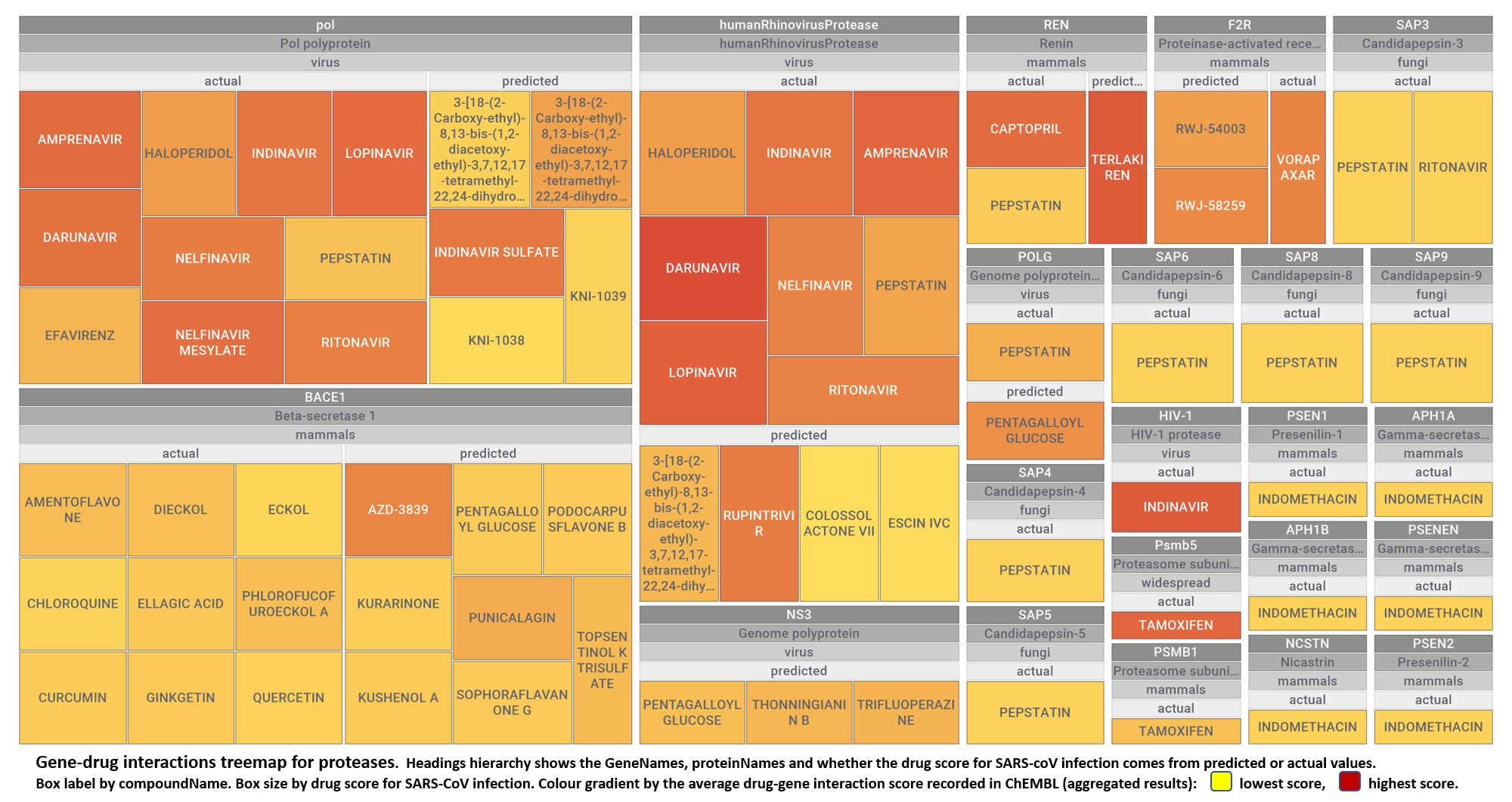
- Proteases
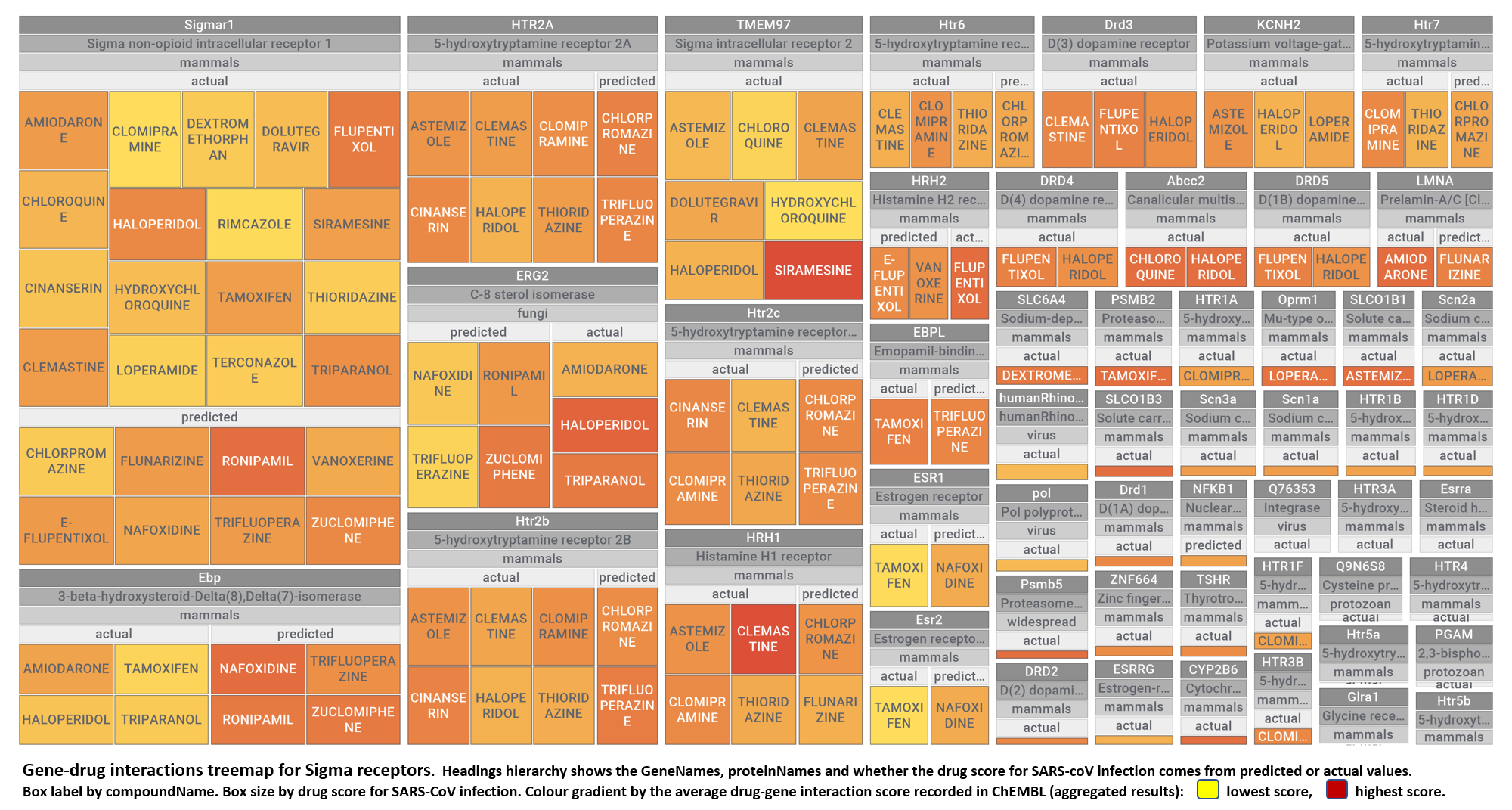
- Sigma receptors
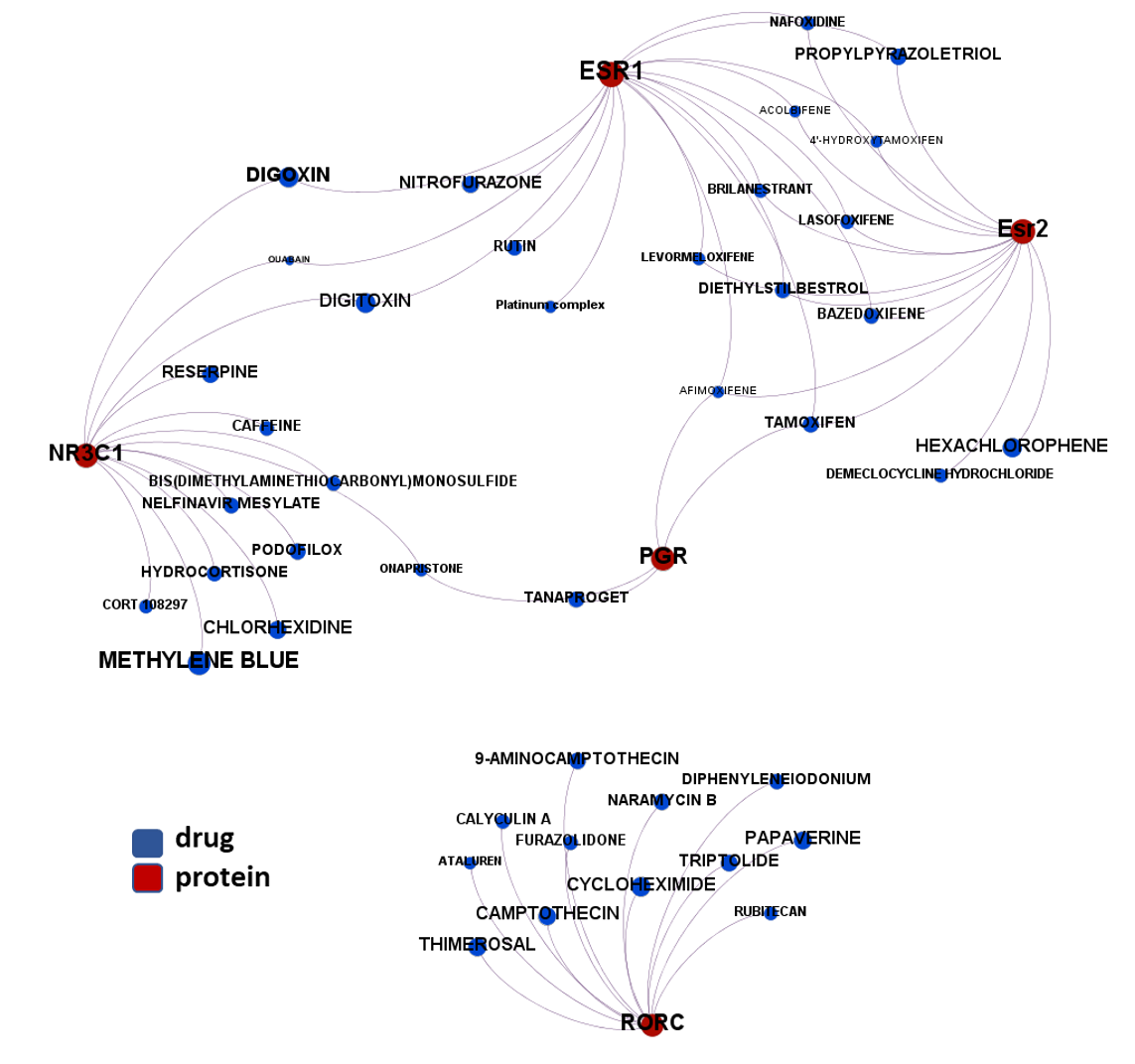
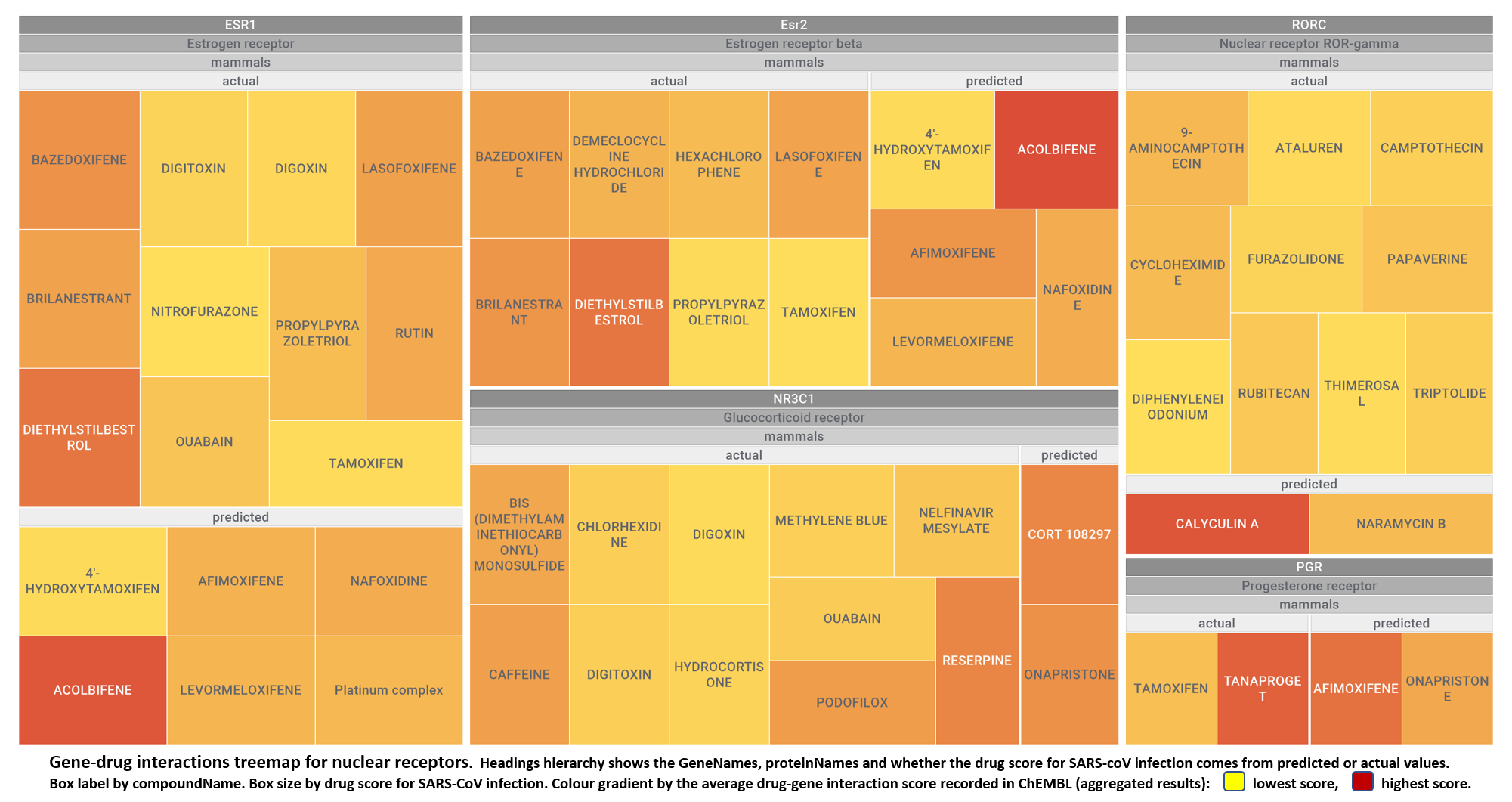
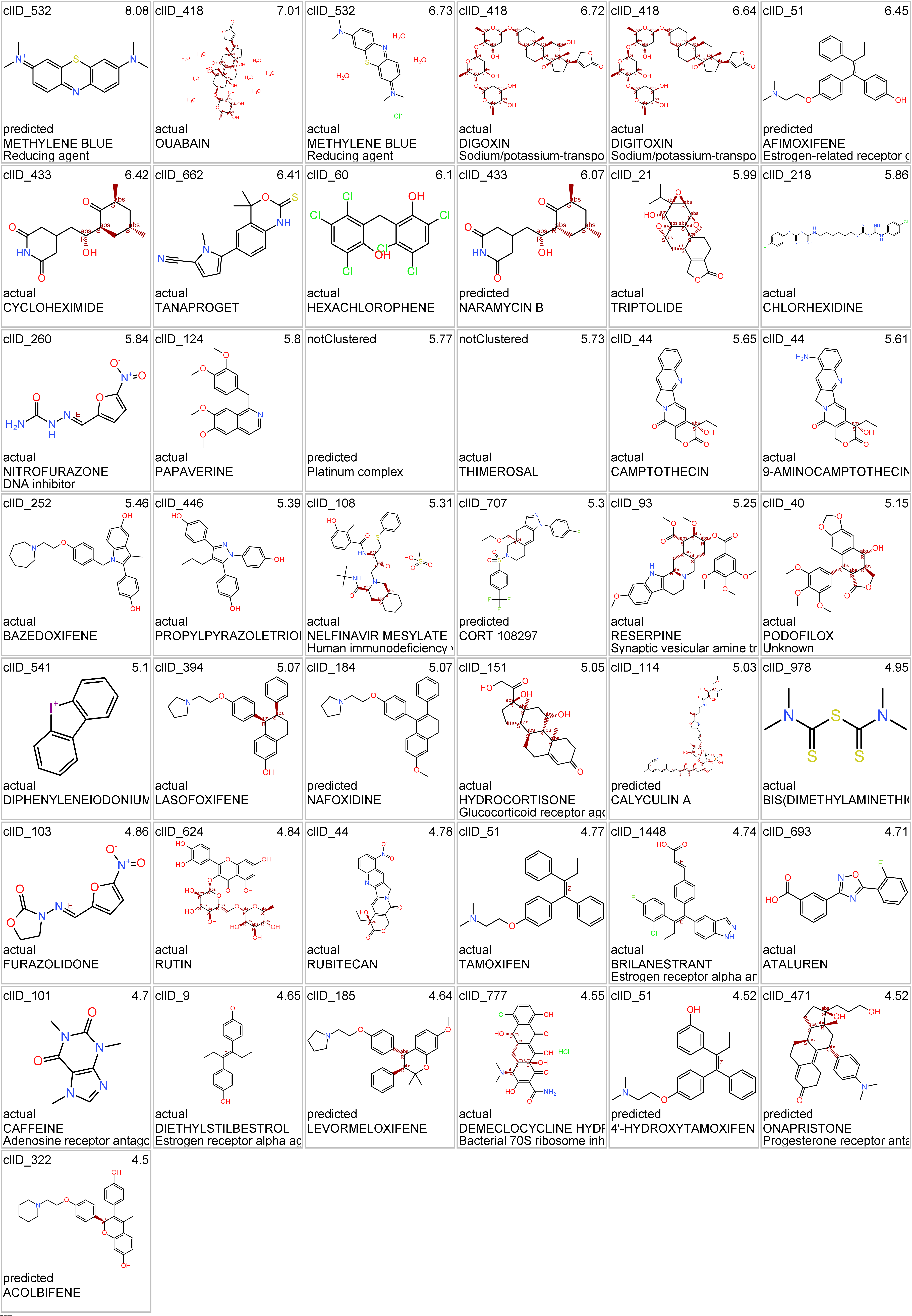
- Nuclear receptors
- Heat shock proteins
- BCL2 protein family
- Potential therapeutics for SARS from Reactome
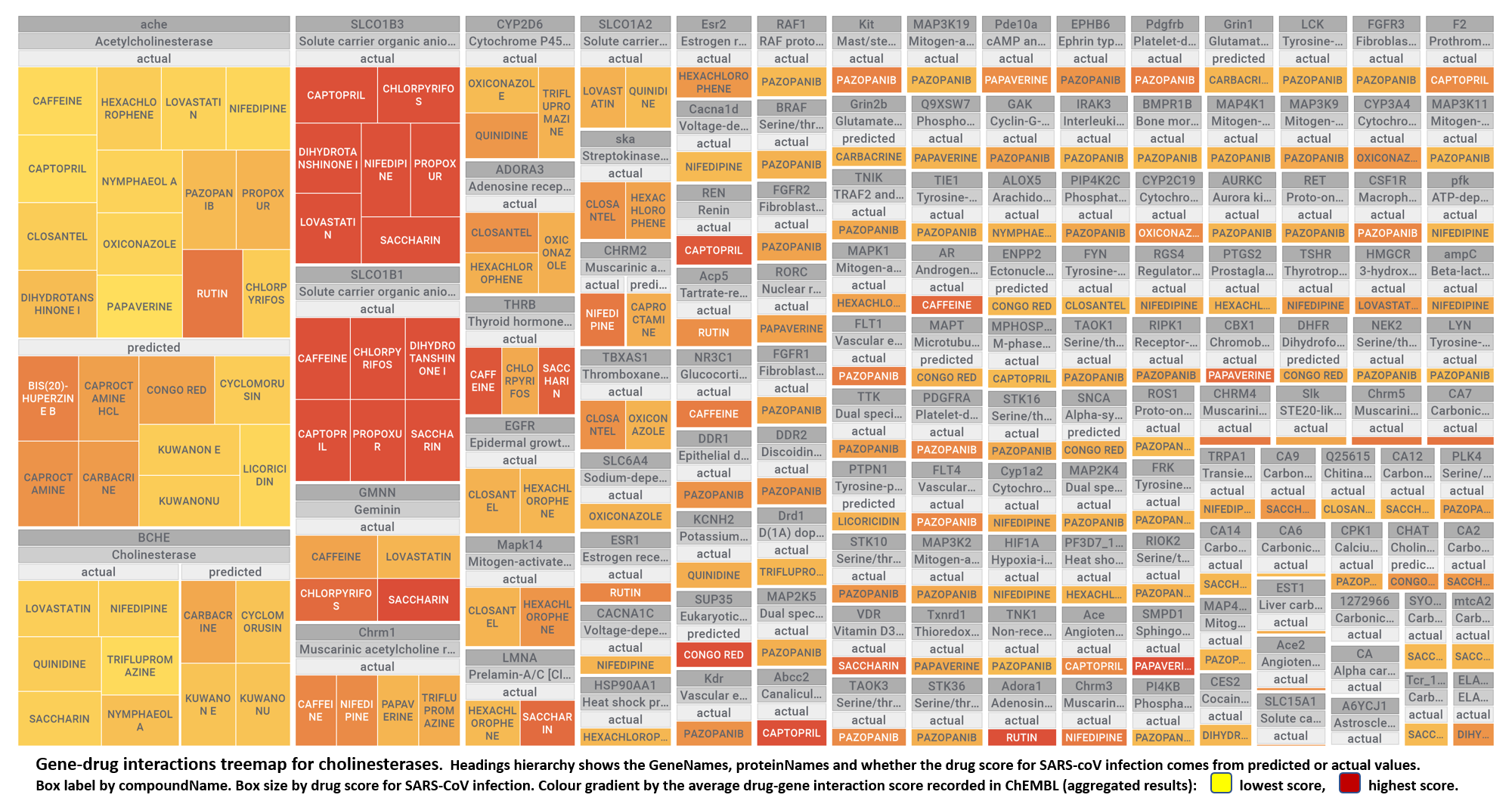
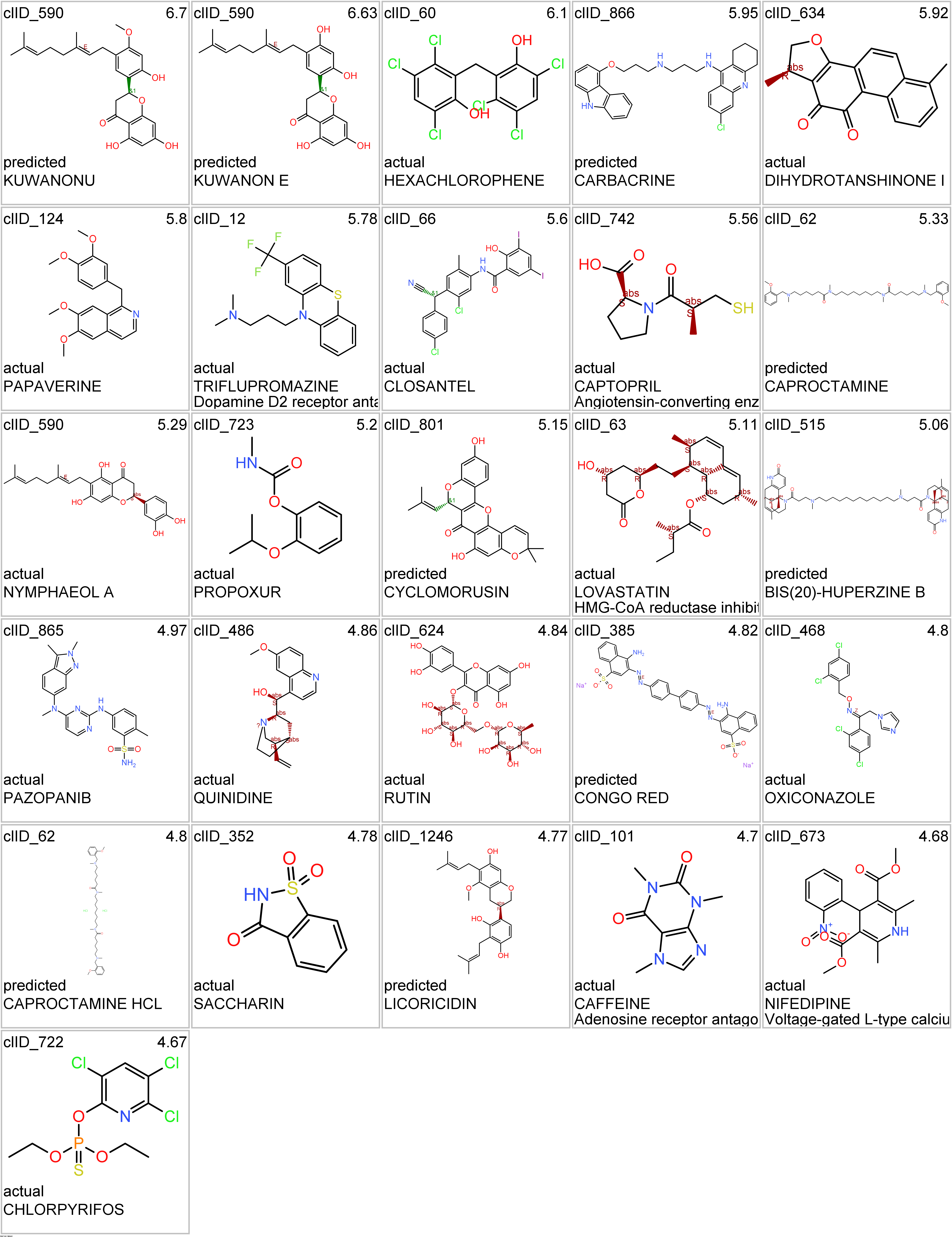
- Cholinesterases
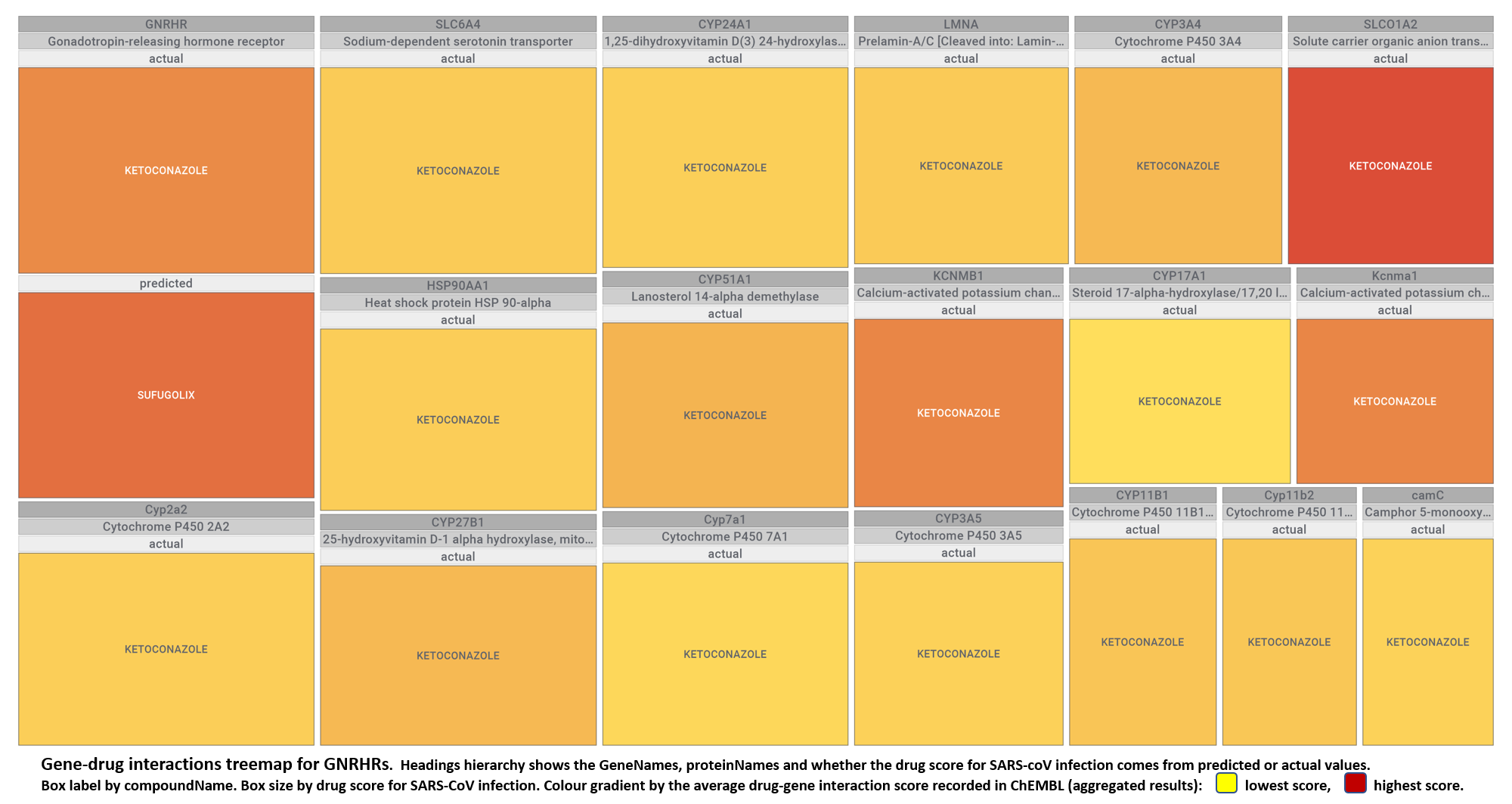
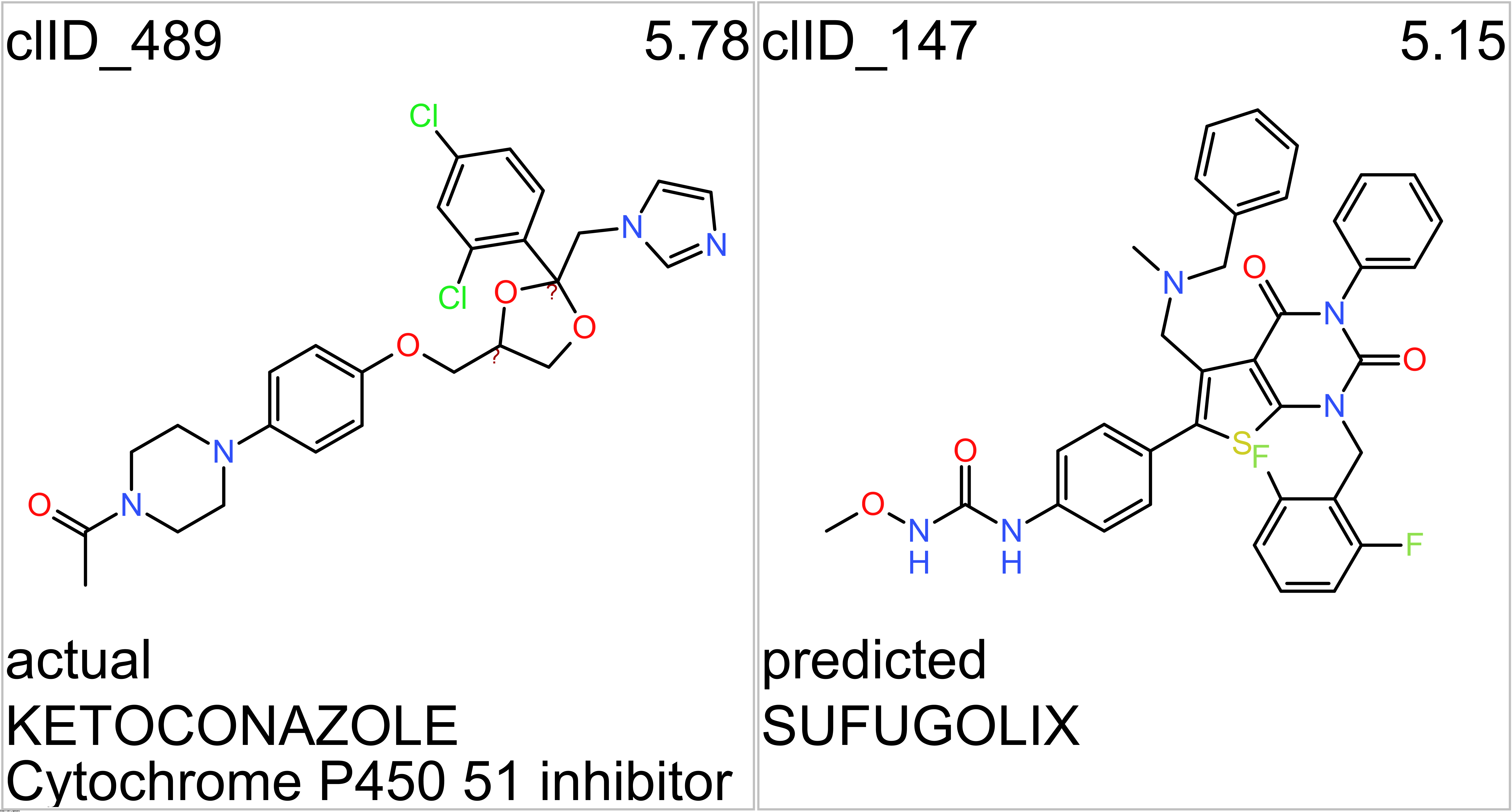
- Gonadotropin-releasing hormone receptors
- ATPase ion pumps
- Histone deacetylases
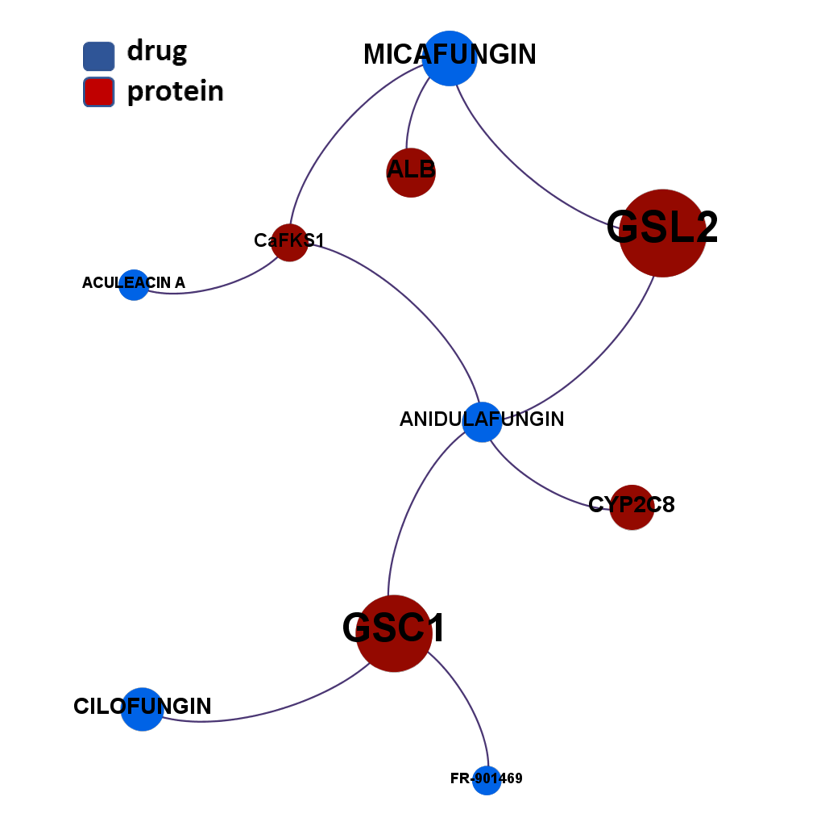
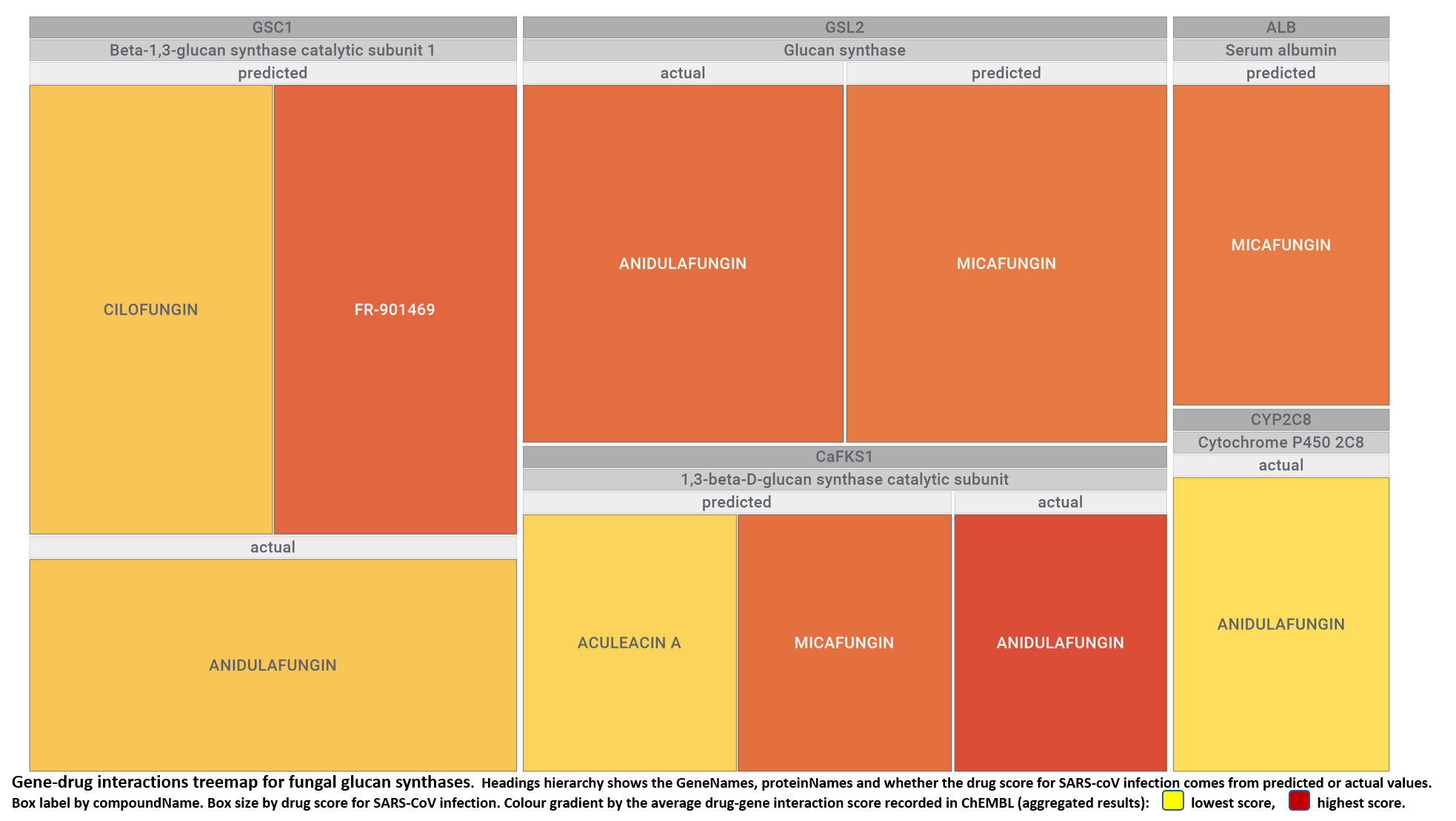
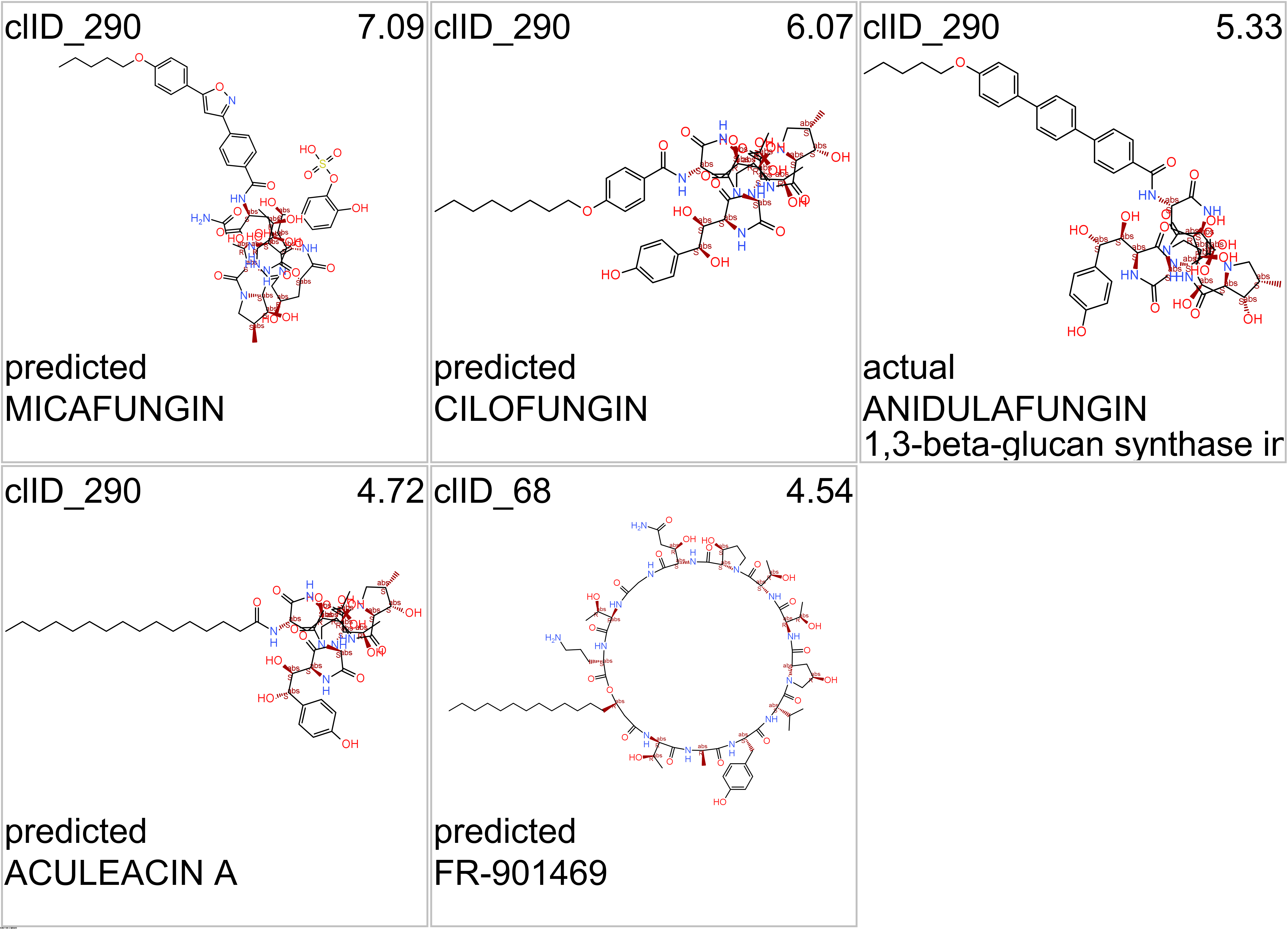
- Fungal glucan synthases
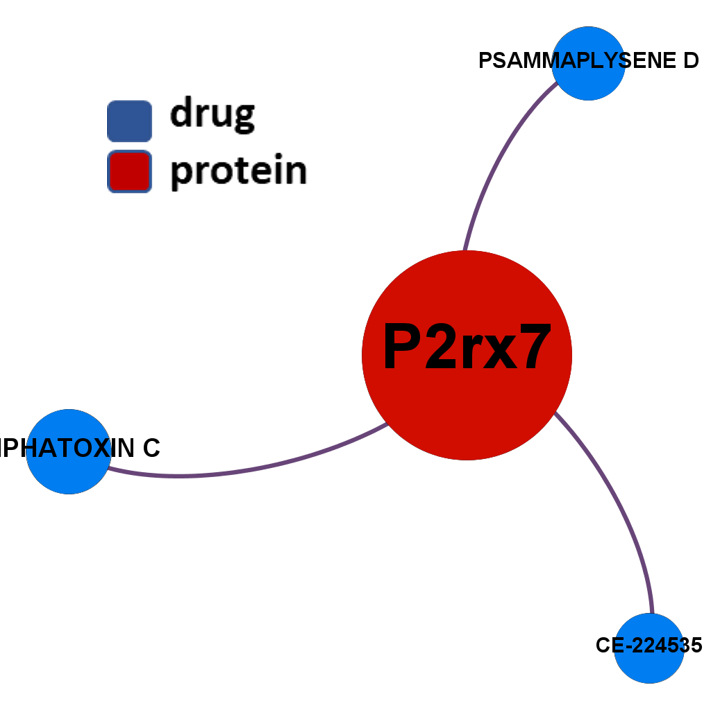
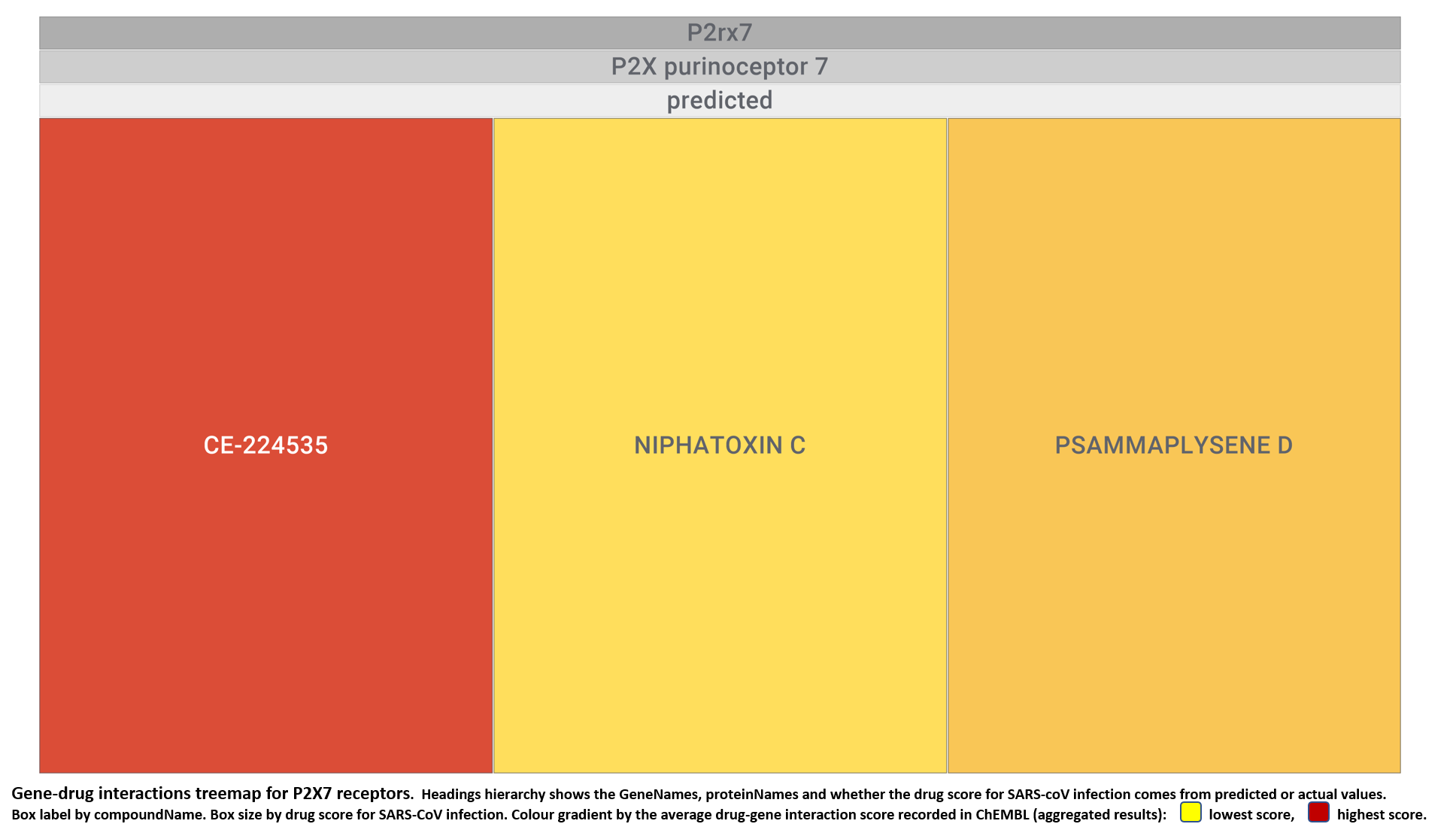
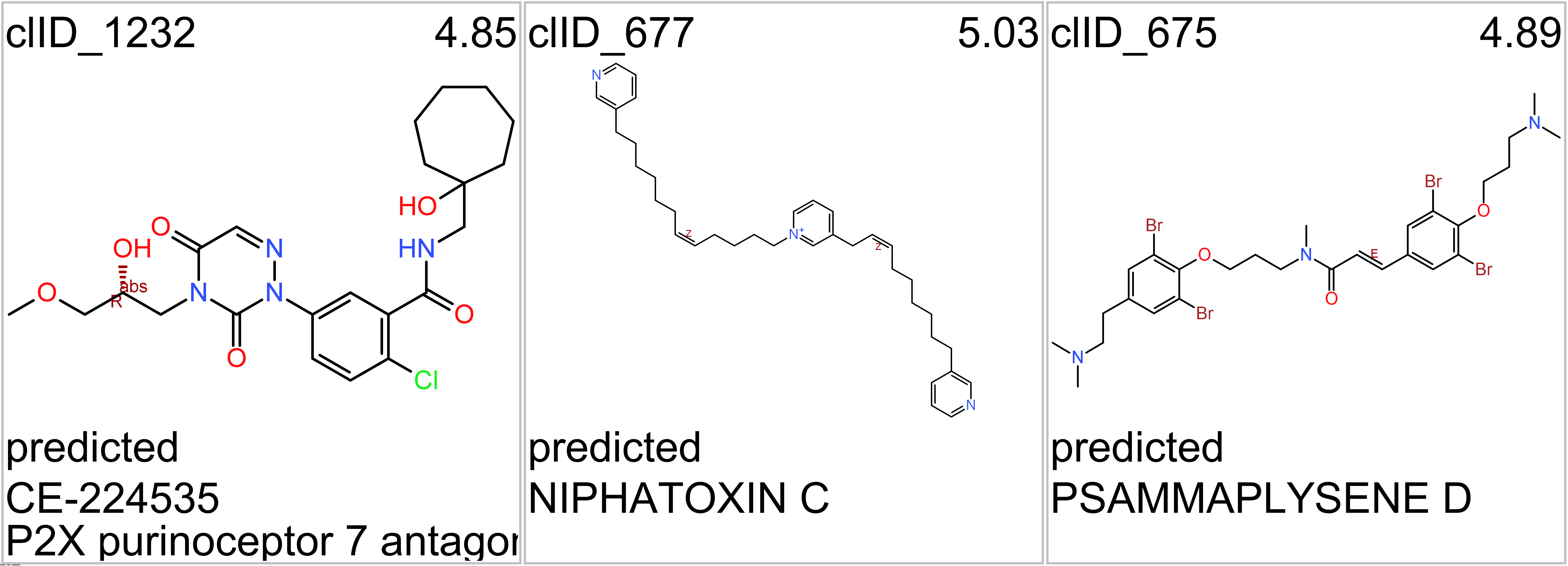
- Purinergic P2X7 receptors
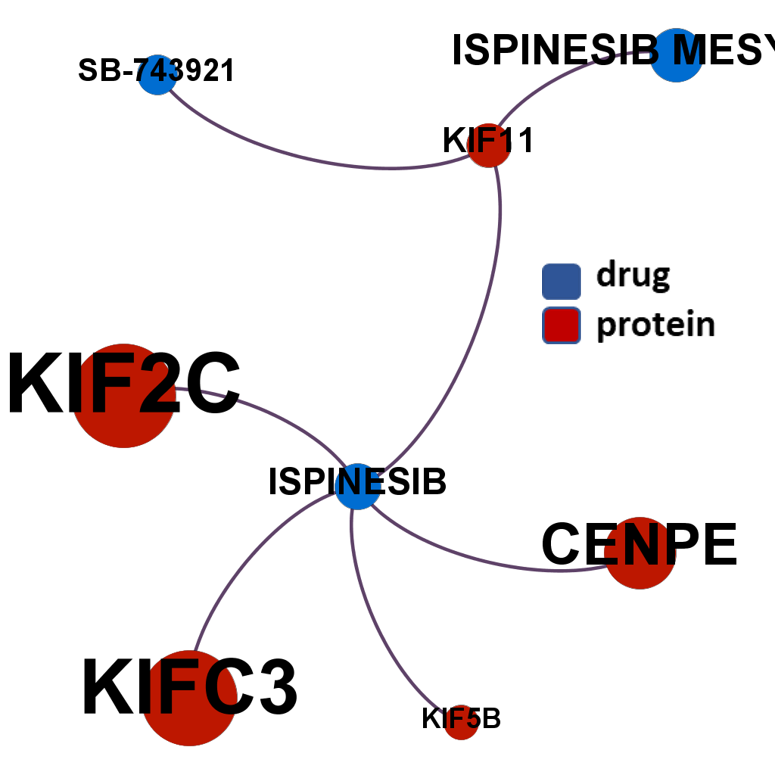
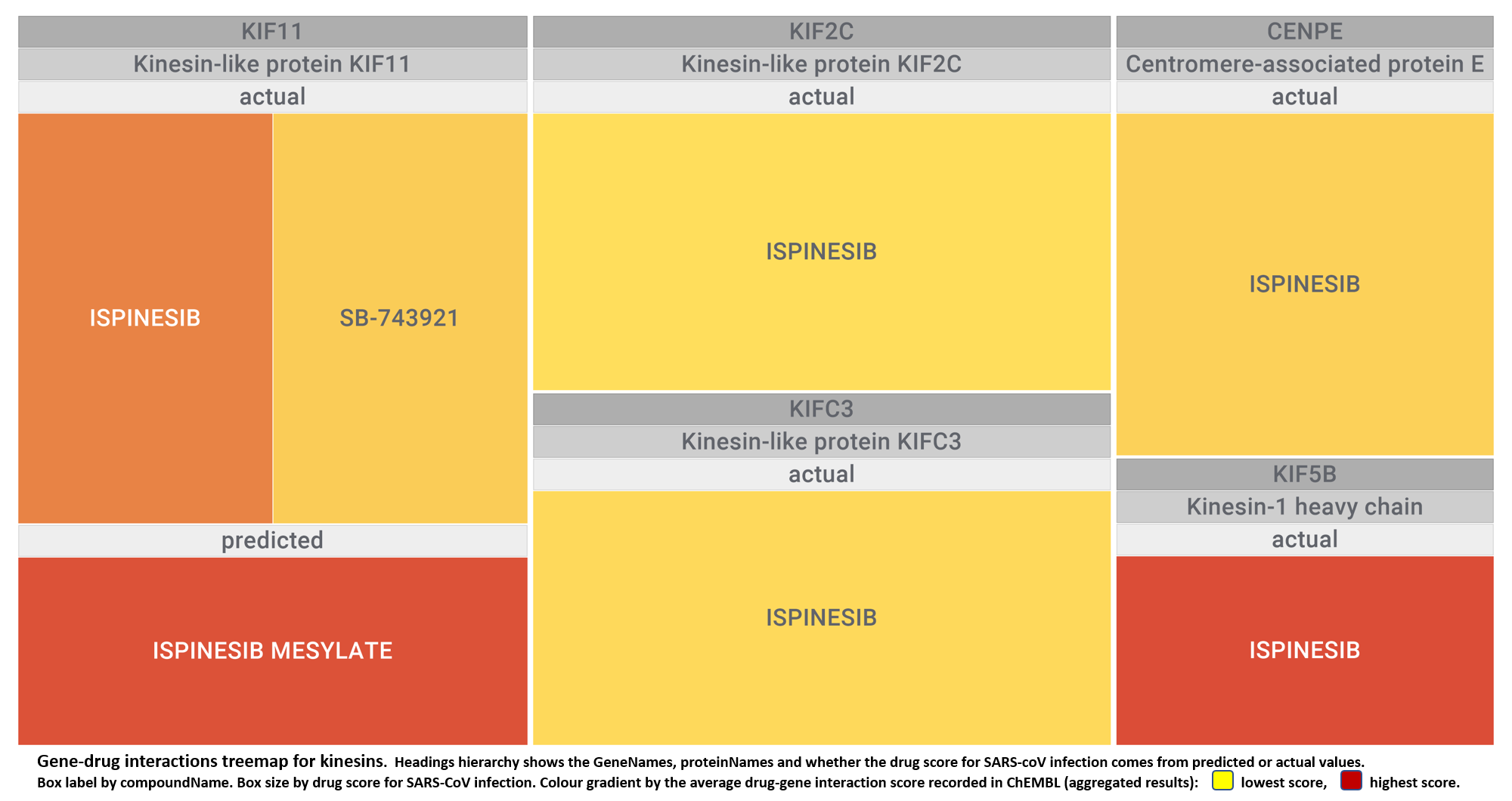
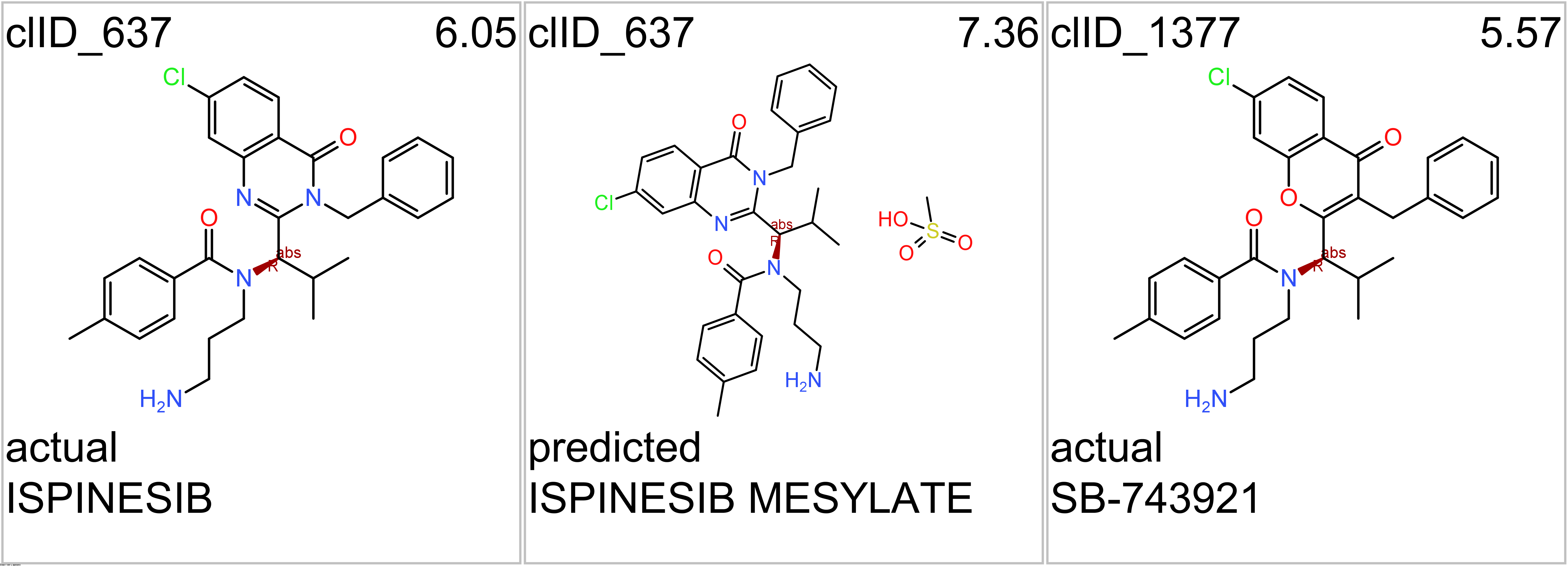
- Kinesins
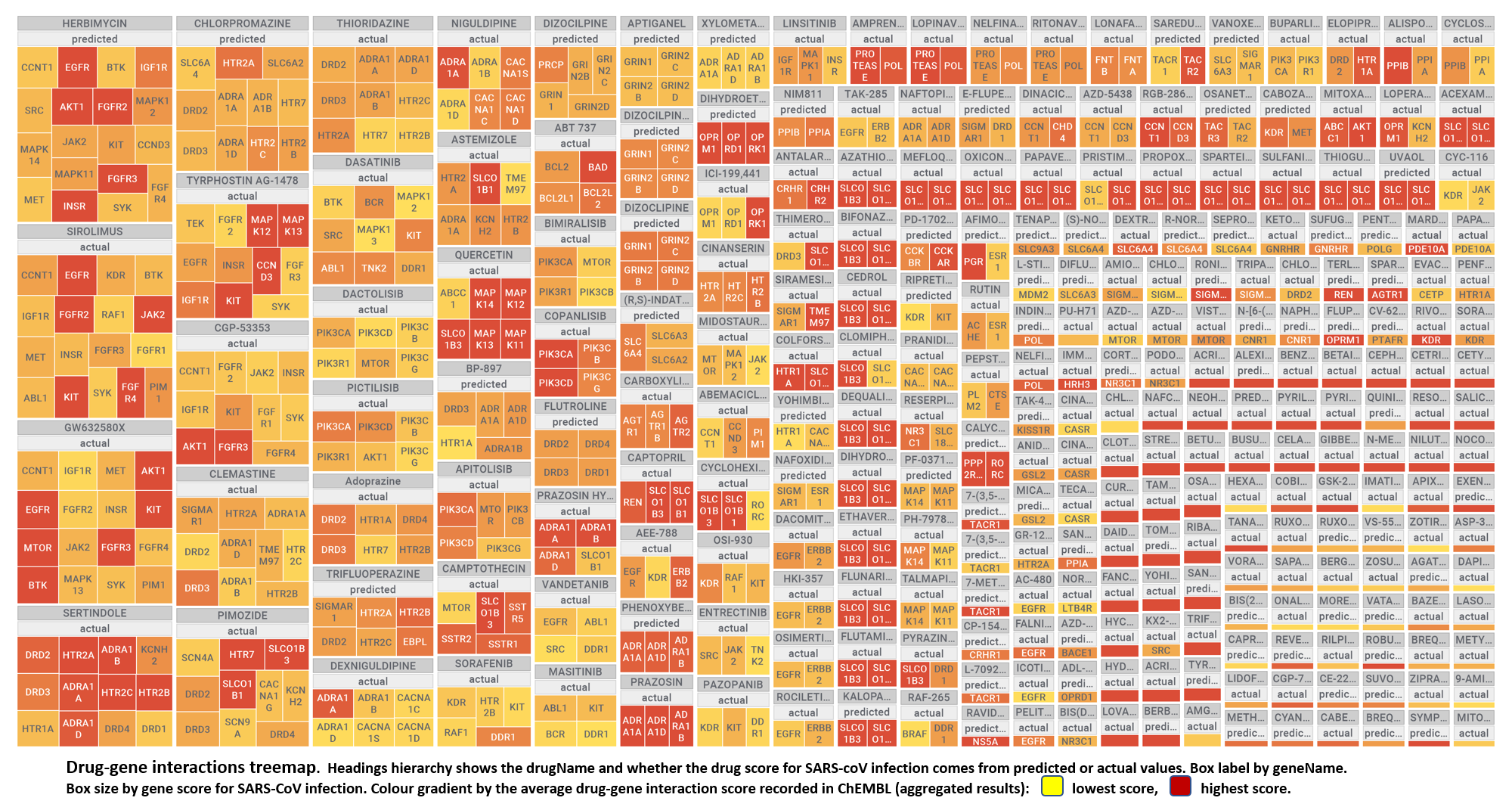
Use search function in table to view values for specific interactions
| molName | geneName | protName | sarsScoreMolecule | geneScore | interactionScore | organismType | kindOfValue | molecularCluster |
|---|---|---|---|---|---|---|---|---|
| AMG-548 | Mapk14 | Mitogen-activated protein kinase 14 | 5.906668 | 8.00432 | 7.290435 | mammals | actual | clID_1182 |
| APITOLISIB | MTOR | Serine/threonine-protein kinase mTOR | 9 | 7.18155 | 7.947535 | mammals | actual | clID_1863 |
| APITOLISIB | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | 9 | 8.414227 | 8.532107 | mammals | actual | clID_1863 |
| APITOLISIB | PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform | 9 | 7.674333 | 7.852496 | mammals | actual | clID_1863 |
| APITOLISIB | PIK3CD | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | 9 | 7.906179 | 8.270104 | mammals | actual | clID_1863 |
| AZD-2014 | MTOR | Serine/threonine-protein kinase mTOR | 5.60587 | 7.18155 | 7.070031 | mammals | actual | clID_100 |
| AZD-8055 | MTOR | Serine/threonine-protein kinase mTOR | 9 | 7.18155 | 7.412212 | mammals | actual | clID_100 |
| BIMIRALISIB | MTOR | Serine/threonine-protein kinase mTOR | 6.532598 | 7.18155 | 7.216821 | mammals | actual | clID_414 |
| BIMIRALISIB | Pik3r1 | Phosphatidylinositol 3-kinase regulatory subunit alpha | 6.532598 | 8.165486 | 7.769551 | mammals | actual | clID_414 |
| BIMIRALISIB | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | 6.532598 | 8.414227 | 7.961124 | mammals | actual | clID_414 |
| BIMIRALISIB | PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform | 6.532598 | 7.674333 | 7.069203 | mammals | actual | clID_414 |
| BUPARLISIB | Pik3r1 | Phosphatidylinositol 3-kinase regulatory subunit alpha | 6.614835 | 8.165486 | 7.285605 | mammals | actual | clID_414 |
| BUPARLISIB | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | 6.614835 | 8.414227 | 7.52825 | mammals | actual | clID_414 |
| CAMPTOTHECIN | MTOR | Serine/threonine-protein kinase mTOR | 5.654142 | 7.18155 | 7.157757 | mammals | actual | clID_1835 |
| CEP-32496 | BCR | Breakpoint cluster region protein | 4.675034 | 8.406418 | 7.387216 | mammals | actual | clID_10342 |
| CEP-32496 | RAF1 | RAF proto-oncogene serine/threonine-protein kinase | 4.675034 | 7.668057 | 7.408935 | mammals | actual | clID_10342 |
| CGP-53353 | AKT1 | RAC-alpha serine/threonine-protein kinase | 5.792715 | 7.667406 | 9 | mammals | actual | clID_670 |
| CGP-53353 | PRKCI | Protein kinase C iota type | 5.792715 | 8.003787 | 7.502399 | mammals | actual | clID_670 |
| DACTOLISIB | MTOR | Serine/threonine-protein kinase mTOR | 6.046152 | 7.18155 | 7.604691 | mammals | actual | clID_918 |
| DACTOLISIB | Pik3r1 | Phosphatidylinositol 3-kinase regulatory subunit alpha | 6.046152 | 8.165486 | 7.126506 | mammals | actual | clID_918 |
| DACTOLISIB | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | 6.046152 | 8.414227 | 7.694048 | mammals | actual | clID_918 |
| DACTOLISIB | PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform | 6.046152 | 7.674333 | 7.20915 | mammals | actual | clID_918 |
| DACTOLISIB | PIK3CD | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | 6.046152 | 7.906179 | 7.130912 | mammals | actual | clID_918 |
| DACTOLISIB | RPTOR | Regulatory-associated protein of mTOR | 6.046152 | 9.155781 | 7.356547 | mammals | actual | clID_918 |
| DASATINIB | BCR | Breakpoint cluster region protein | 5.047298 | 8.406418 | 8.052028 | mammals | actual | clID_5901 |
| DASATINIB | Mapk12 | Mitogen-activated protein kinase 12 | 5.047298 | 8.124694 | 7 | mammals | actual | clID_5901 |
| DASATINIB | MAPK13 | Mitogen-activated protein kinase 13 | 5.047298 | 8.087315 | 7 | mammals | actual | clID_5901 |
| FIMEPINOSTAT | Pik3r1 | Phosphatidylinositol 3-kinase regulatory subunit alpha | 4.771095 | 8.165486 | 7.505594 | mammals | actual | clID_11717 |
| FIMEPINOSTAT | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | 4.771095 | 8.414227 | 7.721246 | mammals | actual | clID_11717 |
| FIMEPINOSTAT | PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform | 4.771095 | 7.674333 | 7.267606 | mammals | actual | clID_11717 |
| FIMEPINOSTAT | PIK3CD | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | 4.771095 | 7.906179 | 7.408935 | mammals | actual | clID_11717 |
| GSK-2636771 | PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform | 5.870258 | 7.674333 | 9.05061 | mammals | actual | clID_1261 |
| GW632580X | AKT1 | RAC-alpha serine/threonine-protein kinase | 4.861247 | 7.667406 | 9 | mammals | actual | clID_780 |
| GW632580X | PRKCI | Protein kinase C iota type | 4.861247 | 8.003787 | 7.754825 | mammals | actual | clID_780 |
| GW632580X | MAPK13 | Mitogen-activated protein kinase 13 | 4.861247 | 8.087315 | 7.502399 | mammals | actual | clID_780 |
| GW632580X | MTOR | Serine/threonine-protein kinase mTOR | 4.861247 | 7.18155 | 9 | mammals | actual | clID_780 |
| HERBIMYCIN | AKT1 | RAC-alpha serine/threonine-protein kinase | 5.122579 | 7.667406 | 9 | mammals | predicted | clID_2407 |
| HERBIMYCIN | MAPK11 | Mitogen-activated protein kinase 11 | 5.122579 | 8.168793 | 7.845098 | mammals | predicted | clID_2407 |
| HERBIMYCIN | Mapk12 | Mitogen-activated protein kinase 12 | 5.122579 | 8.124694 | 7.997818 | mammals | predicted | clID_2407 |
| HERBIMYCIN | Mapk14 | Mitogen-activated protein kinase 14 | 5.122579 | 8.00432 | 7.639377 | mammals | predicted | clID_2407 |
| HEXACHLOROPHENE | Mapk14 | Mitogen-activated protein kinase 14 | 6.099502 | 8.00432 | 7.057496 | mammals | actual | clID_842 |
| IMATINIB MESYLATE | BCR | Breakpoint cluster region protein | 4.92584 | 8.406418 | 7.045757 | mammals | actual | clID_2757 |
| KENPAULLONE | AKT1 | RAC-alpha serine/threonine-protein kinase | 4.771468 | 7.667406 | 8.092918 | mammals | actual | clID_253 |
| KENPAULLONE | PRKCI | Protein kinase C iota type | 4.771468 | 8.003787 | 9 | mammals | actual | clID_253 |
| KENPAULLONE | MTOR | Serine/threonine-protein kinase mTOR | 4.771468 | 7.18155 | 7.534376 | mammals | actual | clID_253 |
| LINSITINIB | MAPK11 | Mitogen-activated protein kinase 11 | 5.151793 | 8.168793 | 8 | mammals | actual | clID_1409 |
| MASITINIB | BCR | Breakpoint cluster region protein | 5.355638 | 8.406418 | 7.107905 | mammals | actual | clID_3395 |
| MIDOSTAURIN | Mapk12 | Mitogen-activated protein kinase 12 | 5.198496 | 8.124694 | 7.412788 | mammals | actual | clID_3146 |
| MIDOSTAURIN | MTOR | Serine/threonine-protein kinase mTOR | 5.198496 | 7.18155 | 7.491846 | mammals | actual | clID_3146 |
| MITOXANTRONE | AKT1 | RAC-alpha serine/threonine-protein kinase | 4.874347 | 7.667406 | 9 | mammals | actual | clID_7603 |
| OSI-930 | RAF1 | RAF proto-oncogene serine/threonine-protein kinase | 9 | 7.668057 | 7.387216 | mammals | actual | clID_99 |
| PF-03715455 | MAPK11 | Mitogen-activated protein kinase 11 | 5.10551 | 8.168793 | 7.638272 | mammals | predicted | clID_5526 |
| PF-03715455 | Mapk14 | Mitogen-activated protein kinase 14 | 5.10551 | 8.00432 | 7.50103 | mammals | predicted | clID_5526 |
| PH-797804 | MAPK11 | Mitogen-activated protein kinase 11 | 5.413946 | 8.168793 | 7.19467 | mammals | actual | clID_2996 |
| PH-797804 | Mapk14 | Mitogen-activated protein kinase 14 | 5.413946 | 8.00432 | 8.249973 | mammals | actual | clID_2996 |
| PICTILISIB | AKT1 | RAC-alpha serine/threonine-protein kinase | 5.564751 | 7.667406 | 7.440627 | mammals | actual | clID_2221 |
| PICTILISIB | Pik3r1 | Phosphatidylinositol 3-kinase regulatory subunit alpha | 5.564751 | 8.165486 | 7.587144 | mammals | actual | clID_2221 |
| PICTILISIB | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | 5.564751 | 8.414227 | 8.319976 | mammals | actual | clID_2221 |
| PICTILISIB | PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform | 5.564751 | 7.674333 | 7.391049 | mammals | actual | clID_2221 |
| PICTILISIB | PIK3CD | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | 5.564751 | 7.906179 | 8.061408 | mammals | actual | clID_2221 |
| PONATINIB | BCR | Breakpoint cluster region protein | 4.627256 | 8.406418 | 9.096265 | mammals | actual | clID_4006 |
| QUERCETIN | MAPK11 | Mitogen-activated protein kinase 11 | 4.893839 | 8.168793 | 9 | mammals | actual | clID_459 |
| QUERCETIN | Mapk12 | Mitogen-activated protein kinase 12 | 4.893839 | 8.124694 | 9 | mammals | actual | clID_459 |
| QUERCETIN | MAPK13 | Mitogen-activated protein kinase 13 | 4.893839 | 8.087315 | 9 | mammals | actual | clID_459 |
| QUERCETIN | Mapk14 | Mitogen-activated protein kinase 14 | 4.893839 | 8.00432 | 9 | mammals | actual | clID_459 |
| RAF-265 | BRAF | Serine/threonine-protein kinase B-raf | 6.138279 | 7.59131 | 7.030823 | mammals | actual | clID_781 |
| SIROLIMUS | RAF1 | RAF proto-oncogene serine/threonine-protein kinase | 5.512884 | 7.668057 | 7.21598 | mammals | actual | clID_265 |
| TALMAPIMOD | MAPK11 | Mitogen-activated protein kinase 11 | 6.076272 | 8.168793 | 7.416341 | mammals | actual | clID_874 |
| TALMAPIMOD | Mapk14 | Mitogen-activated protein kinase 14 | 6.076272 | 8.00432 | 7.633576 | mammals | actual | clID_874 |
| TYRPHOSTIN AG-1478 | Mapk12 | Mitogen-activated protein kinase 12 | 5.452454 | 8.124694 | 9 | mammals | actual | clID_681 |
| TYRPHOSTIN AG-1478 | MAPK13 | Mitogen-activated protein kinase 13 | 5.452454 | 8.087315 | 9 | mammals | actual | clID_681 |
| VISTUSERTIB | MTOR | Serine/threonine-protein kinase mTOR | 9 | 7.18155 | 7.822205 | mammals | actual | clID_3010 |
| WORTMANNIN | Mapk14 | Mitogen-activated protein kinase 14 | 4.758394 | 8.00432 | 7.394185 | mammals | actual | clID_1672 |
| WORTMANNIN | MTOR | Serine/threonine-protein kinase mTOR | 4.758394 | 7.18155 | 7.144717 | mammals | actual | clID_1672 |
| WORTMANNIN | Pik3r1 | Phosphatidylinositol 3-kinase regulatory subunit alpha | 4.758394 | 8.165486 | 7.546149 | mammals | actual | clID_1672 |
| WORTMANNIN | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | 4.758394 | 8.414227 | 7.609936 | mammals | actual | clID_1672 |
| WORTMANNIN | PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform | 4.758394 | 7.674333 | 7.949242 | mammals | actual | clID_1672 |
| WORTMANNIN | PIK3CD | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | 4.758394 | 7.906179 | 8.187273 | mammals | actual | clID_1672 |
Use search function in table to view values for specific interactions
| molName | geneName | protname | sarsScoreMolecule | geneScore | interactionScore | organismType | kindOfValue | molecularCluster |
|---|---|---|---|---|---|---|---|---|
| (R,S)-INDATRALINE | Slc6a2 | Sodium-dependent noradrenaline transporter | 4.804672 | 8.291978 | 7.740743 | mammals | predicted | clID_1783 |
| (R,S)-INDATRALINE | Slc6a3 | Sodium-dependent dopamine transporter | 4.804672 | 9.054049 | 7.603454 | mammals | predicted | clID_1783 |
| (R,S)-INDATRALINE | SLC6A4 | Sodium-dependent serotonin transporter | 4.804672 | 9.398949 | 8.309894 | mammals | predicted | clID_1783 |
| (S)-NORDULOXETINE | SLC6A4 | Sodium-dependent serotonin transporter | 5.443214 | 9.398949 | 7.668244 | mammals | predicted | clID_2375 |
| 3-OXOURSAN (28-13)OLIDE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.511262 | 7.5973 | 9 | mammals | predicted | clID_2585 |
| 4-AMINO-3- (5-CHLOROTHIEN-2-YL)BUTANOIC ACID | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.506539 | 7.5973 | 9 | mammals | predicted | clID_5467 |
| 4-AMINO-3- (5-CHLOROTHIEN-2-YL)BUTANOIC ACID | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.506539 | 7.484017 | 7.521574 | mammals | predicted | clID_5467 |
| ACEXAMIC ACID | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.091012 | 7.5973 | 9 | mammals | actual | clID_5706 |
| ACEXAMIC ACID | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.091012 | 7.484017 | 9 | mammals | actual | clID_5706 |
| ACRIFLAVINE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.476427 | 7.5973 | 9 | mammals | actual | clID_2669 |
| ACRISORCIN | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.206535 | 7.484017 | 9 | mammals | actual | clID_4535 |
| AGMATINE SULFATE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.551143 | 7.5973 | 9 | mammals | actual | clID_10130 |
| AGMATINE SULFATE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.551143 | 7.484017 | 9 | mammals | actual | clID_10130 |
| ALEXIDINE HYDROCHLORIDE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 7.663791 | 7.484017 | 9 | mammals | predicted | clID_181 |
| ASTEMIZOLE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.958607 | 7.5973 | 9 | mammals | actual | clID_1081 |
| AZATHIOPRINE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.068937 | 7.5973 | 9 | mammals | actual | clID_5945 |
| AZATHIOPRINE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.068937 | 7.484017 | 9 | mammals | actual | clID_5945 |
| BENZALKONIUM CHLORIDE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.83546 | 7.484017 | 9 | mammals | actual | clID_2984 |
| BENZOCAINE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.5298 | 7.5973 | 9 | mammals | actual | clID_7652 |
| BENZOCAINE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.5298 | 7.484017 | 9 | mammals | actual | clID_7652 |
| BERBAMINE HYDROCHLORIDE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 6.574623 | 7.5973 | 9 | mammals | predicted | clID_186 |
| BERGAPTEN | ABCB1 | ATP-dependent translocase ABCB1 | 4.885027 | 8.720249 | 7.30103 | widespread | actual | clID_7721 |
| BETAINE HYDROCHLORIDE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 6.344288 | 7.484017 | 9 | mammals | actual | clID_581 |
| BETHANECHOL CHLORIDE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.731346 | 7.5973 | 9 | mammals | actual | clID_9239 |
| BETHANECHOL CHLORIDE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.731346 | 7.484017 | 9 | mammals | actual | clID_9239 |
| BETULIN | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.046427 | 7.5973 | 9 | mammals | actual | clID_195 |
| BIFONAZOLE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.383601 | 7.5973 | 9 | mammals | actual | clID_3195 |
| BIFONAZOLE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.383601 | 7.484017 | 9 | mammals | actual | clID_3195 |
| BUSULFAN | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.999131 | 7.5973 | 9 | mammals | actual | clID_6797 |
| CAFFEINE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.703587 | 7.5973 | 9 | mammals | actual | clID_6550 |
| CAMPTOTHECIN | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.654142 | 7.484017 | 9 | mammals | actual | clID_1835 |
| CAPTOPRIL | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.558465 | 7.5973 | 9 | mammals | actual | clID_2251 |
| CAPTOPRIL | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.558465 | 7.484017 | 9 | mammals | actual | clID_2251 |
| CATECHIN PENTABENZOATE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.653227 | 7.5973 | 9 | mammals | predicted | clID_4206 |
| CATECHIN PENTABENZOATE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.653227 | 7.484017 | 9 | mammals | predicted | clID_4206 |
| CEDROL | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.828067 | 7.5973 | 9 | mammals | actual | clID_491 |
| CEDROL | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.828067 | 7.484017 | 9 | mammals | actual | clID_491 |
| CEFACLOR | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 7.284031 | 7.484017 | 9 | mammals | predicted | clID_3701 |
| CEFADROXIL | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.670843 | 7.484017 | 9 | mammals | actual | clID_3701 |
| CELASTROL | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.376751 | 7.5973 | 9 | mammals | actual | clID_2475 |
| CELECOXIB | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.665836 | 7.5973 | 9 | mammals | actual | clID_7657 |
| CELECOXIB | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.665836 | 7.484017 | 9 | mammals | actual | clID_7657 |
| CELLBIOSE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.633284 | 7.484017 | 9 | mammals | predicted | clID_283 |
| CEPHARANTHINE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.930572 | 7.484017 | 9 | mammals | actual | clID_186 |
| CETRIMONIUM BROMIDE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.088203 | 7.484017 | 9 | mammals | actual | clID_5733 |
| CETYLPYRIDINIUM CHLORIDE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 8.301902 | 7.484017 | 9 | mammals | actual | clID_5869 |
| CHLOROQUINE PHOSPHATE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.766781 | 7.484017 | 7.08261 | mammals | actual | clID_1740 |
| CHLORPROMAZINE | Slc6a2 | Sodium-dependent noradrenaline transporter | 6.908068 | 8.291978 | 7.721246 | mammals | predicted | clID_302 |
| CHLORPROMAZINE | SLC6A4 | Sodium-dependent serotonin transporter | 6.908068 | 9.398949 | 7.301876 | mammals | predicted | clID_302 |
| CHLORPROMAZINE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 6.908068 | 7.5973 | 7.290209 | mammals | predicted | clID_302 |
| CHLORPYRIFOS | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.671842 | 7.5973 | 9 | mammals | actual | clID_9389 |
| CHLORPYRIFOS | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.671842 | 7.484017 | 9 | mammals | actual | clID_9389 |
| CILOSTAZOL | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.639648 | 7.5973 | 9 | mammals | actual | clID_7834 |
| CLEBOPRIDE MALEATE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.722346 | 7.484017 | 9 | mammals | actual | clID_10245 |
| CLOMIPHENE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.386156 | 7.5973 | 7.226578 | mammals | actual | clID_2790 |
| CLOMIPHENE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.386156 | 7.484017 | 9 | mammals | actual | clID_2790 |
| CLOMIPRAMINE | Slc6a2 | Sodium-dependent noradrenaline transporter | 4.522627 | 8.291978 | 7.006628 | mammals | actual | clID_1362 |
| CLOMIPRAMINE | SLC6A4 | Sodium-dependent serotonin transporter | 4.522627 | 9.398949 | 7.96122 | mammals | actual | clID_1362 |
| CLOTRIMAZOLE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.6489 | 7.484017 | 9 | mammals | actual | clID_1854 |
| COLFORSIN | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.920624 | 7.5973 | 9 | mammals | actual | clID_2585 |
| CURCUMIN | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.319754 | 7.484017 | 9 | mammals | actual | clID_3628 |
| CYCLOHEXIMIDE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 6.421559 | 7.5973 | 9 | mammals | actual | clID_525 |
| CYCLOHEXIMIDE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 6.421559 | 7.484017 | 9 | mammals | actual | clID_525 |
| Copper complex | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.665163 | 7.484017 | 9 | mammals | predicted | copper complex |
| DAIDZEIN | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.96663 | 7.484017 | 9 | mammals | actual | clID_7199 |
| DEMECLOCYCLINE HYDROCHLORIDE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.554055 | 7.5973 | 9 | mammals | actual | clID_1527 |
| DEQUALINIUM CHLORIDE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.281308 | 7.5973 | 9 | mammals | actual | clID_914 |
| DEQUALINIUM CHLORIDE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.281308 | 7.484017 | 9 | mammals | actual | clID_914 |
| DEXTROMETHORPHAN | SLC6A4 | Sodium-dependent serotonin transporter | 5.527467 | 9.398949 | 8.704334 | mammals | actual | clID_2960 |
| DIFLUOROBENZTROPINE | Slc6a3 | Sodium-dependent dopamine transporter | 4.850031 | 9.054049 | 7.186786 | mammals | predicted | clID_601 |
| DIHYDROTANSHINONE I | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.920819 | 7.5973 | 9 | mammals | actual | clID_1151 |
| DIHYDROTANSHINONE I | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.920819 | 7.484017 | 9 | mammals | actual | clID_1151 |
| ESCITALOPRAM | SLC6A4 | Sodium-dependent serotonin transporter | 4.64129 | 9.398949 | 7.810003 | mammals | actual | clID_6878 |
| ETHAVERINE HYDROCHLORIDE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 6.973989 | 7.5973 | 9 | mammals | actual | clID_284 |
| ETHAVERINE HYDROCHLORIDE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 6.973989 | 7.484017 | 9 | mammals | actual | clID_284 |
| FANCHININ | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.562726 | 7.484017 | 9 | mammals | actual | clID_186 |
| FLOPROPIONE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.627256 | 7.484017 | 9 | mammals | actual | clID_10249 |
| FLOXURIDINE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.52865 | 7.5973 | 9 | mammals | actual | clID_7153 |
| FLOXURIDINE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.52865 | 7.484017 | 9 | mammals | actual | clID_7153 |
| FLUNARIZINE HYDROCHLORIDE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.364535 | 7.5973 | 9 | mammals | actual | clID_355 |
| FLUNARIZINE HYDROCHLORIDE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.364535 | 7.484017 | 9 | mammals | actual | clID_355 |
| FLUTAMIDE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.910393 | 7.5973 | 9 | mammals | actual | clID_7616 |
| FLUTAMIDE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.910393 | 7.484017 | 9 | mammals | actual | clID_7616 |
| GIBBERELLIN A3 | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.831856 | 7.5973 | 9 | mammals | actual | clID_2585 |
| GLAFENINE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.797686 | 7.5973 | 9 | mammals | actual | clID_8575 |
| GLAFENINE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.797686 | 7.484017 | 9 | mammals | actual | clID_8575 |
| HALOPERIDOL | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.799189 | 7.484017 | 9 | mammals | actual | clID_1001 |
| HYCANTHONE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.179351 | 7.484017 | 9 | mammals | actual | clID_4801 |
| HYDROCORTISONE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.050958 | 7.484017 | 9 | mammals | actual | clID_1633 |
| KALOPANAXSAPONIN B | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.812206 | 7.5973 | 9 | mammals | predicted | clID_2585 |
| KALOPANAXSAPONIN B | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.812206 | 7.484017 | 9 | mammals | predicted | clID_2585 |
| LONIDAMINE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.52796 | 7.5973 | 9 | mammals | actual | clID_438 |
| LONIDAMINE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.52796 | 7.484017 | 9 | mammals | actual | clID_438 |
| LOVASTATIN | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.114456 | 7.484017 | 9 | mammals | actual | clID_5420 |
| LUFENURON | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.754076 | 7.5973 | 9 | mammals | actual | clID_10175 |
| MEFLOQUINE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.363512 | 7.5973 | 9 | mammals | actual | clID_1601 |
| MEFLOQUINE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.363512 | 7.484017 | 9 | mammals | actual | clID_1601 |
| MITOXANTRONE | Abcc1 | Multidrug resistance-associated protein 1 | 4.874347 | 9.003589 | 8.789688 | mammals | actual | clID_7603 |
| MYCOPHENOLIC ACID | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.582474 | 7.5973 | 9 | mammals | actual | clID_7897 |
| MYCOPHENOLIC ACID | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.582474 | 7.484017 | 9 | mammals | actual | clID_7897 |
| N-METHYL-D-ASPARTIC ACID (NMDA) | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.810476 | 7.5973 | 9 | mammals | actual | clID_7880 |
| NAFCILLIN SODIUM | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.828247 | 7.484017 | 9 | mammals | actual | clID_6931 |
| NEOHESPERIDIN DIHYDROCHALCONE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.138218 | 7.484017 | 9 | mammals | actual | clID_197 |
| NIFEDIPINE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.683767 | 7.484017 | 9 | mammals | actual | clID_6999 |
| NILUTAMIDE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.984016 | 7.5973 | 9 | mammals | actual | clID_6978 |
| NOCODAZOLE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.290777 | 7.5973 | 9 | mammals | actual | clID_3850 |
| OLEANDRIN | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.996178 | 7.5973 | 9 | mammals | actual | clID_364 |
| OSAJIN | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.412289 | 7.5973 | 9 | mammals | actual | clID_3006 |
| OUABAIN | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 7.013228 | 7.5973 | 7.004343 | mammals | actual | clID_364 |
| OXICONAZOLE NITRATE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.444295 | 7.5973 | 9 | mammals | actual | clID_2831 |
| OXICONAZOLE NITRATE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.444295 | 7.484017 | 9 | mammals | actual | clID_2831 |
| PAPAVERINE HYDROCHLORIDE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.932229 | 7.5973 | 9 | mammals | actual | clID_1423 |
| PAPAVERINE HYDROCHLORIDE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.932229 | 7.484017 | 9 | mammals | actual | clID_1423 |
| PEFLOXACIN MESYLATE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.578777 | 7.5973 | 9 | mammals | actual | clID_8377 |
| PENTAMIDINE ISETHIONATE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.946776 | 7.5973 | 7.583896 | mammals | actual | clID_1557 |
| PENTAMIDINE ISETHIONATE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.946776 | 7.484017 | 9 | mammals | actual | clID_1557 |
| PENTOLINIUM TARTRATE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.630575 | 7.484017 | 9 | mammals | actual | clID_10163 |
| PIMOZIDE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.765854 | 7.5973 | 9 | mammals | actual | clID_4167 |
| PIMOZIDE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.765854 | 7.484017 | 9 | mammals | actual | clID_4167 |
| PRAZOSIN HYDROCHLORIDE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.140535 | 7.5973 | 7.374923 | mammals | actual | clID_5167 |
| PREDNISOLONE ACETATE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.17591 | 7.484017 | 9 | mammals | actual | clID_3226 |
| PRISTIMERIN | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.508638 | 7.5973 | 9 | mammals | actual | clID_2475 |
| PRISTIMERIN | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.508638 | 7.484017 | 7.257242 | mammals | actual | clID_2475 |
| PROPOXUR | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.197005 | 7.5973 | 9 | mammals | actual | clID_4636 |
| PROPOXUR | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.197005 | 7.484017 | 9 | mammals | actual | clID_4636 |
| PYRAZINAMIDE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.900549 | 7.484017 | 9 | mammals | actual | clID_7678 |
| PYRILAMINE MALEATE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.936305 | 7.484017 | 9 | mammals | actual | clID_10043 |
| PYRIMETHAMINE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.13359 | 7.484017 | 9 | mammals | actual | clID_5236 |
| QUERCETIN | Abcc1 | Multidrug resistance-associated protein 1 | 4.893839 | 9.003589 | 7.177826 | mammals | actual | clID_459 |
| QUERCETIN | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.893839 | 7.484017 | 9 | mammals | actual | clID_459 |
| QUININE SULFATE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 6.410159 | 7.484017 | 7.885492 | mammals | predicted | clID_530 |
| R-NORDULOXETINE | SLC6A4 | Sodium-dependent serotonin transporter | 5.531204 | 9.398949 | 8.124206 | mammals | predicted | clID_2375 |
| RESERPINE | Slc18a2 | Synaptic vesicular amine transporter | 5.253841 | 8.078708 | 7.640538 | mammals | actual | clID_4255 |
| RESORCINOL | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.848207 | 7.484017 | 9 | mammals | actual | clID_7732 |
| RIBAVIRIN | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.951013 | 7.5973 | 9 | mammals | actual | clID_7406 |
| SACCHARIN | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.783147 | 7.5973 | 9 | mammals | actual | clID_8142 |
| SACCHARIN | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.783147 | 7.484017 | 9 | mammals | actual | clID_8142 |
| SALICYL ALCOHOL | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.87032 | 7.484017 | 9 | mammals | actual | clID_7749 |
| SANGUINARINE SULFATE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 6.616845 | 7.5973 | 9 | mammals | predicted | clID_993 |
| SEMUSTINE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.718885 | 7.5973 | 9 | mammals | actual | clID_7630 |
| SEMUSTINE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.718885 | 7.484017 | 9 | mammals | actual | clID_7630 |
| SEPROXETINE | SLC6A4 | Sodium-dependent serotonin transporter | 7.342066 | 9.398949 | 7.319136 | mammals | predicted | clID_208 |
| SISOMICIN SULFATE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.796402 | 7.5973 | 7.307671 | mammals | actual | clID_12135 |
| SISOMICIN SULFATE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.796402 | 7.484017 | 9 | mammals | actual | clID_12135 |
| SPARTEINE SULFATE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.95932 | 7.5973 | 9 | mammals | actual | clID_7292 |
| SPARTEINE SULFATE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.95932 | 7.484017 | 9 | mammals | actual | clID_7292 |
| STREPTOZOSIN | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.972717 | 7.484017 | 9 | mammals | actual | clID_7116 |
| SULFANILAMIDE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.066839 | 7.5973 | 9 | mammals | actual | clID_5969 |
| SULFANILAMIDE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.066839 | 7.484017 | 9 | mammals | actual | clID_5969 |
| TAMOXIFEN CITRATE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.889279 | 7.484017 | 9 | mammals | actual | clID_10039 |
| TENAPANOR HYDROCHLORIDE | SLC9A3 | Sodium/hydrogen exchanger 3 | 4.832468 | 8.993804 | 7.974985 | mammals | predicted | clID_1886 |
| THIMEROSAL | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.725507 | 7.484017 | 9 | mammals | actual | organomercury |
| THIOGUANINE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 7.514173 | 7.5973 | 9 | mammals | actual | clID_194 |
| THIOGUANINE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 7.514173 | 7.484017 | 9 | mammals | actual | clID_194 |
| TIAPRIDE HYDROCHLORIDE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.681391 | 7.5973 | 9 | mammals | actual | clID_10133 |
| TOMATINE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 6.639829 | 7.484017 | 9 | mammals | predicted | clID_407 |
| TOTAROL | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.710937 | 7.5973 | 9 | mammals | predicted | clID_3387 |
| TOTAROL | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.710937 | 7.484017 | 9 | mammals | predicted | clID_3387 |
| TRIFLUOPERAZINE HYDROCHLORIDE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.705533 | 7.5973 | 9 | mammals | actual | clID_4249 |
| TRIFLUPROMAZINE HYDROCHLORIDE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.899668 | 7.5973 | 9 | mammals | actual | clID_1458 |
| TYROTHRICIN | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.156203 | 7.5973 | 9 | mammals | predicted | clID_939 |
| UBIDECARENONE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.679606 | 7.5973 | 9 | mammals | actual | clID_271 |
| UBIDECARENONE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.679606 | 7.484017 | 9 | mammals | actual | clID_271 |
| URETHANE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.644975 | 7.5973 | 9 | mammals | actual | clID_9418 |
| URETHANE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.644975 | 7.484017 | 9 | mammals | actual | clID_9418 |
| UVAOL | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 6.373663 | 7.5973 | 9 | mammals | predicted | clID_1633 |
| UVAOL | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 6.373663 | 7.484017 | 9 | mammals | predicted | clID_1633 |
| VALDECOXIB | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.799518 | 7.5973 | 9 | mammals | actual | clID_7892 |
| VANOXERINE | Slc6a3 | Sodium-dependent dopamine transporter | 6.527334 | 9.054049 | 7.144667 | mammals | predicted | clID_462 |
| XYLOMETAZOLINE HYDROCHLORIDE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.666037 | 7.484017 | 7.077302 | mammals | actual | clID_2569 |
| YOHIMBINE HYDROCHLORIDE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.151434 | 7.484017 | 9 | mammals | actual | clID_4255 |
| ZOSUQUIDAR TRIHYDROCHLORIDE | ABCB1 | ATP-dependent translocase ABCB1 | 5.663266 | 8.720249 | 7.266351 | widespread | actual | clID_1807 |
Use search function in table to view values for specific interactions
| molName | geneName | protname | sarsScoreMolecule | geneScore | interactionScore | organismType | kindOfValue | molecularCluster |
|---|---|---|---|---|---|---|---|---|
| 7-(3,5-BIS-TRIFLUOROMETHYL-BENZYL)-5-P-TOLYL-8,9,10,11-TETRAHYDRO-7H-1,7,11A-TRIAZA-CYCLOOCTA[B]NAPHTHALENE-6,12-DIONE (ATROPISOMERIC MIX) | Tacr1 | Substance-P receptor | 5.16581 | 9.709555 | 8.91215 | mammals | predicted | clID_4408 |
| 7-(3,5-BIS-TRIFLUOROMETHYL-BENZYL)-9-METHYL-5-P-TOLYL-8,9,10,11-TETRAHYDRO-7H-1,7,11A-TRIAZA-CYCLOOCTA[B]NAPHTHALENE-6,12-DIONE (ATROPISOMERIC MIX) | Tacr1 | Substance-P receptor | 5.223811 | 9.709555 | 7.267011 | mammals | predicted | clID_4408 |
| 7-METHYL-8-OXO-5-P-TOLYL-7,8-DIHYDRO-[1,7]NAPHTHYRIDINE-6-CARBOXYLIC ACID (3,5-BIS-TRIFLUOROMETHYL-BENZYL)-METHYL-AMIDE (STRUCTURAL MIX) | Tacr1 | Substance-P receptor | 5.34932 | 9.709555 | 9.092575 | mammals | predicted | clID_3441 |
| ADL-5747 | Oprd1 | Delta-type opioid receptor | 5.291525 | 7.770012 | 7.797754 | mammals | predicted | clID_3845 |
| ANGIOTENSIN II | AGTR1 | Type-1 angiotensin II receptor | 4.50648 | 8.425964 | 9.159495 | mammals | predicted | clID_2005 |
| ANGIOTENSIN II | Agtr2 | Type-2 angiotensin II receptor | 4.50648 | 8.58023 | 8.852604 | mammals | predicted | clID_2005 |
| ANGIOTENSIN II | Agtr1 | Type-1A angiotensin II receptor | 4.50648 | 8.497722 | 9.004202 | mammals | predicted | clID_2005 |
| ANGIOTENSIN II | Agtr1b | Type-1B angiotensin II receptor | 4.50648 | 8.480755 | 9.015087 | mammals | predicted | clID_2005 |
| ANTALARMIN | CRHR1 | Corticotropin-releasing factor receptor 1 | 5.15753 | 9.228861 | 8.477266 | mammals | actual | clID_5004 |
| ASTEMIZOLE | HTR2A | 5-hydroxytryptamine receptor 2A | 5.958607 | 7.818593 | 7.976363 | mammals | actual | clID_1081 |
| ASTEMIZOLE | Htr2b | 5-hydroxytryptamine receptor 2B | 5.958607 | 7.37631 | 8.023255 | mammals | actual | clID_1081 |
| ASTEMIZOLE | ADRA1A | Alpha-1A adrenergic receptor | 5.958607 | 7.742958 | 7.812055 | mammals | actual | clID_1081 |
| AZILSARTAN | AGTR1 | Type-1 angiotensin II receptor | 4.777781 | 8.425964 | 7.376751 | mammals | actual | clID_6141 |
| AZILSARTAN | Agtr2 | Type-2 angiotensin II receptor | 4.777781 | 8.58023 | 9 | mammals | actual | clID_6141 |
| AZILSARTAN | Agtr1 | Type-1A angiotensin II receptor | 4.777781 | 8.497722 | 9 | mammals | actual | clID_6141 |
| AZILSARTAN | Agtr1b | Type-1B angiotensin II receptor | 4.777781 | 8.480755 | 9 | mammals | actual | clID_6141 |
| Adoprazine | HTR1A | 5-hydroxytryptamine receptor 1A | 5.412654 | 7.930581 | 7.722033 | mammals | actual | clID_5683 |
| Adoprazine | Htr2b | 5-hydroxytryptamine receptor 2B | 5.412654 | 7.37631 | 7.899974 | mammals | actual | clID_5683 |
| Adoprazine | DRD2 | D(2) dopamine receptor | 5.412654 | 8.280379 | 8.400008 | mammals | actual | clID_5683 |
| Adoprazine | Drd3 | D(3) dopamine receptor | 5.412654 | 8.166222 | 8.400008 | mammals | actual | clID_5683 |
| Adoprazine | DRD4 | D(4) dopamine receptor | 5.412654 | 7.451626 | 8 | mammals | actual | clID_5683 |
| BENPERIDOL | HTR2A | 5-hydroxytryptamine receptor 2A | 4.591547 | 7.818593 | 8.920819 | mammals | actual | clID_1956 |
| BENPERIDOL | DRD2 | D(2) dopamine receptor | 4.591547 | 8.280379 | 10.568636 | mammals | actual | clID_1956 |
| BENPERIDOL | Drd3 | D(3) dopamine receptor | 4.591547 | 8.166222 | 9.537602 | mammals | actual | clID_1956 |
| BENPERIDOL | DRD4 | D(4) dopamine receptor | 4.591547 | 7.451626 | 10.180456 | mammals | actual | clID_1956 |
| BP-897 | HTR1A | 5-hydroxytryptamine receptor 1A | 4.846732 | 7.930581 | 7.00455 | mammals | predicted | clID_1447 |
| BP-897 | ADRA1A | Alpha-1A adrenergic receptor | 4.846732 | 7.742958 | 7.871212 | mammals | predicted | clID_1447 |
| BP-897 | Adra1b | Alpha-1B adrenergic receptor | 4.846732 | 7.738535 | 7.951545 | mammals | predicted | clID_1447 |
| BP-897 | Adra1d | Alpha-1D adrenergic receptor | 4.846732 | 7.713266 | 7.951545 | mammals | predicted | clID_1447 |
| BP-897 | Drd3 | D(3) dopamine receptor | 4.846732 | 8.166222 | 7.478496 | mammals | predicted | clID_1447 |
| BREXPIPRAZOLE | HTR2A | 5-hydroxytryptamine receptor 2A | 4.612161 | 7.818593 | 8.638272 | mammals | actual | clID_10512 |
| BREXPIPRAZOLE | DRD2 | D(2) dopamine receptor | 4.612161 | 8.280379 | 9.69897 | mammals | actual | clID_10512 |
| CABERGOLINE | Htr2b | 5-hydroxytryptamine receptor 2B | 4.966456 | 7.37631 | 8.579976 | mammals | actual | clID_2232 |
| CAMPTOTHECIN | Sstr1 | Somatostatin receptor type 1 | 5.654142 | 8.134288 | 8.655608 | mammals | actual | clID_1835 |
| CAMPTOTHECIN | Sstr2 | Somatostatin receptor type 2 | 5.654142 | 8.185393 | 8.655608 | mammals | actual | clID_1835 |
| CAMPTOTHECIN | Sstr4 | Somatostatin receptor type 4 | 5.654142 | 8.167809 | 8.655608 | mammals | actual | clID_1835 |
| CARBOXYLIC ACID METABOLITE | AGTR1 | Type-1 angiotensin II receptor | 4.876353 | 8.425964 | 8.55898 | mammals | actual | clID_774 |
| CARBOXYLIC ACID METABOLITE | Agtr2 | Type-2 angiotensin II receptor | 4.876353 | 8.58023 | 8.453694 | mammals | actual | clID_774 |
| CARBOXYLIC ACID METABOLITE | Agtr1 | Type-1A angiotensin II receptor | 4.876353 | 8.497722 | 8.107917 | mammals | actual | clID_774 |
| CARBOXYLIC ACID METABOLITE | Agtr1b | Type-1B angiotensin II receptor | 4.876353 | 8.480755 | 8.100372 | mammals | actual | clID_774 |
| CARPERITIDE | NPR1 | Atrial natriuretic peptide receptor 1 | 4.737933 | 8.853192 | 9.5 | mammals | predicted | clID_2005 |
| CASOPITANT | Tacr1 | Substance-P receptor | 4.670516 | 9.709555 | 8.160005 | mammals | predicted | clID_1663 |
| CELECOXIB | DRD2 | D(2) dopamine receptor | 4.665836 | 8.280379 | 7.690106 | mammals | actual | clID_7657 |
| CHLORHEXIDINE | Npy1r | Neuropeptide Y receptor type 1 | 5.855721 | 7.979235 | 7.314866 | mammals | actual | clID_1295 |
| CHLOROHALOPERIDOL | Mchr1 | Melanin-concentrating hormone receptor 1 | 4.533161 | 8.59381 | 7.250845 | mammals | predicted | clID_1001 |
| CHLORPROMAZINE | HTR2A | 5-hydroxytryptamine receptor 2A | 6.908068 | 7.818593 | 8.500919 | mammals | predicted | clID_302 |
| CHLORPROMAZINE | Htr2b | 5-hydroxytryptamine receptor 2B | 6.908068 | 7.37631 | 8.199774 | mammals | predicted | clID_302 |
| CHLORPROMAZINE | Htr2c | 5-hydroxytryptamine receptor 2C | 6.908068 | 7.439004 | 8.298354 | mammals | predicted | clID_302 |
| CHLORPROMAZINE | ADRA1A | Alpha-1A adrenergic receptor | 6.908068 | 7.742958 | 7.463372 | mammals | predicted | clID_302 |
| CHLORPROMAZINE | Adra1b | Alpha-1B adrenergic receptor | 6.908068 | 7.738535 | 7.452877 | mammals | predicted | clID_302 |
| CHLORPROMAZINE | Adra1d | Alpha-1D adrenergic receptor | 6.908068 | 7.713266 | 7.497263 | mammals | predicted | clID_302 |
| CHLORPROMAZINE | Drd1 | D(1A) dopamine receptor | 6.908068 | 7.436039 | 7.07018 | mammals | predicted | clID_302 |
| CHLORPROMAZINE | DRD2 | D(2) dopamine receptor | 6.908068 | 8.280379 | 7.388453 | mammals | predicted | clID_302 |
| CHLORPROMAZINE | Drd3 | D(3) dopamine receptor | 6.908068 | 8.166222 | 7.269156 | mammals | predicted | clID_302 |
| CHLORPROMAZINE | DRD4 | D(4) dopamine receptor | 6.908068 | 7.451626 | 7.100513 | mammals | predicted | clID_302 |
| CHLORPROMAZINE HYDROCHLORIDE | DRD2 | D(2) dopamine receptor | 5.451548 | 8.280379 | 7.451545 | mammals | actual | clID_302 |
| CINACALCET | CASR | Extracellular calcium-sensing receptor | 5.063412 | 8.205451 | 7.09691 | mammals | actual | clID_6420 |
| CINACALCET HYDROCHLORIDE | CASR | Extracellular calcium-sensing receptor | 5.027804 | 8.205451 | 7.814928 | mammals | actual | clID_6420 |
| CINALUKAST | CYSLTR1 | Cysteinyl leukotriene receptor 1 | 4.936829 | 8.121852 | 8.19382 | mammals | actual | clID_7693 |
| CINANSERIN | HTR2A | 5-hydroxytryptamine receptor 2A | 5.30103 | 7.818593 | 8.285963 | mammals | actual | clID_3770 |
| CINANSERIN | Htr2b | 5-hydroxytryptamine receptor 2B | 5.30103 | 7.37631 | 8.473198 | mammals | actual | clID_3770 |
| CINANSERIN | Htr2c | 5-hydroxytryptamine receptor 2C | 5.30103 | 7.439004 | 8.342811 | mammals | actual | clID_3770 |
| CLEMASTINE | HTR2A | 5-hydroxytryptamine receptor 2A | 5.75225 | 7.818593 | 7.765626 | mammals | actual | clID_1525 |
| CLEMASTINE | Htr2b | 5-hydroxytryptamine receptor 2B | 5.75225 | 7.37631 | 7.540985 | mammals | actual | clID_1525 |
| CLEMASTINE | Htr2c | 5-hydroxytryptamine receptor 2C | 5.75225 | 7.439004 | 7.144181 | mammals | actual | clID_1525 |
| CLEMASTINE | ADRA1A | Alpha-1A adrenergic receptor | 5.75225 | 7.742958 | 7.476752 | mammals | actual | clID_1525 |
| CLEMASTINE | Adra1b | Alpha-1B adrenergic receptor | 5.75225 | 7.738535 | 7.258992 | mammals | actual | clID_1525 |
| CLEMASTINE | Adra1d | Alpha-1D adrenergic receptor | 5.75225 | 7.713266 | 7.659379 | mammals | actual | clID_1525 |
| CLEMASTINE | DRD2 | D(2) dopamine receptor | 5.75225 | 8.280379 | 7.106624 | mammals | actual | clID_1525 |
| CLEMASTINE | Drd3 | D(3) dopamine receptor | 5.75225 | 8.166222 | 8.369113 | mammals | actual | clID_1525 |
| CLOMIPRAMINE | HTR1A | 5-hydroxytryptamine receptor 1A | 4.522627 | 7.930581 | 7.634363 | mammals | actual | clID_1362 |
| CLOMIPRAMINE | HTR2A | 5-hydroxytryptamine receptor 2A | 4.522627 | 7.818593 | 8.17644 | mammals | actual | clID_1362 |
| CLOMIPRAMINE | Htr2b | 5-hydroxytryptamine receptor 2B | 4.522627 | 7.37631 | 8.099839 | mammals | actual | clID_1362 |
| CLOMIPRAMINE | Htr2c | 5-hydroxytryptamine receptor 2C | 4.522627 | 7.439004 | 8.193181 | mammals | actual | clID_1362 |
| CLOMIPRAMINE | ADRA1A | Alpha-1A adrenergic receptor | 4.522627 | 7.742958 | 7.695844 | mammals | actual | clID_1362 |
| CLOMIPRAMINE | Adra1b | Alpha-1B adrenergic receptor | 4.522627 | 7.738535 | 7.078184 | mammals | actual | clID_1362 |
| CLOMIPRAMINE | Adra1d | Alpha-1D adrenergic receptor | 4.522627 | 7.713266 | 7.035012 | mammals | actual | clID_1362 |
| CLOMIPRAMINE | Drd3 | D(3) dopamine receptor | 4.522627 | 8.166222 | 7.092444 | mammals | actual | clID_1362 |
| COLFORSIN | HTR1A | 5-hydroxytryptamine receptor 1A | 4.920624 | 7.930581 | 9 | mammals | actual | clID_2585 |
| CP-154526 | CRHR1 | Corticotropin-releasing factor receptor 1 | 5.284437 | 9.228861 | 8.358958 | mammals | predicted | clID_5004 |
| CP-99994 | Tacr1 | Substance-P receptor | 4.758394 | 9.709555 | 9.275808 | mammals | actual | clID_1699 |
| CV-6209 | PTAFR | Platelet-activating factor receptor | 5.663989 | 8.367622 | 7.725216 | mammals | predicted | clID_2685 |
| DEXNIGULDIPINE | ADRA1A | Alpha-1A adrenergic receptor | 6.60371 | 7.742958 | 8.287983 | mammals | actual | clID_802 |
| DEXNIGULDIPINE | Adra1b | Alpha-1B adrenergic receptor | 6.60371 | 7.738535 | 7.335299 | mammals | actual | clID_802 |
| DEXNIGULDIPINE | Adra1d | Alpha-1D adrenergic receptor | 6.60371 | 7.713266 | 7.128256 | mammals | actual | clID_802 |
| DIHYDROETORPHINE | Oprd1 | Delta-type opioid receptor | 5.543485 | 7.770012 | 9.69897 | mammals | predicted | clID_4751 |
| DIHYDROETORPHINE | Oprk1 | Kappa-type opioid receptor | 5.543485 | 7.699064 | 9.886057 | mammals | predicted | clID_4751 |
| DIHYDROETORPHINE | Oprm1 | Mu-type opioid receptor | 5.543485 | 7.860685 | 9.721246 | mammals | predicted | clID_4751 |
| DP[TYR(ME)2]AVP | Avpr1a | Vasopressin V1a receptor | 4.786488 | 7.7146 | 7.960189 | mammals | predicted | clID_2005 |
| E-FLUPENTIXOL | Drd1 | D(1A) dopamine receptor | 5.538428 | 7.436039 | 7.583618 | mammals | predicted | clID_267 |
| ELEDOISIN | Tacr1 | Substance-P receptor | 4.510241 | 9.709555 | 9 | mammals | predicted | clID_2005 |
| ELOPIPRAZOLE | HTR1A | 5-hydroxytryptamine receptor 1A | 5.958607 | 7.930581 | 8.823909 | mammals | actual | clID_4127 |
| EXENATIDE | Glp1r | Glucagon-like peptide 1 receptor | 5.188882 | 7.163203 | 9.843176 | mammals | predicted | clID_2926 |
| FLUNARIZINE | Drd3 | D(3) dopamine receptor | 4.518593 | 8.166222 | 7.247357 | mammals | predicted | clID_355 |
| FLUPENTIXOL | Drd1 | D(1A) dopamine receptor | 4.760268 | 7.436039 | 8.290481 | mammals | actual | clID_267 |
| FLUPENTIXOL | DRD2 | D(2) dopamine receptor | 4.760268 | 8.280379 | 7.952399 | mammals | actual | clID_267 |
| FLUPENTIXOL | Drd3 | D(3) dopamine receptor | 4.760268 | 8.166222 | 8.522879 | mammals | actual | clID_267 |
| FLUPENTIXOL | DRD4 | D(4) dopamine receptor | 4.760268 | 7.451626 | 8.522879 | mammals | actual | clID_267 |
| FLUPERAMIDE | Oprm1 | Mu-type opioid receptor | 5.925308 | 7.860685 | 9.958607 | mammals | predicted | clID_6355 |
| FLUTROLINE | Drd1 | D(1A) dopamine receptor | 5.45869 | 7.436039 | 7.876188 | mammals | predicted | clID_6792 |
| FLUTROLINE | DRD2 | D(2) dopamine receptor | 5.45869 | 8.280379 | 7.876188 | mammals | predicted | clID_6792 |
| FLUTROLINE | Drd3 | D(3) dopamine receptor | 5.45869 | 8.166222 | 7.876188 | mammals | predicted | clID_6792 |
| FLUTROLINE | DRD4 | D(4) dopamine receptor | 5.45869 | 7.451626 | 7.876188 | mammals | predicted | clID_6792 |
| Fe complex | HTR1A | 5-hydroxytryptamine receptor 1A | 4.317289 | 7.930581 | 7.568636 | mammals | predicted | iron complex |
| Fe complex | ADRA1A | Alpha-1A adrenergic receptor | 4.317289 | 7.742958 | 7.136677 | mammals | predicted | iron complex |
| Fe complex | Adra1b | Alpha-1B adrenergic receptor | 4.317289 | 7.738535 | 7.136677 | mammals | predicted | iron complex |
| Fe complex | Adra1d | Alpha-1D adrenergic receptor | 4.317289 | 7.713266 | 7.136677 | mammals | predicted | iron complex |
| Fe complex | DRD2 | D(2) dopamine receptor | 4.317289 | 8.280379 | 7.614942 | mammals | predicted | iron complex |
| Fe complex | Drd3 | D(3) dopamine receptor | 4.317289 | 8.166222 | 7.824876 | mammals | predicted | iron complex |
| GR-127935 | HTR2A | 5-hydroxytryptamine receptor 2A | 5.064917 | 7.818593 | 7.584771 | mammals | actual | clID_1796 |
| HALOPERIDOL | HTR2A | 5-hydroxytryptamine receptor 2A | 4.799189 | 7.818593 | 7.182611 | mammals | actual | clID_1001 |
| HALOPERIDOL | Htr2b | 5-hydroxytryptamine receptor 2B | 4.799189 | 7.37631 | 7.226378 | mammals | actual | clID_1001 |
| HALOPERIDOL | ADRA1A | Alpha-1A adrenergic receptor | 4.799189 | 7.742958 | 7.663944 | mammals | actual | clID_1001 |
| HALOPERIDOL | Adra1b | Alpha-1B adrenergic receptor | 4.799189 | 7.738535 | 7.676666 | mammals | actual | clID_1001 |
| HALOPERIDOL | Adra1d | Alpha-1D adrenergic receptor | 4.799189 | 7.713266 | 7.656753 | mammals | actual | clID_1001 |
| HALOPERIDOL | Drd1 | D(1A) dopamine receptor | 4.799189 | 7.436039 | 7.641172 | mammals | actual | clID_1001 |
| HALOPERIDOL | DRD2 | D(2) dopamine receptor | 4.799189 | 8.280379 | 8.339276 | mammals | actual | clID_1001 |
| HALOPERIDOL | Drd3 | D(3) dopamine receptor | 4.799189 | 8.166222 | 8.098005 | mammals | actual | clID_1001 |
| HALOPERIDOL | DRD4 | D(4) dopamine receptor | 4.799189 | 7.451626 | 8.10952 | mammals | actual | clID_1001 |
| HONOKIOL | Cnr1 | Cannabinoid receptor 1 | 4.539116 | 8.101802 | 7.459845 | mammals | actual | clID_228 |
| ICI-199,441 | Oprd1 | Delta-type opioid receptor | 5.149823 | 7.770012 | 7.153809 | mammals | actual | clID_4076 |
| ICI-199,441 | Oprk1 | Kappa-type opioid receptor | 5.149823 | 7.699064 | 9.067038 | mammals | actual | clID_4076 |
| ICI-199,441 | Oprm1 | Mu-type opioid receptor | 5.149823 | 7.860685 | 7.125119 | mammals | actual | clID_4076 |
| L-709210 | Tacr1 | Substance-P receptor | 5.97954 | 9.709555 | 8.493825 | mammals | predicted | clID_1042 |
| LOPERAMIDE | Oprm1 | Mu-type opioid receptor | 5.03292 | 7.860685 | 8.876082 | mammals | actual | clID_6355 |
| LOSARTAN POTASSIUM | AGTR1 | Type-1 angiotensin II receptor | 4.55495 | 8.425964 | 7.974806 | mammals | actual | clID_774 |
| METHSERGIDE | Htr2c | 5-hydroxytryptamine receptor 2C | 5.162939 | 7.439004 | 8.508638 | mammals | predicted | clID_4958 |
| METYROSINE | Drd1 | D(1A) dopamine receptor | 4.953051 | 7.436039 | 7.636388 | mammals | actual | clID_529 |
| N-(6-(4-CHLOROPHENYL)-7-(2,4-DICHLOROPHENYL)-2,2-DIMETHYL-3,4-DIHYDRO-2H-PYRANO[2,3-B]PYRIDIN-4-YL)-3-HYDROXYPROPANAMIDE (ENANTIOMERIC MIX) | Cnr1 | Cannabinoid receptor 1 | 4.762402 | 8.101802 | 8.311712 | mammals | predicted | clID_7142 |
| N-(7'-(2-CHLOROPHENYL)-6'-(4-CHLOROPHENYL)-3',4'-DIHYDROSPIRO[CYCLOHEXANE-1,2'-PYRANO[2,3-B]PYRIDINE]-4'-YL)-2-HYDROXY-2-METHYLPROPANAMIDE (ENANTIOMERIC MIX) | Cnr1 | Cannabinoid receptor 1 | 4.60926 | 8.101802 | 8.449815 | mammals | predicted | clID_864 |
| N-[6-(4-CHLOROPHENYL)-7-(2,4-DICHLOROPHENYL)-2,2-DIMETHYL-3,4-DIHYDRO-2H-PYRANO[2,3-B]PYRIDINE-4-YL]-4,4,4-TRIFLUORO-3-HYDROXYBUTANAMIDE (DIASTEREOMERIC MIX) | Cnr1 | Cannabinoid receptor 1 | 4.865752 | 8.101802 | 7.031113 | mammals | predicted | clID_7142 |
| NAFTOPIDIL | ADRA1A | Alpha-1A adrenergic receptor | 5.982981 | 7.742958 | 7.547345 | mammals | actual | clID_1036 |
| NAFTOPIDIL | Adra1d | Alpha-1D adrenergic receptor | 5.982981 | 7.713266 | 7.690368 | mammals | actual | clID_1036 |
| NAPHTHYRIDINONE | Cnr1 | Cannabinoid receptor 1 | 6.317734 | 8.101802 | 8.124939 | mammals | predicted | clID_1215 |
| NIGULDIPINE | ADRA1A | Alpha-1A adrenergic receptor | 6.122052 | 7.742958 | 8.577469 | mammals | actual | clID_802 |
| NIGULDIPINE | Adra1b | Alpha-1B adrenergic receptor | 6.122052 | 7.738535 | 7.288614 | mammals | actual | clID_802 |
| NIGULDIPINE | Adra1d | Alpha-1D adrenergic receptor | 6.122052 | 7.713266 | 7.245142 | mammals | actual | clID_802 |
| NORDIHYDROGUAIARETIC ACID | Ltb4r | Leukotriene B4 receptor 1 | 5.74487 | 8.370347 | 7.070581 | mammals | actual | clID_1544 |
| OLANZAPINE | HTR2A | 5-hydroxytryptamine receptor 2A | 4.747065 | 7.818593 | 8.17339 | mammals | actual | clID_7742 |
| OLANZAPINE | Htr2b | 5-hydroxytryptamine receptor 2B | 4.747065 | 7.37631 | 7.755592 | mammals | actual | clID_7742 |
| OLANZAPINE | Htr2c | 5-hydroxytryptamine receptor 2C | 4.747065 | 7.439004 | 7.907071 | mammals | actual | clID_7742 |
| OLANZAPINE | ADRA1A | Alpha-1A adrenergic receptor | 4.747065 | 7.742958 | 7.562341 | mammals | actual | clID_7742 |
| OLANZAPINE | Adra1b | Alpha-1B adrenergic receptor | 4.747065 | 7.738535 | 7.583029 | mammals | actual | clID_7742 |
| OLANZAPINE | Adra1d | Alpha-1D adrenergic receptor | 4.747065 | 7.713266 | 7.632195 | mammals | actual | clID_7742 |
| OLANZAPINE | Drd1 | D(1A) dopamine receptor | 4.747065 | 7.436039 | 7.195404 | mammals | actual | clID_7742 |
| OLANZAPINE | DRD2 | D(2) dopamine receptor | 4.747065 | 8.280379 | 7.588121 | mammals | actual | clID_7742 |
| OLANZAPINE | Drd3 | D(3) dopamine receptor | 4.747065 | 8.166222 | 7.402076 | mammals | actual | clID_7742 |
| OLANZAPINE | DRD4 | D(4) dopamine receptor | 4.747065 | 7.451626 | 7.398145 | mammals | actual | clID_7742 |
| ORVEPITANT | Tacr1 | Substance-P receptor | 4.572556 | 9.709555 | 10.350004 | mammals | predicted | clID_1663 |
| OSANETANT | TACR2 | Substance-K receptor | 6.03207 | 8.47386 | 7.623215 | mammals | predicted | clID_1341 |
| OXYTOCIN | Avpr1a | Vasopressin V1a receptor | 4.984981 | 7.7146 | 7.752034 | mammals | predicted | clID_939 |
| PD-170292 | Cckar | Cholecystokinin receptor type A | 6.002482 | 8.320266 | 8.920819 | mammals | predicted | clID_1077 |
| PD-170292 | CCKBR | Gastrin/cholecystokinin type B receptor | 6.002482 | 8.357212 | 8.173925 | mammals | predicted | clID_1077 |
| PENFLURIDOL | HTR1A | 5-hydroxytryptamine receptor 1A | 5.663352 | 7.930581 | 7.724275 | mammals | actual | clID_1001 |
| PENFLURIDOL | Drd1 | D(1A) dopamine receptor | 5.663352 | 7.436039 | 7.044455 | mammals | actual | clID_1001 |
| PENFLURIDOL | Drd3 | D(3) dopamine receptor | 5.663352 | 8.166222 | 7.09162 | mammals | actual | clID_1001 |
| PHENOXYBENZAMINE | ADRA1A | Alpha-1A adrenergic receptor | 6.024627 | 7.742958 | 8.744727 | mammals | predicted | clID_956 |
| PHENOXYBENZAMINE | Adra1b | Alpha-1B adrenergic receptor | 6.024627 | 7.738535 | 8.744727 | mammals | predicted | clID_956 |
| PHENOXYBENZAMINE | Adra1d | Alpha-1D adrenergic receptor | 6.024627 | 7.713266 | 8.744727 | mammals | predicted | clID_956 |
| PHYSALAEMIN | Tacr1 | Substance-P receptor | 4.708915 | 9.709555 | 9 | mammals | predicted | clID_2005 |
| PIMOZIDE | Drd1 | D(1A) dopamine receptor | 5.765854 | 7.436039 | 7.493901 | mammals | actual | clID_4167 |
| PIMOZIDE | DRD2 | D(2) dopamine receptor | 5.765854 | 8.280379 | 7.789236 | mammals | actual | clID_4167 |
| PIMOZIDE | Drd3 | D(3) dopamine receptor | 5.765854 | 8.166222 | 7.716479 | mammals | actual | clID_4167 |
| PIMOZIDE | DRD4 | D(4) dopamine receptor | 5.765854 | 7.451626 | 7.716479 | mammals | actual | clID_4167 |
| PRAZOSIN | ADRA1A | Alpha-1A adrenergic receptor | 4.978609 | 7.742958 | 9.003865 | mammals | actual | clID_5167 |
| PRAZOSIN | Adra1b | Alpha-1B adrenergic receptor | 4.978609 | 7.738535 | 9.000473 | mammals | actual | clID_5167 |
| PRAZOSIN | Adra1d | Alpha-1D adrenergic receptor | 4.978609 | 7.713266 | 8.996776 | mammals | actual | clID_5167 |
| PRAZOSIN HYDROCHLORIDE | ADRA1A | Alpha-1A adrenergic receptor | 5.140535 | 7.742958 | 8.740393 | mammals | actual | clID_5167 |
| PRAZOSIN HYDROCHLORIDE | Adra1b | Alpha-1B adrenergic receptor | 5.140535 | 7.738535 | 8.758404 | mammals | actual | clID_5167 |
| PRAZOSIN HYDROCHLORIDE | Adra1d | Alpha-1D adrenergic receptor | 5.140535 | 7.713266 | 8.59793 | mammals | actual | clID_5167 |
| PYRAZINAMIDE | Drd1 | D(1A) dopamine receptor | 4.900549 | 7.436039 | 8.036212 | mammals | actual | clID_7678 |
| Ruthenium complex | HTR1A | 5-hydroxytryptamine receptor 1A | 4.73044 | 7.930581 | 7.113509 | mammals | predicted | ruthenium complex |
| Ruthenium complex | ADRA1A | Alpha-1A adrenergic receptor | 4.73044 | 7.742958 | 7.585027 | mammals | predicted | ruthenium complex |
| Ruthenium complex | Adra1b | Alpha-1B adrenergic receptor | 4.73044 | 7.738535 | 7.585027 | mammals | predicted | ruthenium complex |
| Ruthenium complex | Adra1d | Alpha-1D adrenergic receptor | 4.73044 | 7.713266 | 7.585027 | mammals | predicted | ruthenium complex |
| Ruthenium complex | DRD2 | D(2) dopamine receptor | 4.73044 | 8.280379 | 7.643675 | mammals | predicted | ruthenium complex |
| Ruthenium complex | Drd3 | D(3) dopamine receptor | 4.73044 | 8.166222 | 7.372226 | mammals | predicted | ruthenium complex |
| SAREDUTANT | Tacr1 | Substance-P receptor | 5.775297 | 9.709555 | 7.205428 | mammals | predicted | clID_1341 |
| SAREDUTANT | TACR2 | Substance-K receptor | 5.775297 | 8.47386 | 8.898499 | mammals | predicted | clID_1341 |
| SELEPRESSIN | Avpr1a | Vasopressin V1a receptor | 5.206955 | 7.7146 | 8.939857 | mammals | predicted | clID_939 |
| SERTINDOLE | HTR1A | 5-hydroxytryptamine receptor 1A | 5.969909 | 7.930581 | 7.481486 | mammals | actual | clID_6299 |
| SERTINDOLE | HTR2A | 5-hydroxytryptamine receptor 2A | 5.969909 | 7.818593 | 9.377314 | mammals | actual | clID_6299 |
| SERTINDOLE | Htr2b | 5-hydroxytryptamine receptor 2B | 5.969909 | 7.37631 | 9.386391 | mammals | actual | clID_6299 |
| SERTINDOLE | Htr2c | 5-hydroxytryptamine receptor 2C | 5.969909 | 7.439004 | 9.344634 | mammals | actual | clID_6299 |
| SERTINDOLE | ADRA1A | Alpha-1A adrenergic receptor | 5.969909 | 7.742958 | 8.829211 | mammals | actual | clID_6299 |
| SERTINDOLE | Adra1b | Alpha-1B adrenergic receptor | 5.969909 | 7.738535 | 8.856975 | mammals | actual | clID_6299 |
| SERTINDOLE | Adra1d | Alpha-1D adrenergic receptor | 5.969909 | 7.713266 | 8.752961 | mammals | actual | clID_6299 |
| SERTINDOLE | Drd1 | D(1A) dopamine receptor | 5.969909 | 7.436039 | 7.2993 | mammals | actual | clID_6299 |
| SERTINDOLE | DRD2 | D(2) dopamine receptor | 5.969909 | 8.280379 | 8.678579 | mammals | actual | clID_6299 |
| SERTINDOLE | Drd3 | D(3) dopamine receptor | 5.969909 | 8.166222 | 8.335606 | mammals | actual | clID_6299 |
| SERTINDOLE | DRD4 | D(4) dopamine receptor | 5.969909 | 7.451626 | 7.818194 | mammals | actual | clID_6299 |
| SORAFENIB | Htr2b | 5-hydroxytryptamine receptor 2B | 5.191589 | 7.37631 | 7.251812 | mammals | actual | clID_4696 |
| SPARSENTAN | AGTR1 | Type-1 angiotensin II receptor | 4.829475 | 8.425964 | 9.09691 | mammals | predicted | clID_8749 |
| SPERGUALIN | Tacr1 | Substance-P receptor | 4.719462 | 9.709555 | 8.999995 | mammals | actual | clID_3362 |
| SRX246 | Avpr1a | Vasopressin V1a receptor | 4.80161 | 7.7146 | 8.896196 | mammals | predicted | clID_6134 |
| TECALCET HYDROCHLORIDE | CASR | Extracellular calcium-sensing receptor | 4.846058 | 8.205451 | 7.09691 | mammals | actual | clID_10518 |
| THIMEROSAL | Drd3 | D(3) dopamine receptor | 5.725507 | 8.166222 | 7.304092 | mammals | actual | organomercury |
| THIORIDAZINE | HTR2A | 5-hydroxytryptamine receptor 2A | 5.174574 | 7.818593 | 7.770472 | mammals | actual | clID_4843 |
| THIORIDAZINE | Htr2b | 5-hydroxytryptamine receptor 2B | 5.174574 | 7.37631 | 7.359416 | mammals | actual | clID_4843 |
| THIORIDAZINE | Htr2c | 5-hydroxytryptamine receptor 2C | 5.174574 | 7.439004 | 7.486819 | mammals | actual | clID_4843 |
| THIORIDAZINE | ADRA1A | Alpha-1A adrenergic receptor | 5.174574 | 7.742958 | 7.582719 | mammals | actual | clID_4843 |
| THIORIDAZINE | Adra1b | Alpha-1B adrenergic receptor | 5.174574 | 7.738535 | 7.576849 | mammals | actual | clID_4843 |
| THIORIDAZINE | Adra1d | Alpha-1D adrenergic receptor | 5.174574 | 7.713266 | 7.563576 | mammals | actual | clID_4843 |
| THIORIDAZINE | DRD2 | D(2) dopamine receptor | 5.174574 | 8.280379 | 7.474398 | mammals | actual | clID_4843 |
| THIORIDAZINE | Drd3 | D(3) dopamine receptor | 5.174574 | 8.166222 | 7.404469 | mammals | actual | clID_4843 |
| TRIFLUOPERAZINE | HTR2A | 5-hydroxytryptamine receptor 2A | 5.241725 | 7.818593 | 8.398967 | mammals | predicted | clID_4249 |
| TRIFLUOPERAZINE | Htr2b | 5-hydroxytryptamine receptor 2B | 5.241725 | 7.37631 | 8.632856 | mammals | predicted | clID_4249 |
| TRIFLUOPERAZINE | Htr2c | 5-hydroxytryptamine receptor 2C | 5.241725 | 7.439004 | 8.257419 | mammals | predicted | clID_4249 |
| TRIFLUOPERAZINE | DRD2 | D(2) dopamine receptor | 5.241725 | 8.280379 | 7.880815 | mammals | predicted | clID_4249 |
| VASOPRESSIN | Avpr1a | Vasopressin V1a receptor | 4.686851 | 7.7146 | 9.127829 | mammals | predicted | clID_2005 |
| VASOPRESSIN | AVPR2 | Vasopressin V2 receptor | 4.686851 | 7.879176 | 9.094843 | mammals | predicted | clID_2005 |
| VIGABATRIN | Drd1 | D(1A) dopamine receptor | 4.762327 | 7.436039 | 8.431798 | mammals | actual | clID_8177 |
| XYLOMETAZOLINE | ADRA1A | Alpha-1A adrenergic receptor | 5.492548 | 7.742958 | 7.369311 | mammals | predicted | clID_2569 |
| XYLOMETAZOLINE | Adra1b | Alpha-1B adrenergic receptor | 5.492548 | 7.738535 | 7.391201 | mammals | predicted | clID_2569 |
| XYLOMETAZOLINE | Adra1d | Alpha-1D adrenergic receptor | 5.492548 | 7.713266 | 7.391201 | mammals | predicted | clID_2569 |
| YOHIMBINE | HTR1A | 5-hydroxytryptamine receptor 1A | 5.240925 | 7.930581 | 7.021148 | mammals | predicted | clID_4255 |
| ZAFIRLUKAST | CYSLTR1 | Cysteinyl leukotriene receptor 1 | 4.511604 | 8.121852 | 8.869426 | mammals | actual | clID_3257 |
| ZAFIRLUKAST | CYSLTR2 | Cysteinyl leukotriene receptor 2 | 4.511604 | 8.038902 | 8.310957 | mammals | actual | clID_3257 |
| ZIPRASIDONE HYDROCHLORIDE | Drd1 | D(1A) dopamine receptor | 5.059504 | 7.436039 | 7.442668 | mammals | actual | clID_6064 |
Use search function in table to view values for specific interactions
| molName | geneName | protname | sarsScoreMolecule | geneScore | interactionScore | organismType | kindOfValue | molecularCluster |
|---|---|---|---|---|---|---|---|---|
| AEE-788 | EGFR | Epidermal growth factor receptor | 5.999522 | 8.436627 | 8.314924 | mammals | actual | clID_1012 |
| AEE-788 | ERBB2 | Receptor tyrosine-protein kinase erbB-2 | 5.999522 | 8.200475 | 8.69897 | mammals | actual | clID_1012 |
| AEE-788 | Kdr | Vascular endothelial growth factor receptor 2 | 5.999522 | 7.82472 | 7.113509 | mammals | actual | clID_1012 |
| ASP-3026 | Tnk2 | Activated CDC42 kinase 1 | 5.216853 | 7.730424 | 8.30103 | mammals | actual | clID_4452 |
| BIOTIN | ABL1 | Tyrosine-protein kinase ABL1 | 4.646405 | 7.713451 | 7.628291 | mammals | actual | clID_6631 |
| CABOZANTINIB | FGFR1 | Fibroblast growth factor receptor 1 | 4.666839 | 7.460342 | 7.946922 | mammals | actual | clID_4744 |
| CABOZANTINIB | MET | Hepatocyte growth factor receptor | 4.666839 | 7.440635 | 8.233173 | mammals | actual | clID_4744 |
| CABOZANTINIB | TEK | Angiopoietin-1 receptor | 4.666839 | 8.278308 | 7.844664 | mammals | actual | clID_4744 |
| CABOZANTINIB | Kdr | Vascular endothelial growth factor receptor 2 | 4.666839 | 7.82472 | 8.50106 | mammals | actual | clID_4744 |
| CABOZANTINIB S-MALATE | MET | Hepatocyte growth factor receptor | 7.31367 | 7.440635 | 8.077203 | mammals | predicted | clID_213 |
| CABOZANTINIB S-MALATE | Kdr | Vascular endothelial growth factor receptor 2 | 7.31367 | 7.82472 | 8.522879 | mammals | predicted | clID_213 |
| CEP-32496 | ABL1 | Tyrosine-protein kinase ABL1 | 4.675034 | 7.713451 | 7.911954 | mammals | actual | clID_10342 |
| CEP-32496 | EGFR | Epidermal growth factor receptor | 4.675034 | 8.436627 | 7.657577 | mammals | actual | clID_10342 |
| CEP-32496 | Kdr | Vascular endothelial growth factor receptor 2 | 4.675034 | 7.82472 | 8.09691 | mammals | actual | clID_10342 |
| CGP-53353 | FGFR1 | Fibroblast growth factor receptor 1 | 5.792715 | 7.460342 | 7.376664 | mammals | actual | clID_670 |
| CGP-53353 | IGF1R | Insulin-like growth factor 1 receptor | 5.792715 | 7.603517 | 7.369751 | mammals | actual | clID_670 |
| CGP-53353 | INSR | Insulin receptor | 5.792715 | 7.945268 | 7.487076 | mammals | actual | clID_670 |
| CYC-116 | Kdr | Vascular endothelial growth factor receptor 2 | 5.078035 | 7.82472 | 7.072563 | mammals | actual | clID_5848 |
| DACOMITINIB | EGFR | Epidermal growth factor receptor | 5.772001 | 8.436627 | 8.077544 | mammals | actual | clID_992 |
| DACOMITINIB | ERBB2 | Receptor tyrosine-protein kinase erbB-2 | 5.772001 | 8.200475 | 8.069477 | mammals | actual | clID_992 |
| DASATINIB | ABL1 | Tyrosine-protein kinase ABL1 | 5.047298 | 7.713451 | 8.168396 | mammals | actual | clID_5901 |
| DASATINIB | Tnk2 | Activated CDC42 kinase 1 | 5.047298 | 7.730424 | 8.178818 | mammals | actual | clID_5901 |
| DASATINIB | Btk | Tyrosine-protein kinase BTK | 5.047298 | 7.69029 | 7.172445 | mammals | actual | clID_5901 |
| DASATINIB | SRC | Proto-oncogene tyrosine-protein kinase Src | 5.047298 | 7.76828 | 7.752187 | mammals | actual | clID_5901 |
| DOVITINIB | FGFR1 | Fibroblast growth factor receptor 1 | 4.65048 | 7.460342 | 7.242514 | mammals | actual | clID_9575 |
| DOVITINIB | Kdr | Vascular endothelial growth factor receptor 2 | 4.65048 | 7.82472 | 7.407571 | mammals | actual | clID_9575 |
| ENTRECTINIB | Tnk2 | Activated CDC42 kinase 1 | 5.367357 | 7.730424 | 7.010564 | mammals | actual | clID_3315 |
| ENTRECTINIB | SRC | Proto-oncogene tyrosine-protein kinase Src | 5.367357 | 7.76828 | 7.500038 | mammals | actual | clID_3315 |
| ERLOTINIB HYDROCHLORIDE | EGFR | Epidermal growth factor receptor | 4.540989 | 8.436627 | 7.11852 | mammals | actual | clID_3557 |
| FALNIDAMOL | EGFR | Epidermal growth factor receptor | 5.074534 | 8.436627 | 8.522879 | mammals | actual | clID_5219 |
| GW632580X | Btk | Tyrosine-protein kinase BTK | 4.861247 | 7.69029 | 9 | mammals | actual | clID_780 |
| GW632580X | EGFR | Epidermal growth factor receptor | 4.861247 | 8.436627 | 9 | mammals | actual | clID_780 |
| GW632580X | IGF1R | Insulin-like growth factor 1 receptor | 4.861247 | 7.603517 | 7.091793 | mammals | actual | clID_780 |
| GW632580X | INSR | Insulin receptor | 4.861247 | 7.945268 | 7.418024 | mammals | actual | clID_780 |
| GW632580X | MET | Hepatocyte growth factor receptor | 4.861247 | 7.440635 | 7.6 | mammals | actual | clID_780 |
| HERBIMYCIN | Btk | Tyrosine-protein kinase BTK | 5.122579 | 7.69029 | 7.344315 | mammals | predicted | clID_2407 |
| HERBIMYCIN | EGFR | Epidermal growth factor receptor | 5.122579 | 8.436627 | 9 | mammals | predicted | clID_2407 |
| HERBIMYCIN | IGF1R | Insulin-like growth factor 1 receptor | 5.122579 | 7.603517 | 8.345053 | mammals | predicted | clID_2407 |
| HERBIMYCIN | INSR | Insulin receptor | 5.122579 | 7.945268 | 9 | mammals | predicted | clID_2407 |
| HERBIMYCIN | MET | Hepatocyte growth factor receptor | 5.122579 | 7.440635 | 7.329271 | mammals | predicted | clID_2407 |
| HERBIMYCIN | SRC | Proto-oncogene tyrosine-protein kinase Src | 5.122579 | 7.76828 | 7.530349 | mammals | predicted | clID_2407 |
| HKI-357 | EGFR | Epidermal growth factor receptor | 5.543119 | 8.436627 | 7.468521 | mammals | actual | clID_730 |
| HKI-357 | ERBB2 | Receptor tyrosine-protein kinase erbB-2 | 5.543119 | 8.200475 | 7.481486 | mammals | actual | clID_730 |
| ICOTINIB | EGFR | Epidermal growth factor receptor | 5.330156 | 8.436627 | 7.013411 | mammals | actual | clID_3557 |
| JNJ-38877605 | MET | Hepatocyte growth factor receptor | 4.690473 | 7.440635 | 7.51179 | mammals | actual | clID_10534 |
| KENPAULLONE | EGFR | Epidermal growth factor receptor | 4.771468 | 8.436627 | 7.633334 | mammals | actual | clID_253 |
| KENPAULLONE | IGF1R | Insulin-like growth factor 1 receptor | 4.771468 | 7.603517 | 7.59214 | mammals | actual | clID_253 |
| KENPAULLONE | TEK | Angiopoietin-1 receptor | 4.771468 | 8.278308 | 7.845098 | mammals | actual | clID_253 |
| KX2-391 | SRC | Proto-oncogene tyrosine-protein kinase Src | 5.59908 | 7.76828 | 7.69897 | mammals | actual | clID_2071 |
| LINSITINIB | IGF1R | Insulin-like growth factor 1 receptor | 5.151793 | 7.603517 | 7.887617 | mammals | actual | clID_1409 |
| LINSITINIB | INSR | Insulin receptor | 5.151793 | 7.945268 | 7.320681 | mammals | actual | clID_1409 |
| MASITINIB | ABL1 | Tyrosine-protein kinase ABL1 | 5.355638 | 7.713451 | 7.500654 | mammals | actual | clID_3395 |
| OSI-930 | Kdr | Vascular endothelial growth factor receptor 2 | 9 | 7.82472 | 8.173394 | mammals | actual | clID_99 |
| OSIMERTINIB | EGFR | Epidermal growth factor receptor | 5.026632 | 8.436627 | 7.479191 | mammals | actual | clID_6428 |
| OSIMERTINIB | ERBB2 | Receptor tyrosine-protein kinase erbB-2 | 5.026632 | 8.200475 | 7.422816 | mammals | actual | clID_6428 |
| PAZOPANIB | Kdr | Vascular endothelial growth factor receptor 2 | 4.969786 | 7.82472 | 7.15109 | mammals | actual | clID_7154 |
| PD-0166285 | ABL1 | Tyrosine-protein kinase ABL1 | 5.465159 | 7.713451 | 9.299989 | mammals | actual | clID_1477 |
| PD-0166285 | Tnk2 | Activated CDC42 kinase 1 | 5.465159 | 7.730424 | 8.899974 | mammals | actual | clID_1477 |
| PD-0166285 | Btk | Tyrosine-protein kinase BTK | 5.465159 | 7.69029 | 8 | mammals | actual | clID_1477 |
| PD-0166285 | FGFR1 | Fibroblast growth factor receptor 1 | 5.465159 | 7.460342 | 7.447761 | mammals | actual | clID_1477 |
| PD-0166285 | SRC | Proto-oncogene tyrosine-protein kinase Src | 5.465159 | 7.76828 | 8.045757 | mammals | actual | clID_1477 |
| PD-0166285 | Kdr | Vascular endothelial growth factor receptor 2 | 5.465159 | 7.82472 | 8.100015 | mammals | actual | clID_1477 |
| PELITINIB | EGFR | Epidermal growth factor receptor | 5.700744 | 8.436627 | 8.295255 | mammals | actual | clID_730 |
| PONATINIB | ABL1 | Tyrosine-protein kinase ABL1 | 4.627256 | 7.713451 | 8.03767 | mammals | actual | clID_4006 |
| PONATINIB | FGFR1 | Fibroblast growth factor receptor 1 | 4.627256 | 7.460342 | 8.97205 | mammals | actual | clID_4006 |
| PONATINIB | SRC | Proto-oncogene tyrosine-protein kinase Src | 4.627256 | 7.76828 | 8.151508 | mammals | actual | clID_4006 |
| PONATINIB | Kdr | Vascular endothelial growth factor receptor 2 | 4.627256 | 7.82472 | 8.639854 | mammals | actual | clID_4006 |
| RIPRETINIB | Kdr | Vascular endothelial growth factor receptor 2 | 5.078239 | 7.82472 | 7.008774 | mammals | predicted | clID_1121 |
| RIVOCERANIB MESYLATE | Kdr | Vascular endothelial growth factor receptor 2 | 5.386753 | 7.82472 | 9 | mammals | actual | clID_6137 |
| ROCILETINIB | EGFR | Epidermal growth factor receptor | 5.749254 | 8.436627 | 7.443916 | mammals | actual | clID_1532 |
| ROCILETINIB | ERBB2 | Receptor tyrosine-protein kinase erbB-2 | 5.749254 | 8.200475 | 7.578876 | mammals | actual | clID_1532 |
| SGX-523 | MET | Hepatocyte growth factor receptor | 4.607759 | 7.440635 | 7.582089 | mammals | actual | clID_11897 |
| SIROLIMUS | ABL1 | Tyrosine-protein kinase ABL1 | 5.512884 | 7.713451 | 7.502399 | mammals | actual | clID_265 |
| SIROLIMUS | Btk | Tyrosine-protein kinase BTK | 5.512884 | 7.69029 | 7.502399 | mammals | actual | clID_265 |
| SIROLIMUS | EGFR | Epidermal growth factor receptor | 5.512884 | 8.436627 | 9 | mammals | actual | clID_265 |
| SIROLIMUS | FGFR1 | Fibroblast growth factor receptor 1 | 5.512884 | 7.460342 | 7.115569 | mammals | actual | clID_265 |
| SIROLIMUS | IGF1R | Insulin-like growth factor 1 receptor | 5.512884 | 7.603517 | 7.458617 | mammals | actual | clID_265 |
| SIROLIMUS | INSR | Insulin receptor | 5.512884 | 7.945268 | 7.492174 | mammals | actual | clID_265 |
| SIROLIMUS | MET | Hepatocyte growth factor receptor | 5.512884 | 7.440635 | 7.639377 | mammals | actual | clID_265 |
| SIROLIMUS | Kdr | Vascular endothelial growth factor receptor 2 | 5.512884 | 7.82472 | 7.690106 | mammals | actual | clID_265 |
| SORAFENIB | Kdr | Vascular endothelial growth factor receptor 2 | 5.191589 | 7.82472 | 7.375408 | mammals | actual | clID_4696 |
| SORAFENIB TOSYLATE | Kdr | Vascular endothelial growth factor receptor 2 | 5.431908 | 7.82472 | 7.877083 | mammals | actual | clID_4696 |
| TANESPIMYCIN | ERBB2 | Receptor tyrosine-protein kinase erbB-2 | 4.582691 | 8.200475 | 7.508638 | mammals | actual | clID_2407 |
| TYRPHOSTIN AG-1478 | EGFR | Epidermal growth factor receptor | 5.452454 | 8.436627 | 7.793196 | mammals | actual | clID_681 |
| TYRPHOSTIN AG-1478 | IGF1R | Insulin-like growth factor 1 receptor | 5.452454 | 7.603517 | 8.345053 | mammals | actual | clID_681 |
| TYRPHOSTIN AG-1478 | INSR | Insulin receptor | 5.452454 | 7.945268 | 7.997818 | mammals | actual | clID_681 |
| TYRPHOSTIN AG-1478 | TEK | Angiopoietin-1 receptor | 5.452454 | 8.278308 | 7.344315 | mammals | actual | clID_681 |
| VANDETANIB | ABL1 | Tyrosine-protein kinase ABL1 | 5.021545 | 7.713451 | 7.206383 | mammals | actual | clID_1562 |
| VANDETANIB | EGFR | Epidermal growth factor receptor | 5.021545 | 8.436627 | 7.436342 | mammals | actual | clID_1562 |
| VANDETANIB | SRC | Proto-oncogene tyrosine-protein kinase Src | 5.021545 | 7.76828 | 7.033278 | mammals | actual | clID_1562 |
Use search function in table to view values for specific interactions
| molName | geneName | protname | sarsScoreMolecule | geneScore | interactionScore | organismType | kindOfValue | molecularCluster |
|---|---|---|---|---|---|---|---|---|
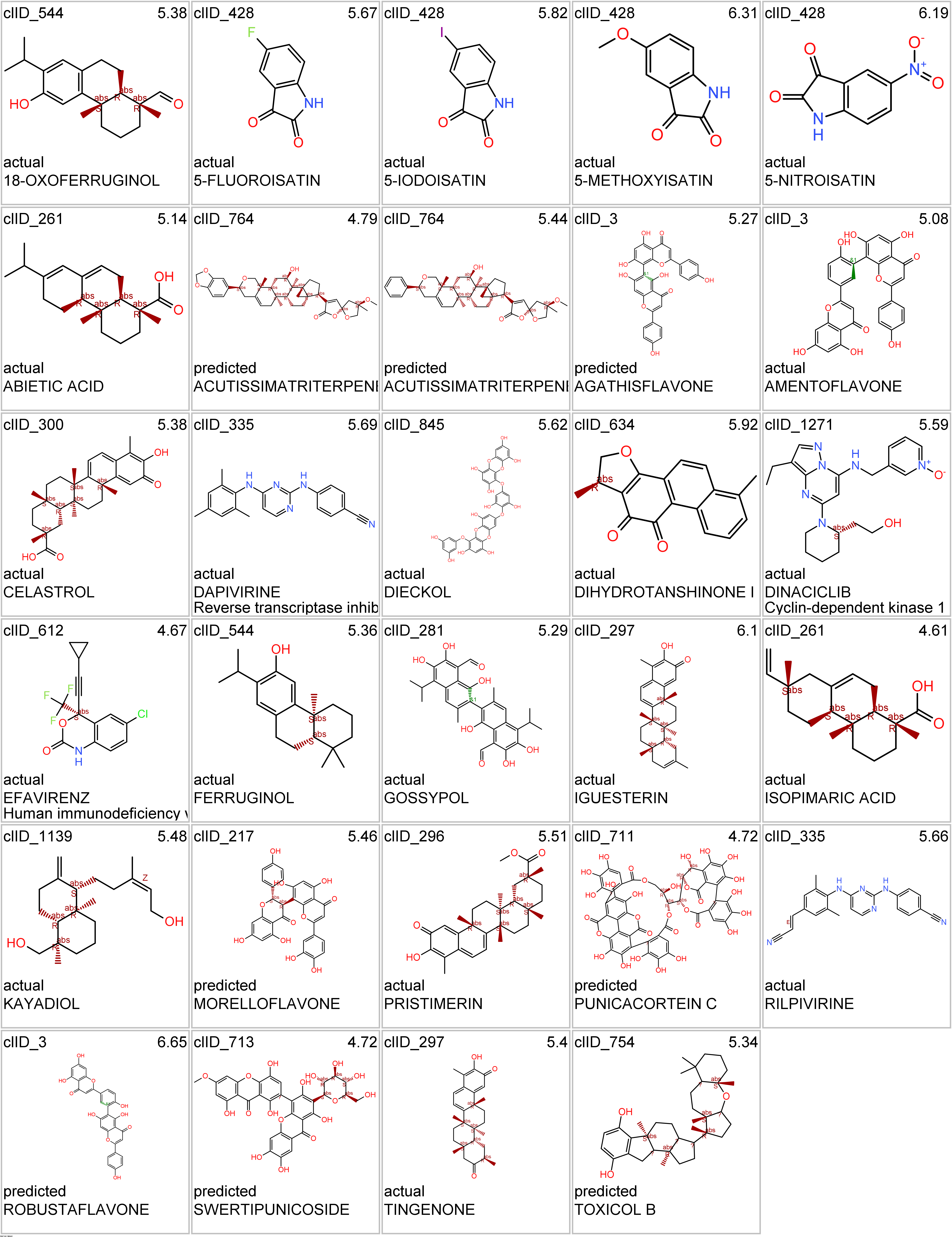
| 18-OXOFERRUGINOL | rep | Replicase polyprotein 1ab | 5.378367 | 12.649926 | 5.378367 | virus | actual | clID_3387 |
| 5-FLUOROISATIN | rep | Replicase polyprotein 1ab | 5.674922 | 12.649926 | 5.674922 | virus | actual | clID_286 |
| 5-IODOISATIN | rep | Replicase polyprotein 1ab | 5.815298 | 12.649926 | 5.815298 | virus | actual | clID_286 |
| 5-METHOXYISATIN | rep | Replicase polyprotein 1ab | 6.308938 | 12.649926 | 6.308938 | virus | actual | clID_286 |
| 5-NITROISATIN | rep | Replicase polyprotein 1ab | 6.191906 | 12.649926 | 6.191906 | virus | actual | clID_286 |
| ABIETIC ACID | rep | Replicase polyprotein 1ab | 5.140179 | 12.649926 | 5.140179 | virus | actual | clID_672 |
| ACUTISSIMATRITERPENE A | reverse | Reverse transcriptase | 4.791265 | 7.682466 | 5.095933 | virus | predicted | clID_7283 |
| ACUTISSIMATRITERPENE B | reverse | Reverse transcriptase | 5.441074 | 7.682466 | 5.286131 | virus | predicted | clID_2585 |
| AGATHISFLAVONE | reverse | Reverse transcriptase | 5.265865 | 7.682466 | 7.698101 | virus | predicted | clID_459 |
| AMENTOFLAVONE | reverse | Reverse transcriptase | 5.080922 | 7.682466 | 6.556749 | virus | actual | clID_459 |
| CELASTROL | rep | Replicase polyprotein 1ab | 5.376751 | 12.649926 | 5.181957 | virus | actual | clID_2475 |
| DAPIVIRINE | reverse | Reverse transcriptase | 5.692874 | 7.682466 | 7.020573 | virus | actual | clID_1716 |
| DIECKOL | rep | Replicase polyprotein 1ab | 5.619789 | 12.649926 | 5.247119 | virus | actual | clID_1978 |
| DIHYDROTANSHINONE I | 1a | Replicase polyprotein 1a | 5.920819 | 12.301604 | 5.161707 | virus | actual | clID_1151 |
| DINACICLIB | CHD4 | Chromodomain-helicase-DNA-binding protein 4 | 5.586219 | 11.473468 | 8.522879 | mammals | actual | clID_2117 |
| EFAVIRENZ | reverse | Reverse transcriptase | 4.674037 | 7.682466 | 7.539352 | virus | actual | clID_5637 |
| FERRUGINOL | rep | Replicase polyprotein 1ab | 5.356695 | 12.649926 | 6.103757 | virus | actual | clID_3387 |
| GOSSYPOL | reverse | Reverse transcriptase | 5.294661 | 7.682466 | 5.472076 | virus | actual | clID_3821 |
| IGUESTERIN | rep | Replicase polyprotein 1ab | 6.09691 | 12.649926 | 5.840968 | virus | actual | clID_3098 |
| ISOPIMARIC ACID | rep | Replicase polyprotein 1ab | 4.609028 | 12.649926 | 4.609028 | virus | actual | clID_672 |
| KAYADIOL | rep | Replicase polyprotein 1ab | 5.481766 | 12.649926 | 5.481766 | virus | actual | clID_2636 |
| MORELLOFLAVONE | reverse | Reverse transcriptase | 5.462412 | 7.682466 | 7.093422 | virus | predicted | clID_4617 |
| PRISTIMERIN | rep | Replicase polyprotein 1ab | 5.508638 | 12.649926 | 5.384138 | virus | actual | clID_2475 |
| PUNICACORTEIN C | reverse | Reverse transcriptase | 4.723579 | 7.682466 | 5.004964 | virus | predicted | clID_348 |
| RILPIVIRINE | reverse | Reverse transcriptase | 5.66149 | 7.682466 | 7.412608 | virus | actual | clID_1716 |
| ROBUSTAFLAVONE | reverse | Reverse transcriptase | 6.652098 | 7.682466 | 6.026448 | virus | predicted | clID_459 |
| SWERTIPUNICOSIDE | reverse | Reverse transcriptase | 4.717274 | 7.682466 | 5.364582 | virus | predicted | clID_9523 |
| TINGENONE | rep | Replicase polyprotein 1ab | 5.39794 | 12.649926 | 5.201152 | virus | actual | clID_3098 |
| TOXICOL B | reverse | Reverse transcriptase | 5.341663 | 7.682466 | 5.118165 | virus | predicted | clID_3492 |
Use search function in table to view values for specific interactions
| molName | geneName | protname | sarsScoreMolecule | geneScore | interactionScore | organismType | kindOfValue | molecularCluster |
|---|---|---|---|---|---|---|---|---|
| AMIODARONE | CACNA1F | Voltage-dependent L-type calcium channel subunit alpha-1F | 5.1595 | 8.55979 | 6.144941 | mammals | actual | clID_6596 |
| AMPEROZIDE HYDROCHLORIDE | KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 4.778337 | 7.946483 | 7.250264 | mammals | actual | clID_827 |
| ANANDAMIDE | Trpv1 | Transient receptor potential cation channel subfamily V member 1 | 5.171247 | 8.158508 | 6.409254 | mammals | predicted | clID_4809 |
| ASTEMIZOLE | KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 5.958607 | 7.946483 | 8.065956 | mammals | actual | clID_1081 |
| CHLORPROMAZINE | Trpv1 | Transient receptor potential cation channel subfamily V member 1 | 6.908068 | 8.158508 | 6.085027 | mammals | predicted | clID_302 |
| CILNIDIPINE | CACNA1C | Voltage-dependent L-type calcium channel subunit alpha-1C | 4.665234 | 8.516207 | 8.25449 | mammals | actual | clID_6999 |
| CILNIDIPINE | Cacna1d | Voltage-dependent L-type calcium channel subunit alpha-1D | 4.665234 | 8.332342 | 8.958607 | mammals | actual | clID_6999 |
| CILNIDIPINE | CACNA1S | Voltage-dependent L-type calcium channel subunit alpha-1S | 4.665234 | 8.453813 | 8.958607 | mammals | actual | clID_6999 |
| CLEMASTINE | KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 5.75225 | 7.946483 | 6.564383 | mammals | actual | clID_1525 |
| CORT 108297 | KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 5.298461 | 7.946483 | 6.09691 | mammals | predicted | clID_3793 |
| CURCUMIN | KCNK2 | Potassium channel subfamily K member 2 | 5.319754 | 10.830631 | 6.031517 | mammals | actual | clID_3628 |
| DEXNIGULDIPINE | CACNA1C | Voltage-dependent L-type calcium channel subunit alpha-1C | 6.60371 | 8.516207 | 7.130119 | mammals | actual | clID_802 |
| DEXNIGULDIPINE | Cacna1d | Voltage-dependent L-type calcium channel subunit alpha-1D | 6.60371 | 8.332342 | 7.130119 | mammals | actual | clID_802 |
| DEXNIGULDIPINE | CACNA1S | Voltage-dependent L-type calcium channel subunit alpha-1S | 6.60371 | 8.453813 | 7.130119 | mammals | actual | clID_802 |
| DRONEDARONE | CACNA1C | Voltage-dependent L-type calcium channel subunit alpha-1C | 5.406714 | 8.516207 | 6.39794 | mammals | actual | clID_3033 |
| DRONEDARONE | KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 5.406714 | 7.946483 | 6.011438 | mammals | actual | clID_3033 |
| ELTROMBOPAG | KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 5.082494 | 7.946483 | 6.180575 | mammals | actual | clID_5799 |
| FANCHININ | CACNA1C | Voltage-dependent L-type calcium channel subunit alpha-1C | 5.562726 | 8.516207 | 6.13 | mammals | actual | clID_186 |
| FANCHININ | Cacna1d | Voltage-dependent L-type calcium channel subunit alpha-1D | 5.562726 | 8.332342 | 6.13 | mammals | actual | clID_186 |
| FANCHININ | CACNA1S | Voltage-dependent L-type calcium channel subunit alpha-1S | 5.562726 | 8.453813 | 6.13 | mammals | actual | clID_186 |
| FANTOFARONE | CACNA1C | Voltage-dependent L-type calcium channel subunit alpha-1C | 5.21134 | 8.516207 | 6.847408 | mammals | predicted | clID_2863 |
| FLUNARIZINE | Cacna2d1 | Voltage-dependent calcium channel subunit alpha-2/delta-1 | 4.518593 | 9.459622 | 7.09691 | mammals | predicted | clID_355 |
| FLUNARIZINE | CACNA1B | Voltage-dependent N-type calcium channel subunit alpha-1B | 4.518593 | 8.75671 | 6.767003 | mammals | predicted | clID_355 |
| FLUNARIZINE | CACNA1G | Voltage-dependent T-type calcium channel subunit alpha-1G | 4.518593 | 8.815555 | 6.275724 | mammals | predicted | clID_355 |
| FLUNARIZINE | CACNA1I | Voltage-dependent T-type calcium channel subunit alpha-1I | 4.518593 | 8.867609 | 6.075721 | mammals | predicted | clID_355 |
| FLUNARIZINE | Cacnb1 | Voltage-dependent L-type calcium channel subunit beta-1 | 4.518593 | 9.644421 | 7.09691 | mammals | predicted | clID_355 |
| FLUNARIZINE | KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 4.518593 | 7.946483 | 6.260963 | mammals | predicted | clID_355 |
| FLUNARIZINE HYDROCHLORIDE | KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 5.364535 | 7.946483 | 6.349984 | mammals | actual | clID_355 |
| FLUSPIRILENE | KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 5.356424 | 7.946483 | 6.069155 | mammals | actual | clID_4784 |
| HALOPERIDOL | KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 4.799189 | 7.946483 | 7.180601 | mammals | actual | clID_1001 |
| INDOMETHACIN | Trpv1 | Transient receptor potential cation channel subfamily V member 1 | 4.738732 | 8.158508 | 6.174369 | mammals | actual | clID_7591 |
| LIDOFLAZINE | KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 5.968362 | 7.946483 | 7.799971 | mammals | actual | clID_1059 |
| LOPERAMIDE | KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 5.03292 | 7.946483 | 7.481486 | mammals | actual | clID_6355 |
| LORATADINE | KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 5.79633 | 7.946483 | 6.765331 | mammals | actual | clID_7357 |
| MIBEFRADIL | CACNA1C | Voltage-dependent L-type calcium channel subunit alpha-1C | 6.635835 | 8.516207 | 6.22266 | mammals | predicted | clID_6251 |
| MIBEFRADIL | Cacna1d | Voltage-dependent L-type calcium channel subunit alpha-1D | 6.635835 | 8.332342 | 6.169266 | mammals | predicted | clID_6251 |
| MIBEFRADIL | CACNA1F | Voltage-dependent L-type calcium channel subunit alpha-1F | 6.635835 | 8.55979 | 6.549653 | mammals | predicted | clID_6251 |
| MIBEFRADIL | Cacna1h | Voltage-dependent T-type calcium channel subunit alpha-1H | 6.635835 | 8.203186 | 6.268285 | mammals | predicted | clID_6251 |
| MIBEFRADIL | CACNA1I | Voltage-dependent T-type calcium channel subunit alpha-1I | 6.635835 | 8.867609 | 6.595229 | mammals | predicted | clID_6251 |
| MIBEFRADIL | CACNA1S | Voltage-dependent L-type calcium channel subunit alpha-1S | 6.635835 | 8.453813 | 6.169266 | mammals | predicted | clID_6251 |
| MIBEFRADIL | Scn4a | Sodium channel protein type 4 subunit alpha | 6.635835 | 9.452118 | 6.035343 | mammals | predicted | clID_6251 |
| N-(6-(4-CHLOROPHENYL)-7-(2,4-DICHLOROPHENYL)-2,2-DIMETHYL-3,4-DIHYDRO-2H-PYRANO[2,3-B]PYRIDIN-4-YL)-3-HYDROXYPROPANAMIDE (ENANTIOMERIC MIX) | KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 4.762402 | 7.946483 | 6.420216 | mammals | predicted | clID_7142 |
| NIFEDIPINE | CACNA1C | Voltage-dependent L-type calcium channel subunit alpha-1C | 4.683767 | 8.516207 | 6.193024 | mammals | actual | clID_6999 |
| NIFEDIPINE | Cacna1d | Voltage-dependent L-type calcium channel subunit alpha-1D | 4.683767 | 8.332342 | 6.019461 | mammals | actual | clID_6999 |
| NIGULDIPINE | CACNA1C | Voltage-dependent L-type calcium channel subunit alpha-1C | 6.122052 | 8.516207 | 8.264371 | mammals | actual | clID_802 |
| NIGULDIPINE | Cacna1d | Voltage-dependent L-type calcium channel subunit alpha-1D | 6.122052 | 8.332342 | 8.337242 | mammals | actual | clID_802 |
| NIGULDIPINE | CACNA1S | Voltage-dependent L-type calcium channel subunit alpha-1S | 6.122052 | 8.453813 | 8.337242 | mammals | actual | clID_802 |
| OLANZAPINE | KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 4.747065 | 7.946483 | 6.695497 | mammals | actual | clID_7742 |
| OLVANIL | Trpv1 | Transient receptor potential cation channel subfamily V member 1 | 4.563624 | 8.158508 | 6.979196 | mammals | actual | clID_219 |
| PIMOZIDE | CACNA1C | Voltage-dependent L-type calcium channel subunit alpha-1C | 5.765854 | 8.516207 | 6.705137 | mammals | actual | clID_4167 |
| PIMOZIDE | Cacna1d | Voltage-dependent L-type calcium channel subunit alpha-1D | 5.765854 | 8.332342 | 6.705137 | mammals | actual | clID_4167 |
| PIMOZIDE | CACNA1F | Voltage-dependent L-type calcium channel subunit alpha-1F | 5.765854 | 8.55979 | 6.705137 | mammals | actual | clID_4167 |
| PIMOZIDE | CACNA1G | Voltage-dependent T-type calcium channel subunit alpha-1G | 5.765854 | 8.815555 | 7.39794 | mammals | actual | clID_4167 |
| PIMOZIDE | CACNA1S | Voltage-dependent L-type calcium channel subunit alpha-1S | 5.765854 | 8.453813 | 6.705137 | mammals | actual | clID_4167 |
| PIMOZIDE | KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 5.765854 | 7.946483 | 7.438449 | mammals | actual | clID_4167 |
| PIMOZIDE | Scn4a | Sodium channel protein type 4 subunit alpha | 5.765854 | 9.452118 | 7.267606 | mammals | actual | clID_4167 |
| PRANIDIPINE | CACNA1C | Voltage-dependent L-type calcium channel subunit alpha-1C | 4.882728 | 8.516207 | 7.431798 | mammals | actual | clID_6999 |
| PRANIDIPINE | Cacna1d | Voltage-dependent L-type calcium channel subunit alpha-1D | 4.882728 | 8.332342 | 7.431798 | mammals | actual | clID_6999 |
| QUINIDINE | KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 4.856749 | 7.946483 | 6.364291 | mammals | actual | clID_530 |
| SB-705498 | Trpv1 | Transient receptor potential cation channel subfamily V member 1 | 5.26066 | 8.158508 | 6.85082 | mammals | actual | clID_4112 |
| SERTINDOLE | KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 5.969909 | 7.946483 | 8.0596 | mammals | actual | clID_6299 |
| THIORIDAZINE | KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 5.174574 | 7.946483 | 6.847955 | mammals | actual | clID_4843 |
| VANOXERINE | CACNA1C | Voltage-dependent L-type calcium channel subunit alpha-1C | 6.527334 | 8.516207 | 6.49485 | mammals | predicted | clID_462 |
| VERAPAMIL | CACNA1C | Voltage-dependent L-type calcium channel subunit alpha-1C | 4.715405 | 8.516207 | 7.059805 | mammals | actual | clID_523 |
| VERAPAMIL | Cacna1d | Voltage-dependent L-type calcium channel subunit alpha-1D | 4.715405 | 8.332342 | 7.373075 | mammals | actual | clID_523 |
| VERAPAMIL | CACNA1F | Voltage-dependent L-type calcium channel subunit alpha-1F | 4.715405 | 8.55979 | 6.69897 | mammals | actual | clID_523 |
| VERAPAMIL | CACNA1S | Voltage-dependent L-type calcium channel subunit alpha-1S | 4.715405 | 8.453813 | 7.369483 | mammals | actual | clID_523 |
| VERAPAMIL | KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 4.715405 | 7.946483 | 6.829336 | mammals | actual | clID_523 |
| YOHIMBINE | CACNA1C | Voltage-dependent L-type calcium channel subunit alpha-1C | 5.240925 | 8.516207 | 7.346787 | mammals | predicted | clID_4255 |
| Z160 | Cacna2d1 | Voltage-dependent calcium channel subunit alpha-2/delta-1 | 5.190982 | 9.459622 | 6.276952 | mammals | actual | clID_933 |
| Z160 | CACNA1B | Voltage-dependent N-type calcium channel subunit alpha-1B | 5.190982 | 8.75671 | 6.958607 | mammals | actual | clID_933 |
| Z160 | Cacnb1 | Voltage-dependent L-type calcium channel subunit beta-1 | 5.190982 | 9.644421 | 6.276952 | mammals | actual | clID_933 |
| ZIPRASIDONE HYDROCHLORIDE | KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 5.059504 | 7.946483 | 6.919987 | mammals | actual | clID_6064 |
| ZUCLOMIPHENE | KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 4.635813 | 7.946483 | 6.74 | mammals | predicted | clID_2790 |
Use search function in table to view values for specific interactions
| molName | geneName | protname | sarsScoreMolecule | geneScore | interactionScore | organismType | kindOfValue | molecularCluster |
|---|---|---|---|---|---|---|---|---|
| AMIODARONE | Ebp | 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | 5.1595 | 8.475116 | 7.60206 | mammals | actual | clID_6596 |
| AMIODARONE | ERG2 | C-8 sterol isomerase | 5.1595 | 8.735231 | 7.207608 | fungi | actual | clID_6596 |
| ARTEMINOLIDE | FNTA | Protein farnesyltransferase/geranylgeranyltransferase type-1 subunit alpha | 5.845382 | 8.685947 | 6.443697 | widespread | predicted | clID_2585 |
| ARTEMINOLIDE | RAM1 | Protein farnesyltransferase subunit beta | 5.845382 | 8.675131 | 6.443697 | widespread | predicted | clID_2585 |
| AVASIMIBE | Soat1 | Sterol O-acyltransferase 1 | 5.869584 | 8.158769 | 6.066123 | mammals | actual | clID_977 |
| Daporinad | FNTA | Protein farnesyltransferase/geranylgeranyltransferase type-1 subunit alpha | 5.024153 | 8.685947 | 8.769551 | widespread | actual | clID_4157 |
| Daporinad | RAM1 | Protein farnesyltransferase subunit beta | 5.024153 | 8.675131 | 8.769551 | widespread | actual | clID_4157 |
| EVACETRAPIB | CETP | Cholesteryl ester transfer protein | 6.270594 | 8.45779 | 7.271817 | mammals | actual | clID_635 |
| HALOPERIDOL | Ebp | 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | 4.799189 | 8.475116 | 6.721246 | mammals | actual | clID_1001 |
| HALOPERIDOL | ERG2 | C-8 sterol isomerase | 4.799189 | 8.735231 | 9.30103 | fungi | actual | clID_1001 |
| LONAFARNIB | FNTA | Protein farnesyltransferase/geranylgeranyltransferase type-1 subunit alpha | 8.061413 | 8.685947 | 8.086193 | widespread | actual | clID_150 |
| LONAFARNIB | RAM1 | Protein farnesyltransferase subunit beta | 8.061413 | 8.675131 | 8.246537 | widespread | actual | clID_150 |
| LOVASTATIN | FNTA | Protein farnesyltransferase/geranylgeranyltransferase type-1 subunit alpha | 5.114456 | 8.685947 | 5.954243 | widespread | actual | clID_5420 |
| LOVASTATIN | RAM1 | Protein farnesyltransferase subunit beta | 5.114456 | 8.675131 | 5.954243 | widespread | actual | clID_5420 |
| NAFOXIDINE | Ebp | 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | 5.072686 | 8.475116 | 9.045757 | mammals | predicted | clID_5910 |
| NAFOXIDINE | ERG2 | C-8 sterol isomerase | 5.072686 | 8.735231 | 6.634512 | fungi | predicted | clID_5910 |
| RONIPAMIL | Ebp | 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | 5.409323 | 8.475116 | 8.69897 | mammals | predicted | clID_3021 |
| RONIPAMIL | ERG2 | C-8 sterol isomerase | 5.409323 | 8.735231 | 7.886057 | fungi | predicted | clID_3021 |
| TAMOXIFEN | Ebp | 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | 4.771074 | 8.475116 | 6.262845 | mammals | actual | clID_500 |
| TETRAHYDROQUINOLINE A | CETP | Cholesteryl ester transfer protein | 5.158694 | 8.45779 | 6.581132 | mammals | predicted | clID_4992 |
| TRIFLUOPERAZINE | Ebp | 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | 5.241725 | 8.475116 | 8.09691 | mammals | predicted | clID_4249 |
| TRIFLUOPERAZINE | ERG2 | C-8 sterol isomerase | 5.241725 | 8.735231 | 6.30103 | fungi | predicted | clID_4249 |
| TRIPARANOL | Ebp | 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | 5.277328 | 8.475116 | 6.529304 | mammals | actual | clID_3965 |
| TRIPARANOL | ERG2 | C-8 sterol isomerase | 5.277328 | 8.735231 | 8.69897 | fungi | actual | clID_3965 |
| ZUCLOMIPHENE | Ebp | 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | 4.635813 | 8.475116 | 8.69897 | mammals | predicted | clID_2790 |
| ZUCLOMIPHENE | ERG2 | C-8 sterol isomerase | 4.635813 | 8.735231 | 8.69897 | fungi | predicted | clID_2790 |
Use search function in table to view values for specific interactions
| molName | geneName | protname | sarsScoreMolecule | geneScore | interactionScore | organismType | kindOfValue | molecularCluster |
|---|---|---|---|---|---|---|---|---|
| ANTHRALIN | SIRT5 | NAD-dependent protein deacylase sirtuin-5, mitochondrial | 4.694402 | 8.405938 | 7 | mammals | actual | clID_7432 |
| APABETALONE | BRD4 | Bromodomain-containing protein 4 | 4.93561 | 7.575957 | 6.0655 | mammals | actual | clID_10703 |
| APICIDIN | 1MB.92 | Histone deacetylase | 4.559786 | 10.092065 | 8.329622 | protozoan | actual | clID_1175 |
| APICIDIN | 56k.08 | Histone deacetylase | 4.559786 | 10.372722 | 9 | protozoan | actual | clID_1175 |
| CGM-097 | Mdm2 | E3 ubiquitin-protein ligase Mdm2 | 4.712129 | 8.187967 | 8.036578 | mammals | actual | clID_6248 |
| DINACICLIB | BRD4 | Bromodomain-containing protein 4 | 5.586219 | 7.575957 | 4.721246 | mammals | actual | clID_2117 |
| GW7647 | SIRT5 | NAD-dependent protein deacylase sirtuin-5, mitochondrial | 4.726949 | 8.405938 | 5.102373 | mammals | actual | clID_7685 |
| L-STINGIN | Mdm2 | E3 ubiquitin-protein ligase Mdm2 | 5.205316 | 8.187967 | 7.134956 | mammals | predicted | clID_2926 |
| MANADOSTEROL A | UBE2V1 | Ubiquitin-conjugating enzyme E2 variant 1 | 7.107444 | 13.279684 | 6.772879 | mammals | predicted | clID_2350 |
| MANADOSTEROL B | UBE2V1 | Ubiquitin-conjugating enzyme E2 variant 1 | 6.499874 | 13.279684 | 6.886057 | mammals | predicted | clID_2350 |
Use search function in table to view values for specific interactions
| molName | geneName | protname | sarsScoreMolecule | geneScore | interactionScore | organismType | kindOfValue | molecularCluster |
|---|---|---|---|---|---|---|---|---|
| 3-[18-(2-Carboxy-ethyl)-8,13-bis-(1,2-diacetoxy-ethyl)-3,7,12,17-tetramethyl-22,24-dihydro-porphin-2-yl]-propionic acid;Co complex | pol | Pol polyprotein | 4.836951 | 8.753859 | 5.124939 | virus | predicted | cobalt complex |
| 3-[18-(2-Carboxy-ethyl)-8,13-bis-(1,2-diacetoxy-ethyl)-3,7,12,17-tetramethyl-22,24-dihydro-porphin-2-yl]-propionic acid;Mn complex | humanRhinovirusProtease | humanRhinovirusProtease | 4.742592 | 9.1456 | 6.03786 | virus | predicted | manganese complex |
| 3-[18-(2-Carboxy-ethyl)-8,13-bis-(1,2-diacetoxy-ethyl)-3,7,12,17-tetramethyl-22,24-dihydro-porphin-2-yl]-propionic acid;Mn complex | pol | Pol polyprotein | 4.742592 | 8.753859 | 6.82284 | virus | predicted | manganese complex |
| AMENTOFLAVONE | BACE1 | Beta-secretase 1 | 5.080922 | 7.470756 | 5.812479 | mammals | actual | clID_459 |
| AMPRENAVIR | pol | Pol polyprotein | 4.853395 | 8.753859 | 8.823247 | virus | actual | clID_1897 |
| AMPRENAVIR | humanRhinovirusProtease | humanRhinovirusProtease | 4.853395 | 9.145042 | 9.047128 | virus | actual | clID_1897 |
| AZD-3839 | BACE1 | Beta-secretase 1 | 4.850839 | 7.470756 | 7.867264 | mammals | predicted | clID_3750 |
| CAPTOPRIL | REN | Renin | 5.558465 | 8.255979 | 8.769551 | mammals | actual | clID_2251 |
| CHLOROQUINE | BACE1 | Beta-secretase 1 | 5.684392 | 7.470756 | 5.017033 | mammals | actual | clID_1740 |
| COLOSSOLACTONE VII | humanRhinovirusProtease | humanRhinovirusProtease | 5.335946 | 9.145042 | 4.607346 | virus | predicted | clID_2585 |
| CURCUMIN | BACE1 | Beta-secretase 1 | 5.319754 | 7.470756 | 5.259925 | mammals | actual | clID_3628 |
| DARUNAVIR | pol | Pol polyprotein | 4.79897 | 8.753859 | 8.574985 | virus | actual | clID_1897 |
| DARUNAVIR | humanRhinovirusProtease | humanRhinovirusProtease | 4.79897 | 9.145042 | 9.917553 | virus | actual | clID_1897 |
| DIECKOL | BACE1 | Beta-secretase 1 | 5.619789 | 7.470756 | 5.739758 | mammals | actual | clID_1978 |
| ECKOL | BACE1 | Beta-secretase 1 | 5.1 | 7.470756 | 4.885313 | mammals | actual | clID_1978 |
| EFAVIRENZ | pol | Pol polyprotein | 4.674037 | 8.753859 | 6.11912 | virus | actual | clID_5637 |
| ELLAGIC ACID | BACE1 | Beta-secretase 1 | 7.647277 | 7.470756 | 5.408935 | mammals | actual | clID_459 |
| ESCIN IVC | humanRhinovirusProtease | humanRhinovirusProtease | 5.183109 | 9.145042 | 4.734968 | virus | predicted | clID_2585 |
| GINKGETIN | BACE1 | Beta-secretase 1 | 4.519993 | 7.470756 | 5.378824 | mammals | actual | clID_459 |
| HALOPERIDOL | humanRhinovirusProtease | humanRhinovirusProtease | 4.799189 | 9.1456 | 7 | virus | actual | clID_1001 |
| HALOPERIDOL | pol | Pol polyprotein | 4.799189 | 8.753859 | 7 | virus | actual | clID_1001 |
| INDINAVIR | humanRhinovirusProtease | humanRhinovirusProtease | 4.621006 | 9.1456 | 8.481486 | virus | actual | clID_939 |
| INDINAVIR | pol | Pol polyprotein | 4.621006 | 8.753859 | 8.430111 | virus | actual | clID_939 |
| INDINAVIR | humanRhinovirusProtease | humanRhinovirusProtease | 4.621006 | 9.145042 | 8.477442 | virus | actual | clID_939 |
| INDINAVIR | HIV-1 | HIV-1 protease | 4.621006 | 10.09572 | 9.420216 | virus | actual | clID_939 |
| INDINAVIR SULFATE | pol | Pol polyprotein | 7.089798 | 8.753859 | 8.344123 | virus | predicted | clID_2034 |
| INDOMETHACIN | APH1A | Gamma-secretase subunit APH-1A | 4.738732 | 8.149194 | 5.052178 | mammals | actual | clID_7591 |
| INDOMETHACIN | APH1B | Gamma-secretase subunit APH-1B | 4.738732 | 8.149194 | 5.052178 | mammals | actual | clID_7591 |
| INDOMETHACIN | NCSTN | Nicastrin | 4.738732 | 8.149194 | 5.052178 | mammals | actual | clID_7591 |
| INDOMETHACIN | PSENEN | Gamma-secretase subunit PEN-2 | 4.738732 | 8.14863 | 5.052178 | mammals | actual | clID_7591 |
| INDOMETHACIN | PSEN1 | Presenilin-1 | 4.738732 | 8.167203 | 5.052178 | mammals | actual | clID_7591 |
| INDOMETHACIN | PSEN2 | Presenilin-2 | 4.738732 | 8.148414 | 5.052178 | mammals | actual | clID_7591 |
| KNI-1038 | pol | Pol polyprotein | 4.569109 | 8.753859 | 4.768861 | virus | predicted | clID_939 |
| KNI-1039 | pol | Pol polyprotein | 4.870674 | 8.753859 | 5.08715 | virus | predicted | clID_939 |
| KURARINONE | BACE1 | Beta-secretase 1 | 4.63155 | 7.470756 | 5.189381 | mammals | predicted | clID_3834 |
| KUSHENOL A | BACE1 | Beta-secretase 1 | 4.662197 | 7.470756 | 5.354024 | mammals | predicted | clID_3834 |
| LOPINAVIR | pol | Pol polyprotein | 5.518043 | 8.753859 | 8.80835 | virus | actual | clID_2433 |
| LOPINAVIR | humanRhinovirusProtease | humanRhinovirusProtease | 5.518043 | 9.145042 | 9.171299 | virus | actual | clID_2433 |
| NELFINAVIR | pol | Pol polyprotein | 6 | 8.753859 | 8.158252 | virus | actual | clID_7128 |
| NELFINAVIR | humanRhinovirusProtease | humanRhinovirusProtease | 6 | 9.145042 | 7.937857 | virus | actual | clID_7128 |
| NELFINAVIR MESYLATE | pol | Pol polyprotein | 5.305206 | 8.753859 | 8.69897 | virus | actual | clID_7128 |
| PENTAGALLOYL GLUCOSE | NS3 | Genome polyprotein | 5.619692 | 8.852282 | 5.79588 | virus | predicted | clID_1684 |
| PENTAGALLOYL GLUCOSE | BACE1 | Beta-secretase 1 | 5.619692 | 7.470756 | 5.357347 | mammals | predicted | clID_1684 |
| PENTAGALLOYL GLUCOSE | POLG | Genome polyprotein [Cleaved into: Core protein p21 | 5.619692 | 8.888157 | 7.479304 | virus | predicted | clID_1684 |
| PEPSTATIN | SAP3 | Candidapepsin-3 | 5.712467 | 11.030201 | 5.1 | fungi | actual | clID_769 |
| PEPSTATIN | SAP4 | Candidapepsin-4 | 5.712467 | 11.725964 | 5.1 | fungi | actual | clID_769 |
| PEPSTATIN | SAP5 | Candidapepsin-5 | 5.712467 | 11.725964 | 5.1 | fungi | actual | clID_769 |
| PEPSTATIN | SAP6 | Candidapepsin-6 | 5.712467 | 11.725964 | 5.1 | fungi | actual | clID_769 |
| PEPSTATIN | SAP8 | Candidapepsin-8 | 5.712467 | 11.725964 | 5.1 | fungi | actual | clID_769 |
| PEPSTATIN | SAP9 | Candidapepsin-9 | 5.712467 | 11.725964 | 5.1 | fungi | actual | clID_769 |
| PEPSTATIN | POLG | Genome polyprotein [Cleaved into: Core protein p21 | 5.712467 | 8.888157 | 6.30103 | virus | actual | clID_769 |
| PEPSTATIN | pol | Pol polyprotein | 5.712467 | 8.753859 | 5.681255 | virus | actual | clID_769 |
| PEPSTATIN | humanRhinovirusProtease | humanRhinovirusProtease | 5.712467 | 9.145042 | 6.711111 | virus | actual | clID_769 |
| PEPSTATIN | REN | Renin | 5.712467 | 8.255979 | 5.310983 | mammals | actual | clID_769 |
| PHLOROFUCOFUROECKOL A | BACE1 | Beta-secretase 1 | 5.1 | 7.470756 | 5.778839 | mammals | actual | clID_4622 |
| PODOCARPUSFLAVONE B | BACE1 | Beta-secretase 1 | 6.467186 | 7.470756 | 5.375718 | mammals | predicted | clID_459 |
| PUNICALAGIN | BACE1 | Beta-secretase 1 | 5.945038 | 7.470756 | 6.387216 | mammals | predicted | clID_1684 |
| QUERCETIN | BACE1 | Beta-secretase 1 | 4.893839 | 7.470756 | 5.10523 | mammals | actual | clID_459 |
| RITONAVIR | SAP3 | Candidapepsin-3 | 4.840613 | 11.030201 | 5.1 | fungi | actual | clID_3116 |
| RITONAVIR | pol | Pol polyprotein | 4.840613 | 8.753859 | 8.058822 | virus | actual | clID_3116 |
| RITONAVIR | humanRhinovirusProtease | humanRhinovirusProtease | 4.840613 | 9.145042 | 7.943779 | virus | actual | clID_3116 |
| RUPINTRIVIR | humanRhinovirusProtease | humanRhinovirusProtease | 4.588063 | 9.1456 | 8.213806 | virus | predicted | clID_4807 |
| RWJ-54003 | F2R | Proteinase-activated receptor 1 | 6.112733 | 7.866621 | 6.86815 | mammals | predicted | clID_4747 |
| RWJ-58259 | F2R | Proteinase-activated receptor 1 | 4.553302 | 7.866621 | 7.734767 | mammals | predicted | clID_4747 |
| SOPHORAFLAVANONE G | BACE1 | Beta-secretase 1 | 5.069695 | 7.470756 | 5.361923 | mammals | predicted | clID_3834 |
| TAMOXIFEN | PSMB1 | Proteasome subunit beta type-1 | 4.771074 | 8.205737 | 5.753328 | mammals | actual | clID_500 |
| TAMOXIFEN | Psmb5 | Proteasome subunit beta type-5 | 4.771074 | 8.338184 | 9 | widespread | actual | clID_500 |
| TERLAKIREN | REN | Renin | 4.891617 | 8.255979 | 9.419445 | mammals | predicted | clID_3261 |
| THONNINGIANIN B | NS3 | Genome polyprotein | 5.040265 | 8.852282 | 6.09691 | virus | predicted | clID_1684 |
| TOPSENTINOL K TRISULFATE | BACE1 | Beta-secretase 1 | 4.793746 | 7.470756 | 5.920819 | mammals | predicted | clID_3538 |
| TRIFLUOPERAZINE | NS3 | Genome polyprotein | 5.241725 | 8.852282 | 6.154902 | virus | predicted | clID_4249 |
| VORAPAXAR | F2R | Proteinase-activated receptor 1 | 4.846237 | 7.866621 | 7.886995 | mammals | actual | clID_1859 |
Use search function in table to view values for specific interactions
| molName | geneName | protname | sarsScoreMolecule | geneScore | interactionScore | organismType | kindOfValue | molecularCluster |
|---|---|---|---|---|---|---|---|---|
| AMIODARONE | Ebp | 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | 5.1595 | 8.475116 | 7.60206 | mammals | actual | clID_6596 |
| AMIODARONE | ERG2 | C-8 sterol isomerase | 5.1595 | 8.735231 | 7.207608 | fungi | actual | clID_6596 |
| AMIODARONE | LMNA | Prelamin-A/C [Cleaved into: Lamin-A/C | 5.1595 | 7.05737 | 9.045757 | mammals | actual | clID_6596 |
| AMIODARONE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.1595 | 8.320941 | 7.800148 | mammals | actual | clID_6596 |
| ASTEMIZOLE | HTR2A | 5-hydroxytryptamine receptor 2A | 5.958607 | 7.818593 | 7.976363 | mammals | actual | clID_1081 |
| ASTEMIZOLE | Htr2b | 5-hydroxytryptamine receptor 2B | 5.958607 | 7.37631 | 8.023255 | mammals | actual | clID_1081 |
| ASTEMIZOLE | Glra1 | Glycine receptor subunit alpha-1 | 5.958607 | 5.677586 | 7.188375 | mammals | actual | clID_1081 |
| ASTEMIZOLE | HRH1 | Histamine H1 receptor | 5.958607 | 7.280988 | 8.034474 | mammals | actual | clID_1081 |
| ASTEMIZOLE | KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 5.958607 | 7.946483 | 8.065956 | mammals | actual | clID_1081 |
| ASTEMIZOLE | TMEM97 | Sigma intracellular receptor 2 | 5.958607 | 8.169457 | 7.021137 | mammals | actual | clID_1081 |
| ASTEMIZOLE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.958607 | 7.5973 | 9 | mammals | actual | clID_1081 |
| CHLOROQUINE | CYP2B6 | Cytochrome P450 2B6 | 5.684392 | 6.293727 | 9 | mammals | actual | clID_1740 |
| CHLOROQUINE | Abcc2 | Canalicular multispecific organic anion transporter 1 | 5.684392 | 7.3938 | 9 | mammals | actual | clID_1740 |
| CHLOROQUINE | Q9N6S8 | Cysteine proteinase falcipain 2a | 5.684392 | 5.97719 | 7.69897 | protozoan | actual | clID_1740 |
| CHLOROQUINE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.684392 | 8.320941 | 7.100015 | mammals | actual | clID_1740 |
| CHLOROQUINE | TMEM97 | Sigma intracellular receptor 2 | 5.684392 | 8.169457 | 6.299998 | mammals | actual | clID_1740 |
| CHLORPROMAZINE | HTR2A | 5-hydroxytryptamine receptor 2A | 6.908068 | 7.818593 | 8.500919 | mammals | predicted | clID_302 |
| CHLORPROMAZINE | Htr2b | 5-hydroxytryptamine receptor 2B | 6.908068 | 7.37631 | 8.199774 | mammals | predicted | clID_302 |
| CHLORPROMAZINE | Htr2c | 5-hydroxytryptamine receptor 2C | 6.908068 | 7.439004 | 8.298354 | mammals | predicted | clID_302 |
| CHLORPROMAZINE | Htr6 | 5-hydroxytryptamine receptor 6 | 6.908068 | 6.485523 | 7.467133 | mammals | predicted | clID_302 |
| CHLORPROMAZINE | Htr7 | 5-hydroxytryptamine receptor 7 | 6.908068 | 6.987997 | 7.502758 | mammals | predicted | clID_302 |
| CHLORPROMAZINE | HRH1 | Histamine H1 receptor | 6.908068 | 7.280988 | 8.014076 | mammals | predicted | clID_302 |
| CHLORPROMAZINE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 6.908068 | 8.320941 | 6.534681 | mammals | predicted | clID_302 |
| CINANSERIN | HTR2A | 5-hydroxytryptamine receptor 2A | 5.30103 | 7.818593 | 8.285963 | mammals | actual | clID_3770 |
| CINANSERIN | Htr2b | 5-hydroxytryptamine receptor 2B | 5.30103 | 7.37631 | 8.473198 | mammals | actual | clID_3770 |
| CINANSERIN | Htr2c | 5-hydroxytryptamine receptor 2C | 5.30103 | 7.439004 | 8.342811 | mammals | actual | clID_3770 |
| CINANSERIN | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.30103 | 8.320941 | 6.74 | mammals | actual | clID_3770 |
| CLEMASTINE | HTR2A | 5-hydroxytryptamine receptor 2A | 5.75225 | 7.818593 | 7.765626 | mammals | actual | clID_1525 |
| CLEMASTINE | Htr2b | 5-hydroxytryptamine receptor 2B | 5.75225 | 7.37631 | 7.540985 | mammals | actual | clID_1525 |
| CLEMASTINE | Htr2c | 5-hydroxytryptamine receptor 2C | 5.75225 | 7.439004 | 7.144181 | mammals | actual | clID_1525 |
| CLEMASTINE | Htr6 | 5-hydroxytryptamine receptor 6 | 5.75225 | 6.485523 | 7.278603 | mammals | actual | clID_1525 |
| CLEMASTINE | Drd3 | D(3) dopamine receptor | 5.75225 | 8.166222 | 8.369113 | mammals | actual | clID_1525 |
| CLEMASTINE | HRH1 | Histamine H1 receptor | 5.75225 | 7.280988 | 9.842761 | mammals | actual | clID_1525 |
| CLEMASTINE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.75225 | 8.320941 | 7.774131 | mammals | actual | clID_1525 |
| CLEMASTINE | TMEM97 | Sigma intracellular receptor 2 | 5.75225 | 8.169457 | 7.59998 | mammals | actual | clID_1525 |
| CLOMIPRAMINE | HTR1A | 5-hydroxytryptamine receptor 1A | 4.522627 | 7.930581 | 7.634363 | mammals | actual | clID_1362 |
| CLOMIPRAMINE | HTR1B | 5-hydroxytryptamine receptor 1B | 4.522627 | 6.726785 | 7.634363 | mammals | actual | clID_1362 |
| CLOMIPRAMINE | HTR1D | 5-hydroxytryptamine receptor 1D | 4.522627 | 6.702207 | 7.634363 | mammals | actual | clID_1362 |
| CLOMIPRAMINE | HTR1F | 5-hydroxytryptamine receptor 1F | 4.522627 | 6.028175 | 7.634363 | mammals | actual | clID_1362 |
| CLOMIPRAMINE | HTR2A | 5-hydroxytryptamine receptor 2A | 4.522627 | 7.818593 | 8.17644 | mammals | actual | clID_1362 |
| CLOMIPRAMINE | Htr2b | 5-hydroxytryptamine receptor 2B | 4.522627 | 7.37631 | 8.099839 | mammals | actual | clID_1362 |
| CLOMIPRAMINE | Htr2c | 5-hydroxytryptamine receptor 2C | 4.522627 | 7.439004 | 8.193181 | mammals | actual | clID_1362 |
| CLOMIPRAMINE | HTR3A | 5-hydroxytryptamine receptor 3A | 4.522627 | 6.129818 | 8.30103 | mammals | actual | clID_1362 |
| CLOMIPRAMINE | HTR3B | 5-hydroxytryptamine receptor 3B | 4.522627 | 6.020968 | 8.30103 | mammals | actual | clID_1362 |
| CLOMIPRAMINE | HTR4 | 5-hydroxytryptamine receptor 4 | 4.522627 | 5.959168 | 8.30103 | mammals | actual | clID_1362 |
| CLOMIPRAMINE | Htr5a | 5-hydroxytryptamine receptor 5A | 4.522627 | 5.845588 | 8.30103 | mammals | actual | clID_1362 |
| CLOMIPRAMINE | Htr5b | 5-hydroxytryptamine receptor 5B | 4.522627 | 4.66724 | 8.30103 | mammals | actual | clID_1362 |
| CLOMIPRAMINE | Htr6 | 5-hydroxytryptamine receptor 6 | 4.522627 | 6.485523 | 8.042302 | mammals | actual | clID_1362 |
| CLOMIPRAMINE | Htr7 | 5-hydroxytryptamine receptor 7 | 4.522627 | 6.987997 | 8.30103 | mammals | actual | clID_1362 |
| CLOMIPRAMINE | HRH1 | Histamine H1 receptor | 4.522627 | 7.280988 | 8.259043 | mammals | actual | clID_1362 |
| CLOMIPRAMINE | PGAM | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase | 4.522627 | 5.45283 | 7.320572 | protozoan | actual | clID_1362 |
| CLOMIPRAMINE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 4.522627 | 8.320941 | 6.054213 | mammals | actual | clID_1362 |
| DEXTROMETHORPHAN | SLC6A4 | Sodium-dependent serotonin transporter | 5.527467 | 9.398949 | 8.704334 | mammals | actual | clID_2960 |
| DEXTROMETHORPHAN | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.527467 | 8.320941 | 6.55152 | mammals | actual | clID_2960 |
| DEXTROMETHORPHAN | TSHR | Thyrotropin receptor | 5.527467 | 6.41967 | 8.200659 | mammals | actual | clID_2960 |
| DOLUTEGRAVIR | Q76353 | Integrase | 4.584705 | 6.282515 | 7.868064 | virus | actual | clID_687 |
| DOLUTEGRAVIR | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 4.584705 | 8.320941 | 6.906141 | mammals | actual | clID_687 |
| DOLUTEGRAVIR | TMEM97 | Sigma intracellular receptor 2 | 4.584705 | 8.169457 | 7.49485 | mammals | actual | clID_687 |
| E-FLUPENTIXOL | HRH2 | Histamine H2 receptor | 5.538428 | 6.157915 | 8.241845 | mammals | predicted | clID_267 |
| E-FLUPENTIXOL | NFKB1 | Nuclear factor NF-kappa-B p105 subunit | 5.538428 | 6.468499 | 7.649752 | mammals | predicted | clID_267 |
| E-FLUPENTIXOL | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.538428 | 8.320941 | 7.830032 | mammals | predicted | clID_267 |
| FLUNARIZINE | HRH1 | Histamine H1 receptor | 4.518593 | 7.280988 | 7.273035 | mammals | predicted | clID_355 |
| FLUNARIZINE | LMNA | Prelamin-A/C [Cleaved into: Lamin-A/C | 4.518593 | 7.05737 | 8.49485 | mammals | predicted | clID_355 |
| FLUNARIZINE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 4.518593 | 8.320941 | 7.80103 | mammals | predicted | clID_355 |
| FLUPENTIXOL | Drd1 | D(1A) dopamine receptor | 4.760268 | 7.436039 | 8.290481 | mammals | actual | clID_267 |
| FLUPENTIXOL | Drd3 | D(3) dopamine receptor | 4.760268 | 8.166222 | 8.522879 | mammals | actual | clID_267 |
| FLUPENTIXOL | DRD4 | D(4) dopamine receptor | 4.760268 | 7.451626 | 8.522879 | mammals | actual | clID_267 |
| FLUPENTIXOL | DRD5 | D(1B) dopamine receptor | 4.760268 | 7.117191 | 8.522879 | mammals | actual | clID_267 |
| FLUPENTIXOL | HRH2 | Histamine H2 receptor | 4.760268 | 6.157915 | 9.065502 | mammals | actual | clID_267 |
| FLUPENTIXOL | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 4.760268 | 8.320941 | 8.649946 | mammals | actual | clID_267 |
| HALOPERIDOL | HTR2A | 5-hydroxytryptamine receptor 2A | 4.799189 | 7.818593 | 7.182611 | mammals | actual | clID_1001 |
| HALOPERIDOL | Htr2b | 5-hydroxytryptamine receptor 2B | 4.799189 | 7.37631 | 7.226378 | mammals | actual | clID_1001 |
| HALOPERIDOL | DRD2 | D(2) dopamine receptor | 4.799189 | 8.280379 | 8.339276 | mammals | actual | clID_1001 |
| HALOPERIDOL | Drd3 | D(3) dopamine receptor | 4.799189 | 8.166222 | 8.098005 | mammals | actual | clID_1001 |
| HALOPERIDOL | DRD4 | D(4) dopamine receptor | 4.799189 | 7.451626 | 8.10952 | mammals | actual | clID_1001 |
| HALOPERIDOL | DRD5 | D(1B) dopamine receptor | 4.799189 | 7.117191 | 8.044783 | mammals | actual | clID_1001 |
| HALOPERIDOL | Ebp | 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | 4.799189 | 8.475116 | 6.721246 | mammals | actual | clID_1001 |
| HALOPERIDOL | ERG2 | C-8 sterol isomerase | 4.799189 | 8.735231 | 9.30103 | fungi | actual | clID_1001 |
| HALOPERIDOL | KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 4.799189 | 7.946483 | 7.180601 | mammals | actual | clID_1001 |
| HALOPERIDOL | Abcc2 | Canalicular multispecific organic anion transporter 1 | 4.799189 | 7.3938 | 9 | mammals | actual | clID_1001 |
| HALOPERIDOL | humanRhinovirusProtease | humanRhinovirusProtease | 4.799189 | 9.1456 | 7 | virus | actual | clID_1001 |
| HALOPERIDOL | pol | Pol polyprotein | 4.799189 | 8.753859 | 7 | virus | actual | clID_1001 |
| HALOPERIDOL | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 4.799189 | 8.320941 | 8.291093 | mammals | actual | clID_1001 |
| HALOPERIDOL | TMEM97 | Sigma intracellular receptor 2 | 4.799189 | 8.169457 | 7.189326 | mammals | actual | clID_1001 |
| HALOPERIDOL | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.799189 | 7.484017 | 9 | mammals | actual | clID_1001 |
| HALOPERIDOL | ZNF664 | Zinc finger protein 664 | 4.799189 | 7.4195 | 8.180456 | mammals | actual | clID_1001 |
| HYDROXYCHLOROQUINE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.92742 | 8.320941 | 6.900009 | mammals | actual | clID_1740 |
| HYDROXYCHLOROQUINE | TMEM97 | Sigma intracellular receptor 2 | 5.92742 | 8.169457 | 6 | mammals | actual | clID_1740 |
| LOPERAMIDE | KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 5.03292 | 7.946483 | 7.481486 | mammals | actual | clID_6355 |
| LOPERAMIDE | Oprm1 | Mu-type opioid receptor | 5.03292 | 7.860685 | 8.876082 | mammals | actual | clID_6355 |
| LOPERAMIDE | Scn1a | Sodium channel protein type 1 subunit alpha | 5.03292 | 7.139546 | 7.784318 | mammals | actual | clID_6355 |
| LOPERAMIDE | Scn2a | Sodium channel protein type 2 subunit alpha | 5.03292 | 7.567507 | 7.784318 | mammals | actual | clID_6355 |
| LOPERAMIDE | Scn3a | Sodium channel protein type 3 subunit alpha | 5.03292 | 7.20794 | 7.784318 | mammals | actual | clID_6355 |
| LOPERAMIDE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.03292 | 8.320941 | 6.390218 | mammals | actual | clID_6355 |
| NAFOXIDINE | Ebp | 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | 5.072686 | 8.475116 | 9.045757 | mammals | predicted | clID_5910 |
| NAFOXIDINE | ERG2 | C-8 sterol isomerase | 5.072686 | 8.735231 | 6.634512 | fungi | predicted | clID_5910 |
| NAFOXIDINE | ESR1 | Estrogen receptor | 5.072686 | 8.667211 | 7.388277 | mammals | predicted | clID_5910 |
| NAFOXIDINE | Esr2 | Estrogen receptor beta | 5.072686 | 8.418744 | 7.388277 | mammals | predicted | clID_5910 |
| NAFOXIDINE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.072686 | 8.320941 | 7.522879 | mammals | predicted | clID_5910 |
| RIMCAZOLE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.278436 | 8.320941 | 6.111734 | mammals | actual | clID_4588 |
| RONIPAMIL | Ebp | 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | 5.409323 | 8.475116 | 8.69897 | mammals | predicted | clID_3021 |
| RONIPAMIL | ERG2 | C-8 sterol isomerase | 5.409323 | 8.735231 | 7.886057 | fungi | predicted | clID_3021 |
| RONIPAMIL | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.409323 | 8.320941 | 9 | mammals | predicted | clID_3021 |
| SIRAMESINE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 9 | 8.320941 | 7.681082 | mammals | actual | clID_91 |
| SIRAMESINE | TMEM97 | Sigma intracellular receptor 2 | 9 | 8.169457 | 9.920819 | mammals | actual | clID_91 |
| TAMOXIFEN | Ebp | 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | 4.771074 | 8.475116 | 6.262845 | mammals | actual | clID_500 |
| TAMOXIFEN | EBPL | Emopamil-binding protein-like | 4.771074 | 8.870553 | 8.552842 | mammals | actual | clID_500 |
| TAMOXIFEN | Esrra | Steroid hormone receptor ERR1 | 4.771074 | 6.067221 | 6.69897 | mammals | actual | clID_500 |
| TAMOXIFEN | ESRRG | Estrogen-related receptor gamma | 4.771074 | 7.329231 | 7.20621 | mammals | actual | clID_500 |
| TAMOXIFEN | ESR1 | Estrogen receptor | 4.771074 | 8.667211 | 6.081137 | mammals | actual | clID_500 |
| TAMOXIFEN | Esr2 | Estrogen receptor beta | 4.771074 | 8.418744 | 6.0265 | mammals | actual | clID_500 |
| TAMOXIFEN | PSMB2 | Proteasome subunit beta type-2 | 4.771074 | 8.126604 | 9 | mammals | actual | clID_500 |
| TAMOXIFEN | Psmb5 | Proteasome subunit beta type-5 | 4.771074 | 8.338184 | 9 | widespread | actual | clID_500 |
| TAMOXIFEN | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 4.771074 | 8.320941 | 7.731165 | mammals | actual | clID_500 |
| TERCONAZOLE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 4.890817 | 8.320941 | 6.481861 | mammals | actual | clID_6688 |
| THIORIDAZINE | HTR2A | 5-hydroxytryptamine receptor 2A | 5.174574 | 7.818593 | 7.770472 | mammals | actual | clID_4843 |
| THIORIDAZINE | Htr2b | 5-hydroxytryptamine receptor 2B | 5.174574 | 7.37631 | 7.359416 | mammals | actual | clID_4843 |
| THIORIDAZINE | Htr2c | 5-hydroxytryptamine receptor 2C | 5.174574 | 7.439004 | 7.486819 | mammals | actual | clID_4843 |
| THIORIDAZINE | Htr6 | 5-hydroxytryptamine receptor 6 | 5.174574 | 6.485523 | 7.315114 | mammals | actual | clID_4843 |
| THIORIDAZINE | Htr7 | 5-hydroxytryptamine receptor 7 | 5.174574 | 6.987997 | 7.154902 | mammals | actual | clID_4843 |
| THIORIDAZINE | HRH1 | Histamine H1 receptor | 5.174574 | 7.280988 | 7.407378 | mammals | actual | clID_4843 |
| THIORIDAZINE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.174574 | 8.320941 | 6.460114 | mammals | actual | clID_4843 |
| TRIFLUOPERAZINE | HTR2A | 5-hydroxytryptamine receptor 2A | 5.241725 | 7.818593 | 8.398967 | mammals | predicted | clID_4249 |
| TRIFLUOPERAZINE | Htr2b | 5-hydroxytryptamine receptor 2B | 5.241725 | 7.37631 | 8.632856 | mammals | predicted | clID_4249 |
| TRIFLUOPERAZINE | Htr2c | 5-hydroxytryptamine receptor 2C | 5.241725 | 7.439004 | 8.257419 | mammals | predicted | clID_4249 |
| TRIFLUOPERAZINE | Ebp | 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | 5.241725 | 8.475116 | 8.09691 | mammals | predicted | clID_4249 |
| TRIFLUOPERAZINE | EBPL | Emopamil-binding protein-like | 5.241725 | 8.870553 | 8.408935 | mammals | predicted | clID_4249 |
| TRIFLUOPERAZINE | ERG2 | C-8 sterol isomerase | 5.241725 | 8.735231 | 6.30103 | fungi | predicted | clID_4249 |
| TRIFLUOPERAZINE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.241725 | 8.320941 | 7.823909 | mammals | predicted | clID_4249 |
| TRIPARANOL | Ebp | 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | 5.277328 | 8.475116 | 6.529304 | mammals | actual | clID_3965 |
| TRIPARANOL | ERG2 | C-8 sterol isomerase | 5.277328 | 8.735231 | 8.69897 | fungi | actual | clID_3965 |
| TRIPARANOL | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.277328 | 8.320941 | 8.09691 | mammals | actual | clID_3965 |
| VANOXERINE | HRH2 | Histamine H2 receptor | 6.527334 | 6.157915 | 8.030118 | mammals | predicted | clID_462 |
| VANOXERINE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 6.527334 | 8.320941 | 7.418757 | mammals | predicted | clID_462 |
| ZUCLOMIPHENE | Ebp | 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | 4.635813 | 8.475116 | 8.69897 | mammals | predicted | clID_2790 |
| ZUCLOMIPHENE | ERG2 | C-8 sterol isomerase | 4.635813 | 8.735231 | 8.69897 | fungi | predicted | clID_2790 |
| ZUCLOMIPHENE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 4.635813 | 8.320941 | 8.30103 | mammals | predicted | clID_2790 |
Use search function in table to view values for specific interactions
| molName | geneName | protname | sarsScoreMolecule | geneScore | interactionScore | organismType | kindOfValue | molecularCluster |
|---|---|---|---|---|---|---|---|---|
| 4'-HYDROXYTAMOXIFEN | ESR1 | Estrogen receptor | 4.516256 | 8.667211 | 6.120304 | mammals | predicted | clID_500 |
| 4'-HYDROXYTAMOXIFEN | Esr2 | Estrogen receptor beta | 4.516256 | 8.418744 | 6.120304 | mammals | predicted | clID_500 |
| 9-AMINOCAMPTOTHECIN | RORC | Nuclear receptor ROR-gamma | 5.610799 | 7.548863 | 7.074493 | mammals | actual | clID_1835 |
| ACOLBIFENE | ESR1 | Estrogen receptor | 4.500612 | 8.667211 | 9.825667 | mammals | predicted | clID_6520 |
| ACOLBIFENE | Esr2 | Estrogen receptor beta | 4.500612 | 8.418744 | 10.121021 | mammals | predicted | clID_6520 |
| AFIMOXIFENE | ESR1 | Estrogen receptor | 6.454651 | 8.667211 | 7.240626 | mammals | predicted | clID_500 |
| AFIMOXIFENE | Esr2 | Estrogen receptor beta | 6.454651 | 8.418744 | 8.080151 | mammals | predicted | clID_500 |
| AFIMOXIFENE | PGR | Progesterone receptor | 6.454651 | 7.946326 | 9.100015 | mammals | predicted | clID_500 |
| ATALUREN | RORC | Nuclear receptor ROR-gamma | 4.710967 | 7.548863 | 6.150028 | mammals | actual | clID_9317 |
| BAZEDOXIFENE | ESR1 | Estrogen receptor | 5.463442 | 8.667211 | 8.148237 | mammals | actual | clID_2729 |
| BAZEDOXIFENE | Esr2 | Estrogen receptor beta | 5.463442 | 8.418744 | 7.745399 | mammals | actual | clID_2729 |
| BIS(DIMETHYLAMINETHIOCARBONYL)MONOSULFIDE | NR3C1 | Glucocorticoid receptor | 4.946601 | 8.072674 | 7.549751 | mammals | actual | clID_7469 |
| BRILANESTRANT | ESR1 | Estrogen receptor | 4.738011 | 8.667211 | 7.693773 | mammals | actual | clID_5201 |
| BRILANESTRANT | Esr2 | Estrogen receptor beta | 4.738011 | 8.418744 | 8.055517 | mammals | actual | clID_5201 |
| CAFFEINE | NR3C1 | Glucocorticoid receptor | 4.703587 | 8.072674 | 7.74958 | mammals | actual | clID_6550 |
| CALYCULIN A | RORC | Nuclear receptor ROR-gamma | 5.030222 | 7.548863 | 9.851865 | mammals | predicted | clID_1087 |
| CAMPTOTHECIN | RORC | Nuclear receptor ROR-gamma | 5.654142 | 7.548863 | 6.393808 | mammals | actual | clID_1835 |
| CHLORHEXIDINE | NR3C1 | Glucocorticoid receptor | 5.855721 | 8.072674 | 6.450016 | mammals | actual | clID_1295 |
| CORT 108297 | NR3C1 | Glucocorticoid receptor | 5.298461 | 8.072674 | 8.460247 | mammals | predicted | clID_3793 |
| CYCLOHEXIMIDE | RORC | Nuclear receptor ROR-gamma | 6.421559 | 7.548863 | 7.005708 | mammals | actual | clID_525 |
| DEMECLOCYCLINE HYDROCHLORIDE | Esr2 | Estrogen receptor beta | 4.554055 | 8.418744 | 7.21169 | mammals | actual | clID_1527 |
| DIETHYLSTILBESTROL | ESR1 | Estrogen receptor | 4.646201 | 8.667211 | 8.978894 | mammals | actual | clID_7593 |
| DIETHYLSTILBESTROL | Esr2 | Estrogen receptor beta | 4.646201 | 8.418744 | 8.994429 | mammals | actual | clID_7593 |
| DIGITOXIN | ESR1 | Estrogen receptor | 6.638272 | 8.667211 | 6.300006 | mammals | actual | clID_364 |
| DIGITOXIN | NR3C1 | Glucocorticoid receptor | 6.638272 | 8.072674 | 6.450016 | mammals | actual | clID_364 |
| DIGOXIN | ESR1 | Estrogen receptor | 6.721246 | 8.667211 | 6.216672 | mammals | actual | clID_364 |
| DIGOXIN | NR3C1 | Glucocorticoid receptor | 6.721246 | 8.072674 | 6.125016 | mammals | actual | clID_364 |
| DIPHENYLENEIODONIUM | RORC | Nuclear receptor ROR-gamma | 5.095581 | 7.548863 | 6 | mammals | actual | clID_5657 |
| FURAZOLIDONE | RORC | Nuclear receptor ROR-gamma | 4.855182 | 7.548863 | 6.299989 | mammals | actual | clID_8219 |
| HEXACHLOROPHENE | Esr2 | Estrogen receptor beta | 6.099502 | 8.418744 | 7.007579 | mammals | actual | clID_842 |
| HYDROCORTISONE | NR3C1 | Glucocorticoid receptor | 5.050958 | 8.072674 | 6.312103 | mammals | actual | clID_1633 |
| LASOFOXIFENE | ESR1 | Estrogen receptor | 5.074184 | 8.667211 | 7.741065 | mammals | actual | clID_5892 |
| LASOFOXIFENE | Esr2 | Estrogen receptor beta | 5.074184 | 8.418744 | 7.637865 | mammals | actual | clID_5892 |
| LEVORMELOXIFENE | ESR1 | Estrogen receptor | 4.644769 | 8.667211 | 7.136677 | mammals | predicted | clID_7365 |
| LEVORMELOXIFENE | Esr2 | Estrogen receptor beta | 4.644769 | 8.418744 | 7.136677 | mammals | predicted | clID_7365 |
| METHYLENE BLUE | NR3C1 | Glucocorticoid receptor | 6.727169 | 8.072674 | 6.400008 | mammals | actual | clID_363 |
| NAFOXIDINE | ESR1 | Estrogen receptor | 5.072686 | 8.667211 | 7.388277 | mammals | predicted | clID_5910 |
| NAFOXIDINE | Esr2 | Estrogen receptor beta | 5.072686 | 8.418744 | 7.388277 | mammals | predicted | clID_5910 |
| NARAMYCIN B | RORC | Nuclear receptor ROR-gamma | 6.074023 | 7.548863 | 7 | mammals | predicted | clID_525 |
| NELFINAVIR MESYLATE | NR3C1 | Glucocorticoid receptor | 5.305206 | 8.072674 | 6.676421 | mammals | actual | clID_7128 |
| NITROFURAZONE | ESR1 | Estrogen receptor | 5.837994 | 8.667211 | 6 | mammals | actual | clID_1343 |
| ONAPRISTONE | NR3C1 | Glucocorticoid receptor | 4.515057 | 8.072674 | 8.183708 | mammals | predicted | clID_5285 |
| ONAPRISTONE | PGR | Progesterone receptor | 4.515057 | 7.946326 | 7.521692 | mammals | predicted | clID_5285 |
| OUABAIN | ESR1 | Estrogen receptor | 7.013228 | 8.667211 | 6.749975 | mammals | actual | clID_364 |
| OUABAIN | NR3C1 | Glucocorticoid receptor | 7.013228 | 8.072674 | 6.550059 | mammals | actual | clID_364 |
| PAPAVERINE | RORC | Nuclear receptor ROR-gamma | 5.795233 | 7.548863 | 6.72642 | mammals | actual | clID_1423 |
| PODOFILOX | NR3C1 | Glucocorticoid receptor | 5.153942 | 8.072674 | 7.853872 | mammals | actual | clID_5043 |
| PROPYLPYRAZOLETRIOL | ESR1 | Estrogen receptor | 5.390549 | 8.667211 | 6.947734 | mammals | actual | clID_3137 |
| PROPYLPYRAZOLETRIOL | Esr2 | Estrogen receptor beta | 5.390549 | 8.418744 | 6.112784 | mammals | actual | clID_3137 |
| Platinum complex | ESR1 | Estrogen receptor | 4.223807 | 8.667211 | 7.075771 | mammals | predicted | platinum complex |
| RESERPINE | NR3C1 | Glucocorticoid receptor | 5.253841 | 8.072674 | 8.49485 | mammals | actual | clID_4255 |
| RUBITECAN | RORC | Nuclear receptor ROR-gamma | 4.778151 | 7.548863 | 6.625046 | mammals | actual | clID_1835 |
| RUTIN | ESR1 | Estrogen receptor | 4.838698 | 8.667211 | 7.49793 | mammals | actual | clID_1263 |
| TAMOXIFEN | ESR1 | Estrogen receptor | 4.771074 | 8.667211 | 6.081137 | mammals | actual | clID_500 |
| TAMOXIFEN | Esr2 | Estrogen receptor beta | 4.771074 | 8.418744 | 6.0265 | mammals | actual | clID_500 |
| TAMOXIFEN | PGR | Progesterone receptor | 4.771074 | 7.946326 | 6.890017 | mammals | actual | clID_500 |
| TANAPROGET | PGR | Progesterone receptor | 6.406969 | 7.946326 | 9.562469 | mammals | actual | clID_533 |
| THIMEROSAL | RORC | Nuclear receptor ROR-gamma | 5.725507 | 7.548863 | 6.074989 | mammals | actual | organomercury |
| TRIPTOLIDE | RORC | Nuclear receptor ROR-gamma | 5.987582 | 7.548863 | 6.450023 | mammals | actual | clID_2585 |
Use search function in table to view values for specific interactions
| molName | geneName | protname | sarsScoreMolecule | geneScore | interactionScore | organismType | kindOfValue | molecularCluster |
|---|---|---|---|---|---|---|---|---|
| BUTOCONAZOLE NITRATE | HSP90AA1 | Heat shock protein HSP 90-alpha | 4.980549 | 8.516022 | 6.709965 | mammals | predicted | clID_3221 |
| CLOTRIMAZOLE | CBX1 | Chromobox protein homolog 1 | 5.6489 | 6.203048 | 8.075721 | mammals | actual | clID_1854 |
| CLOTRIMAZOLE | cyp121 | Mycocyclosin synthase | 5.6489 | 6.60905 | 7.136677 | bacteria | actual | clID_1854 |
| CLOTRIMAZOLE | CYP17A1 | Steroid 17-alpha-hydroxylase/17,20 lyase | 5.6489 | 6.366169 | 7.088842 | mammals | actual | clID_1854 |
| CLOTRIMAZOLE | CYP19A1 | Aromatase | 5.6489 | 5.843948 | 8.744727 | mammals | actual | clID_1854 |
| CLOTRIMAZOLE | HSP90AA1 | Heat shock protein HSP 90-alpha | 5.6489 | 8.516022 | 6.709965 | mammals | actual | clID_1854 |
| CLOTRIMAZOLE | TBXAS1 | Thromboxane-A synthase | 5.6489 | 6.112514 | 7.853872 | mammals | actual | clID_1854 |
| CYCLOHEXIMIDE | HSP90AA1 | Heat shock protein HSP 90-alpha | 6.421559 | 8.516022 | 6.709965 | mammals | actual | clID_525 |
| CYCLOHEXIMIDE | RORC | Nuclear receptor ROR-gamma | 6.421559 | 7.548863 | 7.005708 | mammals | actual | clID_525 |
| CYCLOHEXIMIDE | THRB | Thyroid hormone receptor beta | 6.421559 | 5.722362 | 7.349692 | mammals | actual | clID_525 |
| HEXACHLOROPHENE | Esr2 | Estrogen receptor beta | 6.099502 | 8.418744 | 7.007579 | mammals | actual | clID_842 |
| HEXACHLOROPHENE | HSP90AA1 | Heat shock protein HSP 90-alpha | 6.099502 | 8.516022 | 6.113509 | mammals | actual | clID_842 |
| HEXACHLOROPHENE | Mapk14 | Mitogen-activated protein kinase 14 | 6.099502 | 8.00432 | 7.057496 | mammals | actual | clID_842 |
| KETOCONAZOLE | GNRHR | Gonadotropin-releasing hormone receptor | 5.777396 | 9.328642 | 7.717881 | mammals | actual | clID_6688 |
| KETOCONAZOLE | HSP90AA1 | Heat shock protein HSP 90-alpha | 5.777396 | 8.516022 | 6.361511 | mammals | actual | clID_6688 |
| KETOCONAZOLE | Kcnma1 | Calcium-activated potassium channel subunit alpha-1 | 5.777396 | 5.67589 | 7.818086 | mammals | actual | clID_6688 |
| KETOCONAZOLE | KCNMB1 | Calcium-activated potassium channel subunit beta-1 | 5.777396 | 6.648685 | 7.818086 | mammals | actual | clID_6688 |
| LUMINESPIB | HSP90B1 | Endoplasmin | 7.780795 | 7.22687 | 7.796447 | mammals | actual | clID_176 |
| LUMINESPIB | HSP90AB1 | Heat shock protein HSP 90-beta | 7.780795 | 8.314665 | 6.356689 | mammals | actual | clID_176 |
| LUMINESPIB | TRAP1 | Heat shock protein 75 kDa, mitochondrial | 7.780795 | 8.050957 | 7.162581 | mammals | actual | clID_176 |
| ONALESPIB | HSP90AA1 | Heat shock protein HSP 90-alpha | 8.093037 | 8.516022 | 6.96826 | mammals | actual | clID_146 |
| ONALESPIB | HSP90AB1 | Heat shock protein HSP 90-beta | 8.093037 | 8.314665 | 7.24229 | mammals | actual | clID_146 |
| PU-H71 | HSP90B1 | Endoplasmin | 5.129502 | 7.22687 | 7.553953 | mammals | actual | clID_5279 |
| PU-H71 | HSP90AA1 | Heat shock protein HSP 90-alpha | 5.129502 | 8.516022 | 7.476617 | mammals | actual | clID_5279 |
| PU-H71 | phoQ | Virulence sensor histidine kinase PhoQ | 5.129502 | 7.828643 | 9 | bacteria | actual | clID_5279 |
| SERTACONAZOLE NITRATE | HSP90AA1 | Heat shock protein HSP 90-alpha | 5.904901 | 8.516022 | 6.632644 | mammals | actual | clID_1770 |
| TANESPIMYCIN | HSP90B1 | Endoplasmin | 4.582691 | 7.22687 | 7.207608 | mammals | actual | clID_2407 |
| TANESPIMYCIN | ERBB2 | Receptor tyrosine-protein kinase erbB-2 | 4.582691 | 8.200475 | 7.508638 | mammals | actual | clID_2407 |
| TANESPIMYCIN | GMNN | Geminin | 4.582691 | 7.313646 | 7.175224 | mammals | actual | clID_2407 |
| TANESPIMYCIN | HSP90AA1 | Heat shock protein HSP 90-alpha | 4.582691 | 8.516022 | 6.463329 | mammals | actual | clID_2407 |
| TANESPIMYCIN | HSP90AB1 | Heat shock protein HSP 90-beta | 4.582691 | 8.314665 | 6.178516 | mammals | actual | clID_2407 |
| TANESPIMYCIN | Pmp22 | Peripheral myelin protein 22 | 4.582691 | 7.357957 | 7.142065 | mammals | actual | clID_2407 |
| WORTMANNIN | HSP90AA1 | Heat shock protein HSP 90-alpha | 4.758394 | 8.516022 | 6.440093 | mammals | actual | clID_1672 |
| WORTMANNIN | PRKCA | Protein kinase C alpha type | 4.758394 | 7.556484 | 7.163679 | mammals | actual | clID_1672 |
| WORTMANNIN | RPS6KB1 | Ribosomal protein S6 kinase beta-1 | 4.758394 | 6.84867 | 7.639377 | mammals | actual | clID_1672 |
| WORTMANNIN | MAPKAPK2 | MAP kinase-activated protein kinase 2 | 4.758394 | 6.442401 | 9 | mammals | actual | clID_1672 |
| WORTMANNIN | Mapk3 | Mitogen-activated protein kinase 3 | 4.758394 | 6.702352 | 9 | mammals | actual | clID_1672 |
| WORTMANNIN | Mapk14 | Mitogen-activated protein kinase 14 | 4.758394 | 8.00432 | 7.394185 | mammals | actual | clID_1672 |
| WORTMANNIN | MTOR | Serine/threonine-protein kinase mTOR | 4.758394 | 7.18155 | 7.144717 | mammals | actual | clID_1672 |
| WORTMANNIN | NEK6 | Serine/threonine-protein kinase Nek6 | 4.758394 | 6.792791 | 9 | mammals | actual | clID_1672 |
| WORTMANNIN | NEK7 | Serine/threonine-protein kinase Nek7 | 4.758394 | 6.805889 | 9 | mammals | actual | clID_1672 |
| WORTMANNIN | Pik3r1 | Phosphatidylinositol 3-kinase regulatory subunit alpha | 4.758394 | 8.165486 | 7.546149 | mammals | actual | clID_1672 |
| WORTMANNIN | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | 4.758394 | 8.414227 | 7.609936 | mammals | actual | clID_1672 |
| WORTMANNIN | PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform | 4.758394 | 7.674333 | 7.949242 | mammals | actual | clID_1672 |
| WORTMANNIN | PIK3CD | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | 4.758394 | 7.906179 | 8.187273 | mammals | actual | clID_1672 |
| WORTMANNIN | PIK3CG | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | 4.758394 | 7.447055 | 8.032971 | mammals | actual | clID_1672 |
| WORTMANNIN | SGK1 | Serine/threonine-protein kinase Sgk1 | 4.758394 | 6.443642 | 7.394185 | mammals | actual | clID_1672 |
Use search function in table to view values for specific interactions
| molName | geneName | protname | sarsScoreMolecule | geneScore | interactionScore | organismType | kindOfValue | molecularCluster |
|---|---|---|---|---|---|---|---|---|
| ABT 737 | Bcl2l1 | Bcl-2-like protein 1 | 5.232257 | 11.693024 | 8.107546 | mammals | actual | clID_305 |
| ABT 737 | BCL2L2 | Bcl-2-like protein 2 | 5.232257 | 9.098299 | 8.429733 | mammals | actual | clID_305 |
| ABT 737 | BCL2L10 | Bcl-2-like protein 10 | 5.232257 | 6.950294 | 8.2 | mammals | actual | clID_305 |
| ABT 737 | BCL2L11 | Bcl-2-like protein 11 | 5.232257 | 6.905743 | 8.69897 | mammals | actual | clID_305 |
| ABT 737 | Bcl2a1 | Bcl-2-related protein A1 | 5.232257 | 6.214488 | 5.2 | mammals | actual | clID_305 |
| ABT 737 | BAD | Bcl2-associated agonist of cell death | 5.232257 | 7.878042 | 9.110924 | mammals | actual | clID_305 |
| ABT 737 | BAK1 | Bcl-2 homologous antagonist/killer | 5.232257 | 8.424892 | 8.23299 | mammals | actual | clID_305 |
| ABT 737 | BAX | Apoptosis regulator BAX | 5.232257 | 6.362786 | 7.638272 | mammals | actual | clID_305 |
| ABT 737 | BBC3 | Bcl-2-binding component 3, isoforms 1/2 | 5.232257 | 10.765544 | 9.522879 | mammals | actual | clID_305 |
| ABT 737 | BCL2 | Apoptosis regulator Bcl-2 | 5.232257 | 12.252343 | 7.952957 | mammals | actual | clID_305 |
| ABT 737 | BID | BH3-interacting domain death agonist | 5.232257 | 6.811185 | 5.998097 | mammals | actual | clID_305 |
| ABT 737 | Csn2 | Beta-casein | 5.232257 | 7.417525 | 8.2 | mammals | actual | clID_305 |
| ABT 737 | Mcl1 | Induced myeloid leukemia cell differentiation protein Mcl-1 homolog | 5.232257 | 7.578731 | 5.360646 | mammals | actual | clID_305 |
| ALEXIDINE DIHYDROCHLORIDE | Bcl2l1 | Bcl-2-like protein 1 | 5.705465 | 11.693024 | 4.737549 | mammals | actual | clID_1681 |
| ALEXIDINE DIHYDROCHLORIDE | BAK1 | Bcl-2 homologous antagonist/killer | 5.705465 | 8.424892 | 4.737549 | mammals | actual | clID_1681 |
| ALEXIDINE DIHYDROCHLORIDE | GALC | Galactocerebrosidase | 5.705465 | 4.47278 | 8 | mammals | actual | clID_1681 |
| ALEXIDINE DIHYDROCHLORIDE | CBFB | Core-binding factor subunit beta | 5.705465 | 6.558397 | 5.950007 | mammals | actual | clID_1681 |
| ALEXIDINE DIHYDROCHLORIDE | RUNX1 | Runt-related transcription factor 1 | 5.705465 | 6.566483 | 5.950007 | mammals | actual | clID_1681 |
| ALEXIDINE HYDROCHLORIDE | Bcl2a1 | Bcl-2-related protein A1 | 7.663791 | 6.214488 | 5.225483 | mammals | predicted | clID_181 |
| ALEXIDINE HYDROCHLORIDE | Poli | DNA polymerase iota | 7.663791 | 6.579491 | 5.05 | mammals | predicted | clID_181 |
| ALEXIDINE HYDROCHLORIDE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 7.663791 | 7.5973 | 5.366943 | mammals | predicted | clID_181 |
| ALEXIDINE HYDROCHLORIDE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 7.663791 | 7.484017 | 9 | mammals | predicted | clID_181 |
| ALEXIDINE HYDROCHLORIDE | USP2 | Ubiquitin carboxyl-terminal hydrolase 2 | 7.663791 | 6.388307 | 5.116502 | mammals | predicted | clID_181 |
| GOSSYPOL | MPG | DNA-3-methyladenine glycosylase | 5.294661 | 5.528214 | 5.522879 | mammals | actual | clID_3821 |
| GOSSYPOL | Akr1b1 | Aldo-keto reductase family 1 member B1 | 5.294661 | 4.989436 | 6.30103 | mammals | actual | clID_3821 |
| GOSSYPOL | ATAD5 | ATPase family AAA domain-containing protein 5 | 5.294661 | 7.207699 | 5.086393 | mammals | actual | clID_3821 |
| GOSSYPOL | Bcl2l1 | Bcl-2-like protein 1 | 5.294661 | 11.693024 | 6.274247 | mammals | actual | clID_3821 |
| GOSSYPOL | BCL2L2 | Bcl-2-like protein 2 | 5.294661 | 9.098299 | 4.752027 | mammals | actual | clID_3821 |
| GOSSYPOL | BCL2 | Apoptosis regulator Bcl-2 | 5.294661 | 12.252343 | 6.344437 | mammals | actual | clID_3821 |
| GOSSYPOL | CYP2C19 | Cytochrome P450 2C19 | 5.294661 | 6.646306 | 5.199999 | mammals | actual | clID_3821 |
| GOSSYPOL | HSD17B1 | Estradiol 17-beta-dehydrogenase 1 | 5.294661 | 5.786959 | 5.176091 | mammals | actual | clID_3821 |
| GOSSYPOL | HSD17B2 | Estradiol 17-beta-dehydrogenase 2 | 5.294661 | 5.237317 | 5.265032 | mammals | actual | clID_3821 |
| GOSSYPOL | POLB | DNA polymerase beta | 5.294661 | 6.219344 | 6.250032 | mammals | actual | clID_3821 |
| GOSSYPOL | ESR1 | Estrogen receptor | 5.294661 | 8.667211 | 5.496614 | mammals | actual | clID_3821 |
| GOSSYPOL | FEN1 | Flap endonuclease 1 | 5.294661 | 6.162891 | 5.149214 | mammals | actual | clID_3821 |
| GOSSYPOL | HSD17B10 | 3-hydroxyacyl-CoA dehydrogenase type-2 | 5.294661 | 6.303408 | 5.174999 | mammals | actual | clID_3821 |
| GOSSYPOL | KDM4E | Lysine-specific demethylase 4E | 5.294661 | 6.67228 | 5.2 | mammals | actual | clID_3821 |
| GOSSYPOL | LDHA | L-lactate dehydrogenase A chain | 5.294661 | 4.902878 | 5.721246 | mammals | actual | clID_3821 |
| GOSSYPOL | LDHB | L-lactate dehydrogenase B chain | 5.294661 | 5.315478 | 5.853872 | mammals | actual | clID_3821 |
| GOSSYPOL | lef | Lethal factor | 5.294661 | 6.363322 | 6.59998 | bacteria | actual | clID_3821 |
| GOSSYPOL | LMNA | Prelamin-A/C [Cleaved into: Lamin-A/C | 5.294661 | 7.05737 | 5.650004 | mammals | actual | clID_3821 |
| GOSSYPOL | ALOX12 | Arachidonate 12-lipoxygenase, 12S-type | 5.294661 | 6.703768 | 5.862502 | mammals | actual | clID_3821 |
| GOSSYPOL | ALOX15B | Arachidonate 15-lipoxygenase B | 5.294661 | 6.906258 | 5.250001 | mammals | actual | clID_3821 |
| GOSSYPOL | GAA | Lysosomal alpha-glucosidase | 5.294661 | 6.349346 | 5.05 | mammals | actual | clID_3821 |
| GOSSYPOL | Mcl1 | Induced myeloid leukemia cell differentiation protein Mcl-1 homolog | 5.294661 | 7.578731 | 6.254999 | mammals | actual | clID_3821 |
| GOSSYPOL | MDH1 | Malate dehydrogenase, cytoplasmic | 5.294661 | 6.161278 | 5.236572 | mammals | actual | clID_3821 |
| GOSSYPOL | MDH2 | Malate dehydrogenase, mitochondrial | 5.294661 | 5.953822 | 5.552842 | mammals | actual | clID_3821 |
| GOSSYPOL | MTOR | Serine/threonine-protein kinase mTOR | 5.294661 | 7.18155 | 5.082999 | mammals | actual | clID_3821 |
| GOSSYPOL | CBFB | Core-binding factor subunit beta | 5.294661 | 6.558397 | 5.199998 | mammals | actual | clID_3821 |
| GOSSYPOL | PIN1 | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | 5.294661 | 6.426581 | 5.520984 | mammals | actual | clID_3821 |
| GOSSYPOL | Poli | DNA polymerase iota | 5.294661 | 6.579491 | 5.599998 | mammals | actual | clID_3821 |
| GOSSYPOL | POLK | DNA polymerase kappa | 5.294661 | 6.364127 | 5.38294 | mammals | actual | clID_3821 |
| GOSSYPOL | Pparg | Peroxisome proliferator-activated receptor gamma | 5.294661 | 6.626045 | 5.099999 | mammals | actual | clID_3821 |
| GOSSYPOL | LDH | Lactate dehydrogenase | 5.294661 | 6.826336 | 5.578396 | protozoan | actual | clID_3821 |
| GOSSYPOL | PGAM | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase | 5.294661 | 5.45283 | 7.821023 | protozoan | actual | clID_3821 |
| GOSSYPOL | Q6VVP7 | Malate dehydrogenase | 5.294661 | 7.074408 | 5.692504 | protozoan | actual | clID_3821 |
| GOSSYPOL | reverse | Reverse transcriptase | 5.294661 | 7.682466 | 5.472076 | virus | actual | clID_3821 |
| GOSSYPOL | RGS4 | Regulator of G-protein signaling 4 | 5.294661 | 6.451243 | 5.374235 | mammals | actual | clID_3821 |
| GOSSYPOL | RUNX1 | Runt-related transcription factor 1 | 5.294661 | 6.566483 | 5.199998 | mammals | actual | clID_3821 |
| GOSSYPOL | SMAD3 | Mothers against decapentaplegic homolog 3 | 5.294661 | 6.518109 | 5.950007 | mammals | actual | clID_3821 |
| GOSSYPOL | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.294661 | 7.5973 | 5.525519 | mammals | actual | clID_3821 |
| GOSSYPOL | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.294661 | 7.484017 | 5.315068 | mammals | actual | clID_3821 |
| GOSSYPOL | SNCA | Alpha-synuclein | 5.294661 | 6.323601 | 5.168105 | mammals | actual | clID_3821 |
| GOSSYPOL | TDP1 | Tyrosyl-DNA phosphodiesterase 1 | 5.294661 | 7.013656 | 5.322243 | mammals | actual | clID_3821 |
| GOSSYPOL | USP2 | Ubiquitin carboxyl-terminal hydrolase 2 | 5.294661 | 6.388307 | 5.227665 | mammals | actual | clID_3821 |
| NAVITOCLAX | Bcl2l1 | Bcl-2-like protein 1 | 4.734906 | 11.693024 | 8.593359 | mammals | predicted | clID_1535 |
| NAVITOCLAX | BCL2L11 | Bcl-2-like protein 11 | 4.734906 | 6.905743 | 8.366532 | mammals | predicted | clID_1535 |
| NAVITOCLAX | BAD | Bcl2-associated agonist of cell death | 4.734906 | 7.878042 | 9.150515 | mammals | predicted | clID_1535 |
| NAVITOCLAX | BCL2 | Apoptosis regulator Bcl-2 | 4.734906 | 12.252343 | 8.408452 | mammals | predicted | clID_1535 |
| NAVITOCLAX | Mcl1 | Induced myeloid leukemia cell differentiation protein Mcl-1 homolog | 4.734906 | 7.578731 | 6.280334 | mammals | predicted | clID_1535 |
| NAVITOCLAX | SMAD3 | Mothers against decapentaplegic homolog 3 | 4.734906 | 6.518109 | 5.499997 | mammals | predicted | clID_1535 |
| NAVITOCLAX | TDP1 | Tyrosyl-DNA phosphodiesterase 1 | 4.734906 | 7.013656 | 6.425011 | mammals | predicted | clID_1535 |
| THONZONIUM | GMNN | Geminin | 4.935721 | 7.313646 | 5.094075 | mammals | actual | clID_4368 |
| THONZONIUM | LMNA | Prelamin-A/C [Cleaved into: Lamin-A/C | 4.935721 | 7.05737 | 5.149997 | mammals | actual | clID_4368 |
| THONZONIUM BROMIDE | Bcl2a1 | Bcl-2-related protein A1 | 5.510663 | 6.214488 | 5.191114 | mammals | actual | clID_4368 |
| THONZONIUM BROMIDE | TDP1 | Tyrosyl-DNA phosphodiesterase 1 | 5.510663 | 7.013656 | 5.365142 | mammals | actual | clID_4368 |
Use search function in table to view values for specific interactions
| molName | geneName | protname | sarsScoreMolecule | geneScore | interactionScore | organismType | kindOfValue | molecularCluster |
|---|---|---|---|---|---|---|---|---|
| 18-OXOFERRUGINOL | JAK3 | Tyrosine-protein kinase JAK3 | 5.378367 | 6.782696 | 6.2 | mammals | actual | clID_3387 |
| 5-METHOXYISATIN | rep | Replicase polyprotein 1ab | 6.308938 | 12.649926 | 6.308938 | virus | actual | clID_286 |
| 5-NITROISATIN | rep | Replicase polyprotein 1ab | 6.191906 | 12.649926 | 6.191906 | virus | actual | clID_286 |
| ABEMACICLIB | GSK3A | Glycogen synthase kinase-3 alpha | 5.179142 | 5.878393 | 7.886057 | mammals | actual | clID_4805 |
| ABEMACICLIB | Gsk3b | Glycogen synthase kinase-3 beta | 5.179142 | 6.111173 | 7.886057 | mammals | actual | clID_4805 |
| AMENTOFLAVONE | VEGFA | Vascular endothelial growth factor A | 5.080922 | 6.3527 | 7.782516 | mammals | actual | clID_459 |
| AMIODARONE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.1595 | 8.320941 | 7.800148 | mammals | actual | clID_6596 |
| APABETALONE | BRD4 | Bromodomain-containing protein 4 | 4.93561 | 7.575957 | 6.0655 | mammals | actual | clID_10703 |
| APICIDIN | Hdac2 | Histone deacetylase 2 | 4.559786 | 6.79746 | 7.914313 | mammals | actual | clID_1175 |
| APITOLISIB | JAK1 | Tyrosine-protein kinase JAK1 | 9 | 6.919803 | 7.585027 | mammals | actual | clID_1863 |
| AZD-5438 | GSK3A | Glycogen synthase kinase-3 alpha | 5.310904 | 5.878393 | 6.179819 | mammals | actual | clID_3693 |
| AZD-5438 | Gsk3b | Glycogen synthase kinase-3 beta | 5.310904 | 6.111173 | 6.153804 | mammals | actual | clID_3693 |
| BIS(DIMETHYLAMINETHIOCARBONYL)MONOSULFIDE | NR3C1 | Glucocorticoid receptor | 4.946601 | 8.072674 | 7.549751 | mammals | actual | clID_7469 |
| CAFFEINE | NR3C1 | Glucocorticoid receptor | 4.703587 | 8.072674 | 7.74958 | mammals | actual | clID_6550 |
| CAPTOPRIL | Ace2 | Angiotensin-converting enzyme 2 | 5.558465 | 5.164637 | 7.209829 | mammals | actual | clID_2251 |
| CGP-53353 | JAK2 | Tyrosine-protein kinase JAK2 | 5.792715 | 7.073032 | 7.21598 | mammals | actual | clID_670 |
| CGP-53353 | JAK3 | Tyrosine-protein kinase JAK3 | 5.792715 | 6.782696 | 7.163679 | mammals | actual | clID_670 |
| CGP-53353 | ROCK1 | Rho-associated protein kinase 1 | 5.792715 | 5.991738 | 9 | mammals | actual | clID_670 |
| CGP-53353 | ROCK2 | Rho-associated protein kinase 2 | 5.792715 | 6.479814 | 7.274836 | mammals | actual | clID_670 |
| CHLORHEXIDINE | NR3C1 | Glucocorticoid receptor | 5.855721 | 8.072674 | 6.450016 | mammals | actual | clID_1295 |
| CHLOROQUINE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.684392 | 8.320941 | 7.100015 | mammals | actual | clID_1740 |
| CHLORPROMAZINE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 6.908068 | 8.320941 | 6.534681 | mammals | predicted | clID_302 |
| CINALUKAST | CYSLTR1 | Cysteinyl leukotriene receptor 1 | 4.936829 | 8.121852 | 8.19382 | mammals | actual | clID_7693 |
| CINANSERIN | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.30103 | 8.320941 | 6.74 | mammals | actual | clID_3770 |
| CITRIC ACID | Hdac2 | Histone deacetylase 2 | 4.931063 | 6.79746 | 9 | mammals | actual | clID_8565 |
| CLEMASTINE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.75225 | 8.320941 | 7.774131 | mammals | actual | clID_1525 |
| CLOMIPRAMINE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 4.522627 | 8.320941 | 6.054213 | mammals | actual | clID_1362 |
| CORT 108297 | NR3C1 | Glucocorticoid receptor | 5.298461 | 8.072674 | 8.460247 | mammals | predicted | clID_3793 |
| CYC-116 | JAK2 | Tyrosine-protein kinase JAK2 | 5.078035 | 7.073032 | 7.199971 | mammals | actual | clID_5848 |
| CYC-116 | JAK3 | Tyrosine-protein kinase JAK3 | 5.078035 | 6.782696 | 6.199998 | mammals | actual | clID_5848 |
| CYC-116 | ROCK1 | Rho-associated protein kinase 1 | 5.078035 | 5.991738 | 6.299998 | mammals | actual | clID_5848 |
| CYC-116 | ROCK2 | Rho-associated protein kinase 2 | 5.078035 | 6.479814 | 6.025671 | mammals | actual | clID_5848 |
| CYC-116 | TBK1 | Serine/threonine-protein kinase TBK1 | 5.078035 | 6.967071 | 6.567382 | mammals | actual | clID_5848 |
| CYC-116 | TYK2 | Non-receptor tyrosine-protein kinase TYK2 | 5.078035 | 6.189362 | 6.545495 | mammals | actual | clID_5848 |
| CYCLOSPORINE | FKBP1A | Peptidyl-prolyl cis-trans isomerase FKBP1A | 5.468606 | 7.167158 | 7.69897 | mammals | actual | clID_769 |
| DEXTROMETHORPHAN | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.527467 | 8.320941 | 6.55152 | mammals | actual | clID_2960 |
| DIGITOXIGENIN | ATP1A1 | Sodium/potassium-transporting ATPase subunit alpha-1 | 5.176634 | 7.258587 | 7.11674 | mammals | actual | clID_364 |
| DIGITOXIN | ATP1A1 | Sodium/potassium-transporting ATPase subunit alpha-1 | 6.638272 | 7.258587 | 8.09691 | mammals | actual | clID_364 |
| DIGITOXIN | NR3C1 | Glucocorticoid receptor | 6.638272 | 8.072674 | 6.450016 | mammals | actual | clID_364 |
| DIGOXIN | ATP1A1 | Sodium/potassium-transporting ATPase subunit alpha-1 | 6.721246 | 7.258587 | 6.061936 | mammals | actual | clID_364 |
| DIGOXIN | NR3C1 | Glucocorticoid receptor | 6.721246 | 8.072674 | 6.125016 | mammals | actual | clID_364 |
| DOLUTEGRAVIR | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 4.584705 | 8.320941 | 6.906141 | mammals | actual | clID_687 |
| DOVITINIB | JAK1 | Tyrosine-protein kinase JAK1 | 4.65048 | 6.919803 | 6.428043 | mammals | actual | clID_9575 |
| DOVITINIB | JAK2 | Tyrosine-protein kinase JAK2 | 4.65048 | 7.073032 | 6.013653 | mammals | actual | clID_9575 |
| DOVITINIB | RIPK1 | Receptor-interacting serine/threonine-protein kinase 1 | 4.65048 | 6.203598 | 6.49485 | mammals | actual | clID_9575 |
| DOVITINIB | TBK1 | Serine/threonine-protein kinase TBK1 | 4.65048 | 6.967071 | 6.013057 | mammals | actual | clID_9575 |
| DOVITINIB | TYK2 | Non-receptor tyrosine-protein kinase TYK2 | 4.65048 | 6.189362 | 6.393826 | mammals | actual | clID_9575 |
| E-FLUPENTIXOL | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.538428 | 8.320941 | 7.830032 | mammals | predicted | clID_267 |
| ENTRECTINIB | JAK1 | Tyrosine-protein kinase JAK1 | 5.367357 | 6.919803 | 6.950782 | mammals | actual | clID_3315 |
| ENTRECTINIB | JAK2 | Tyrosine-protein kinase JAK2 | 5.367357 | 7.073032 | 7.548999 | mammals | actual | clID_3315 |
| ENTRECTINIB | JAK3 | Tyrosine-protein kinase JAK3 | 5.367357 | 6.782696 | 6.378586 | mammals | actual | clID_3315 |
| FERRUGINOL | rep | Replicase polyprotein 1ab | 5.356695 | 12.649926 | 6.103757 | virus | actual | clID_3387 |
| FIMEPINOSTAT | Hdac2 | Histone deacetylase 2 | 4.771095 | 6.79746 | 7.946298 | mammals | actual | clID_11717 |
| FLUNARIZINE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 4.518593 | 8.320941 | 7.80103 | mammals | predicted | clID_355 |
| FLUPENTIXOL | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 4.760268 | 8.320941 | 8.649946 | mammals | actual | clID_267 |
| GW632580X | GSK3A | Glycogen synthase kinase-3 alpha | 4.861247 | 5.878393 | 7.533333 | mammals | actual | clID_780 |
| GW632580X | Gsk3b | Glycogen synthase kinase-3 beta | 4.861247 | 6.111173 | 6.400687 | mammals | actual | clID_780 |
| GW632580X | JAK2 | Tyrosine-protein kinase JAK2 | 4.861247 | 7.073032 | 7.6 | mammals | actual | clID_780 |
| GW632580X | JAK3 | Tyrosine-protein kinase JAK3 | 4.861247 | 6.782696 | 7.597488 | mammals | actual | clID_780 |
| GW632580X | ROCK1 | Rho-associated protein kinase 1 | 4.861247 | 5.991738 | 7.533333 | mammals | actual | clID_780 |
| HALOPERIDOL | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 4.799189 | 8.320941 | 8.291093 | mammals | actual | clID_1001 |
| HERBIMYCIN | GSK3A | Glycogen synthase kinase-3 alpha | 5.122579 | 5.878393 | 9 | mammals | predicted | clID_2407 |
| HERBIMYCIN | Gsk3b | Glycogen synthase kinase-3 beta | 5.122579 | 6.111173 | 7.530349 | mammals | predicted | clID_2407 |
| HERBIMYCIN | JAK2 | Tyrosine-protein kinase JAK2 | 5.122579 | 7.073032 | 7.754825 | mammals | predicted | clID_2407 |
| HERBIMYCIN | ROCK1 | Rho-associated protein kinase 1 | 5.122579 | 5.991738 | 9 | mammals | predicted | clID_2407 |
| HERBIMYCIN | ROCK2 | Rho-associated protein kinase 2 | 5.122579 | 6.479814 | 7.30103 | mammals | predicted | clID_2407 |
| HYDROCORTISONE | NR3C1 | Glucocorticoid receptor | 5.050958 | 8.072674 | 6.312103 | mammals | actual | clID_1633 |
| HYDROXYCHLOROQUINE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.92742 | 8.320941 | 6.900009 | mammals | actual | clID_1740 |
| HYDROXYCHLOROQUINE | Tlr7 | Toll-like receptor 7 | 5.92742 | 5.531899 | 6.09691 | mammals | actual | clID_1740 |
| HYDROXYCHLOROQUINE | Tlr9 | Toll-like receptor 9 | 5.92742 | 6.019685 | 6.958607 | mammals | actual | clID_1740 |
| IMATINIB | JAK3 | Tyrosine-protein kinase JAK3 | 4.704932 | 6.782696 | 6.8 | mammals | actual | clID_2757 |
| KENPAULLONE | GSK3A | Glycogen synthase kinase-3 alpha | 4.771468 | 5.878393 | 7.11914 | mammals | actual | clID_253 |
| KENPAULLONE | Gsk3b | Glycogen synthase kinase-3 beta | 4.771468 | 6.111173 | 7.019753 | mammals | actual | clID_253 |
| KENPAULLONE | JAK2 | Tyrosine-protein kinase JAK2 | 4.771468 | 7.073032 | 6.462954 | mammals | actual | clID_253 |
| KENPAULLONE | JAK3 | Tyrosine-protein kinase JAK3 | 4.771468 | 6.782696 | 6.462954 | mammals | actual | clID_253 |
| KENPAULLONE | ROCK1 | Rho-associated protein kinase 1 | 4.771468 | 5.991738 | 7.9 | mammals | actual | clID_253 |
| LOPERAMIDE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.03292 | 8.320941 | 6.390218 | mammals | actual | clID_6355 |
| METHYLENE BLUE | NR3C1 | Glucocorticoid receptor | 6.727169 | 8.072674 | 6.400008 | mammals | actual | clID_363 |
| MIDOSTAURIN | JAK1 | Tyrosine-protein kinase JAK1 | 5.198496 | 6.919803 | 6.173925 | mammals | actual | clID_3146 |
| MIDOSTAURIN | JAK2 | Tyrosine-protein kinase JAK2 | 5.198496 | 7.073032 | 7.026872 | mammals | actual | clID_3146 |
| MIDOSTAURIN | JAK3 | Tyrosine-protein kinase JAK3 | 5.198496 | 6.782696 | 7.920819 | mammals | actual | clID_3146 |
| MIDOSTAURIN | ROCK1 | Rho-associated protein kinase 1 | 5.198496 | 5.991738 | 7.053953 | mammals | actual | clID_3146 |
| MIDOSTAURIN | TBK1 | Serine/threonine-protein kinase TBK1 | 5.198496 | 6.967071 | 7.596292 | mammals | actual | clID_3146 |
| MIDOSTAURIN | TYK2 | Non-receptor tyrosine-protein kinase TYK2 | 5.198496 | 6.189362 | 6.60206 | mammals | actual | clID_3146 |
| MYCOPHENOLIC ACID | IMPDH1 | Inosine-5'-monophosphate dehydrogenase 1 | 4.582474 | 5.293895 | 7.013849 | mammals | actual | clID_7897 |
| MYCOPHENOLIC ACID | IMPDH2 | Inosine-5'-monophosphate dehydrogenase 2 | 4.582474 | 5.452397 | 7.467011 | mammals | actual | clID_7897 |
| NAFOXIDINE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.072686 | 8.320941 | 7.522879 | mammals | predicted | clID_5910 |
| NELFINAVIR MESYLATE | NR3C1 | Glucocorticoid receptor | 5.305206 | 8.072674 | 6.676421 | mammals | actual | clID_7128 |
| ONAPRISTONE | NR3C1 | Glucocorticoid receptor | 4.515057 | 8.072674 | 8.183708 | mammals | predicted | clID_5285 |
| ORVEPITANT | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 4.572556 | 8.320941 | 6.026872 | mammals | predicted | clID_1663 |
| OUABAIN | NR3C1 | Glucocorticoid receptor | 7.013228 | 8.072674 | 6.550059 | mammals | actual | clID_364 |
| PAZOPANIB | RIPK1 | Receptor-interacting serine/threonine-protein kinase 1 | 4.969786 | 6.203598 | 6.585027 | mammals | actual | clID_7154 |
| PD-0166285 | JAK2 | Tyrosine-protein kinase JAK2 | 5.465159 | 7.073032 | 7.400008 | mammals | actual | clID_1477 |
| PD-0166285 | JAK3 | Tyrosine-protein kinase JAK3 | 5.465159 | 6.782696 | 6.399997 | mammals | actual | clID_1477 |
| PELITINIB | JAK3 | Tyrosine-protein kinase JAK3 | 5.700744 | 6.782696 | 7.60206 | mammals | actual | clID_730 |
| PICTILISIB | JAK1 | Tyrosine-protein kinase JAK1 | 5.564751 | 6.919803 | 6.366532 | mammals | actual | clID_2221 |
| PODOFILOX | NR3C1 | Glucocorticoid receptor | 5.153942 | 8.072674 | 7.853872 | mammals | actual | clID_5043 |
| RAF-265 | TYK2 | Non-receptor tyrosine-protein kinase TYK2 | 6.138279 | 6.189362 | 6.387216 | mammals | actual | clID_781 |
| RESERPINE | NR3C1 | Glucocorticoid receptor | 5.253841 | 8.072674 | 8.49485 | mammals | actual | clID_4255 |
| RG-547 | GSK3A | Glycogen synthase kinase-3 alpha | 5.65239 | 5.878393 | 6.552842 | mammals | actual | clID_1847 |
| RG-547 | ROCK2 | Rho-associated protein kinase 2 | 5.65239 | 6.479814 | 6.136677 | mammals | actual | clID_1847 |
| RGB-286638 | GSK3A | Glycogen synthase kinase-3 alpha | 5.371291 | 5.878393 | 8.154902 | mammals | actual | clID_3282 |
| RGB-286638 | Gsk3b | Glycogen synthase kinase-3 beta | 5.371291 | 6.111173 | 8.09691 | mammals | actual | clID_3282 |
| RGB-286638 | JAK1 | Tyrosine-protein kinase JAK1 | 5.371291 | 6.919803 | 6.856985 | mammals | actual | clID_3282 |
| RIMCAZOLE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.278436 | 8.320941 | 6.111734 | mammals | actual | clID_4588 |
| RONIPAMIL | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.409323 | 8.320941 | 9 | mammals | predicted | clID_3021 |
| RUXOLITINIB | JAK1 | Tyrosine-protein kinase JAK1 | 5.319868 | 6.919803 | 7.587198 | mammals | actual | clID_3626 |
| RUXOLITINIB | JAK2 | Tyrosine-protein kinase JAK2 | 5.319868 | 7.073032 | 7.595015 | mammals | actual | clID_3626 |
| RUXOLITINIB | JAK3 | Tyrosine-protein kinase JAK3 | 5.319868 | 6.782696 | 6.784478 | mammals | actual | clID_3626 |
| RUXOLITINIB | ROCK1 | Rho-associated protein kinase 1 | 5.319868 | 5.991738 | 6.654282 | mammals | actual | clID_3626 |
| RUXOLITINIB | ROCK2 | Rho-associated protein kinase 2 | 5.319868 | 6.479814 | 6.254849 | mammals | actual | clID_3626 |
| RUXOLITINIB | TYK2 | Non-receptor tyrosine-protein kinase TYK2 | 5.319868 | 6.189362 | 7.292321 | mammals | actual | clID_3626 |
| RUXOLITINIB PHOSPHATE | JAK1 | Tyrosine-protein kinase JAK1 | 7.659934 | 6.919803 | 8 | mammals | predicted | clID_3626 |
| RUXOLITINIB PHOSPHATE | JAK2 | Tyrosine-protein kinase JAK2 | 7.659934 | 7.073032 | 7.552842 | mammals | predicted | clID_3626 |
| RUXOLITINIB PHOSPHATE | JAK3 | Tyrosine-protein kinase JAK3 | 7.659934 | 6.782696 | 6.091515 | mammals | predicted | clID_3626 |
| RUXOLITINIB PHOSPHATE | TYK2 | Non-receptor tyrosine-protein kinase TYK2 | 7.659934 | 6.189362 | 6.935542 | mammals | predicted | clID_3626 |
| SALICYLIC ACID | Hdac2 | Histone deacetylase 2 | 5.39542 | 6.79746 | 6.060698 | mammals | actual | clID_3107 |
| SILMITASERTIB | TBK1 | Serine/threonine-protein kinase TBK1 | 5.358996 | 6.967071 | 7.455932 | mammals | actual | clID_3377 |
| SIRAMESINE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 9 | 8.320941 | 7.681082 | mammals | actual | clID_91 |
| SIROLIMUS | FKBP1A | Peptidyl-prolyl cis-trans isomerase FKBP1A | 5.512884 | 7.167158 | 6.59157 | mammals | actual | clID_265 |
| SIROLIMUS | GSK3A | Glycogen synthase kinase-3 alpha | 5.512884 | 5.878393 | 9 | mammals | actual | clID_265 |
| SIROLIMUS | Gsk3b | Glycogen synthase kinase-3 beta | 5.512884 | 6.111173 | 6.744209 | mammals | actual | clID_265 |
| SIROLIMUS | JAK2 | Tyrosine-protein kinase JAK2 | 5.512884 | 7.073032 | 9 | mammals | actual | clID_265 |
| SIROLIMUS | JAK3 | Tyrosine-protein kinase JAK3 | 5.512884 | 6.782696 | 9 | mammals | actual | clID_265 |
| SIROLIMUS | ROCK1 | Rho-associated protein kinase 1 | 5.512884 | 5.991738 | 9 | mammals | actual | clID_265 |
| SIROLIMUS | ROCK2 | Rho-associated protein kinase 2 | 5.512884 | 6.479814 | 7.029258 | mammals | actual | clID_265 |
| SNS-314 | ROCK2 | Rho-associated protein kinase 2 | 5.64226 | 6.479814 | 6.199998 | mammals | actual | clID_1882 |
| TAMOXIFEN | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 4.771074 | 8.320941 | 7.731165 | mammals | actual | clID_500 |
| TERCONAZOLE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 4.890817 | 8.320941 | 6.481861 | mammals | actual | clID_6688 |
| THIORIDAZINE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.174574 | 8.320941 | 6.460114 | mammals | actual | clID_4843 |
| TRAPOXIN B | Hdac2 | Histone deacetylase 2 | 4.785863 | 6.79746 | 8.25 | mammals | predicted | clID_939 |
| TRIFLUOPERAZINE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.241725 | 8.320941 | 7.823909 | mammals | predicted | clID_4249 |
| TRIPARANOL | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 5.277328 | 8.320941 | 8.09691 | mammals | actual | clID_3965 |
| TYRPHOSTIN AG-1478 | GSK3A | Glycogen synthase kinase-3 alpha | 5.452454 | 5.878393 | 9 | mammals | actual | clID_681 |
| TYRPHOSTIN AG-1478 | JAK2 | Tyrosine-protein kinase JAK2 | 5.452454 | 7.073032 | 7.394185 | mammals | actual | clID_681 |
| TYRPHOSTIN AG-1478 | JAK3 | Tyrosine-protein kinase JAK3 | 5.452454 | 6.782696 | 9 | mammals | actual | clID_681 |
| TYRPHOSTIN AG-1478 | ROCK1 | Rho-associated protein kinase 1 | 5.452454 | 5.991738 | 7.502399 | mammals | actual | clID_681 |
| TYRPHOSTIN AG-1478 | ROCK2 | Rho-associated protein kinase 2 | 5.452454 | 6.479814 | 7.412788 | mammals | actual | clID_681 |
| VANDETANIB | ROCK2 | Rho-associated protein kinase 2 | 5.021545 | 6.479814 | 6.221849 | mammals | actual | clID_1562 |
| VANOXERINE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 6.527334 | 8.320941 | 7.418757 | mammals | predicted | clID_462 |
| VINBLASTINE SULFATE | TUBB | Tubulin beta chain | 4.711161 | 7.544926 | 6.886057 | mammals | actual | clID_440 |
| VS-5584 | JAK2 | Tyrosine-protein kinase JAK2 | 6.185686 | 7.073032 | 7.585027 | mammals | predicted | clID_720 |
| VS-5584 | JAK3 | Tyrosine-protein kinase JAK3 | 6.185686 | 6.782696 | 8.522879 | mammals | predicted | clID_720 |
| WORTMANNIN | ROCK2 | Rho-associated protein kinase 2 | 4.758394 | 6.479814 | 6.500217 | mammals | actual | clID_1672 |
| ZAFIRLUKAST | CYSLTR1 | Cysteinyl leukotriene receptor 1 | 4.511604 | 8.121852 | 8.869426 | mammals | actual | clID_3257 |
| ZOTIRACICLIB | GSK3A | Glycogen synthase kinase-3 alpha | 5.114266 | 5.878393 | 6.237321 | mammals | actual | clID_5424 |
| ZOTIRACICLIB | Gsk3b | Glycogen synthase kinase-3 beta | 5.114266 | 6.111173 | 6.349692 | mammals | actual | clID_5424 |
| ZOTIRACICLIB | JAK2 | Tyrosine-protein kinase JAK2 | 5.114266 | 7.073032 | 7.014302 | mammals | actual | clID_5424 |
| ZUCLOMIPHENE | Sigmar1 | Sigma non-opioid intracellular receptor 1 | 4.635813 | 8.320941 | 8.30103 | mammals | predicted | clID_2790 |
Use search function in table to view values for specific interactions
| molName | geneName | protname | sarsScoreMolecule | geneScore | interactionScore | organismType | kindOfValue | molecularCluster |
|---|---|---|---|---|---|---|---|---|
| BIS(20)-HUPERZINE B | ache | Acetylcholinesterase | 5.056192 | 7.688367 | 7.598599 | widespread | predicted | clID_2339 |
| CAFFEINE | ache | Acetylcholinesterase | 4.703587 | 7.688367 | 5.139662 | widespread | actual | clID_6550 |
| CAFFEINE | Chrm1 | Muscarinic acetylcholine receptor M1 | 4.703587 | 6.534599 | 7.449772 | mammals | actual | clID_6550 |
| CAFFEINE | AR | Androgen receptor | 4.703587 | 6.822547 | 8.30103 | mammals | actual | clID_6550 |
| CAFFEINE | NR3C1 | Glucocorticoid receptor | 4.703587 | 8.072674 | 7.74958 | mammals | actual | clID_6550 |
| CAFFEINE | GMNN | Geminin | 4.703587 | 7.313646 | 6.6362 | mammals | actual | clID_6550 |
| CAFFEINE | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.703587 | 7.5973 | 9 | mammals | actual | clID_6550 |
| CAFFEINE | THRB | Thyroid hormone receptor beta | 4.703587 | 5.722362 | 8.853872 | mammals | actual | clID_6550 |
| CAPROCTAMINE | ache | Acetylcholinesterase | 5.334903 | 7.688367 | 7.120051 | widespread | predicted | clID_6683 |
| CAPROCTAMINE | CHRM2 | Muscarinic acetylcholine receptor M2 | 5.334903 | 6.324426 | 6.389072 | mammals | predicted | clID_6683 |
| CAPROCTAMINE HCL | ache | Acetylcholinesterase | 4.800956 | 7.688367 | 6.769551 | widespread | predicted | clID_6683 |
| CAPTOPRIL | Ace | Angiotensin-converting enzyme | 5.558465 | 6.20709 | 7.384878 | mammals | actual | clID_2251 |
| CAPTOPRIL | Ace2 | Angiotensin-converting enzyme 2 | 5.558465 | 5.164637 | 7.209829 | mammals | actual | clID_2251 |
| CAPTOPRIL | ache | Acetylcholinesterase | 5.558465 | 7.688367 | 5.113559 | widespread | actual | clID_2251 |
| CAPTOPRIL | MPHOSPH8 | M-phase phosphoprotein 8 | 5.558465 | 6.568582 | 6 | mammals | actual | clID_2251 |
| CAPTOPRIL | Abcc2 | Canalicular multispecific organic anion transporter 1 | 5.558465 | 7.3938 | 9 | mammals | actual | clID_2251 |
| CAPTOPRIL | REN | Renin | 5.558465 | 8.255979 | 8.769551 | mammals | actual | clID_2251 |
| CAPTOPRIL | SLC15A1 | Solute carrier family 15 member 1 | 5.558465 | 5.094052 | 7.106304 | mammals | actual | clID_2251 |
| CAPTOPRIL | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.558465 | 7.5973 | 9 | mammals | actual | clID_2251 |
| CAPTOPRIL | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.558465 | 7.484017 | 9 | mammals | actual | clID_2251 |
| CAPTOPRIL | F2 | Prothrombin | 5.558465 | 7.222099 | 7.318759 | mammals | actual | clID_2251 |
| CARBACRINE | ache | Acetylcholinesterase | 5.947197 | 7.688367 | 6.901198 | widespread | predicted | clID_1110 |
| CARBACRINE | BCHE | Cholinesterase | 5.947197 | 7.593404 | 6.528708 | mammals | predicted | clID_1110 |
| CARBACRINE | Grin2b | Glutamate receptor ionotropic, NMDA 2B | 5.947197 | 7.199885 | 6.130768 | mammals | predicted | clID_1110 |
| CARBACRINE | Grin1 | Glutamate receptor ionotropic, NMDA 1 | 5.947197 | 7.27334 | 6.130768 | mammals | predicted | clID_1110 |
| CHLORPYRIFOS | ache | Acetylcholinesterase | 4.671842 | 7.688367 | 6.008151 | widespread | actual | clID_9389 |
| CHLORPYRIFOS | GMNN | Geminin | 4.671842 | 7.313646 | 8.283997 | mammals | actual | clID_9389 |
| CHLORPYRIFOS | Ache | Carboxylic ester hydrolase | 4.671842 | 4.41675 | 5.021819 | arthropoda | actual | clID_9389 |
| CHLORPYRIFOS | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.671842 | 7.5973 | 9 | mammals | actual | clID_9389 |
| CHLORPYRIFOS | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.671842 | 7.484017 | 9 | mammals | actual | clID_9389 |
| CHLORPYRIFOS | THRB | Thyroid hormone receptor beta | 4.671842 | 5.722362 | 6.500038 | mammals | actual | clID_9389 |
| CLOSANTEL | ADORA3 | Adenosine receptor A3 | 5.600704 | 6.389069 | 7.142668 | mammals | actual | clID_2064 |
| CLOSANTEL | ache | Acetylcholinesterase | 5.600704 | 7.688367 | 5.530031 | widespread | actual | clID_2064 |
| CLOSANTEL | EGFR | Epidermal growth factor receptor | 5.600704 | 8.436627 | 6.248721 | mammals | actual | clID_2064 |
| CLOSANTEL | FYN | Tyrosine-protein kinase Fyn | 5.600704 | 6.452409 | 6.027334 | mammals | actual | clID_2064 |
| CLOSANTEL | Mapk14 | Mitogen-activated protein kinase 14 | 5.600704 | 8.00432 | 6.183096 | mammals | actual | clID_2064 |
| CLOSANTEL | Q25615 | Chitinase | 5.600704 | 5.44795 | 6.062354 | worms | actual | clID_2064 |
| CLOSANTEL | ska | Streptokinase A | 5.600704 | 6.6046 | 7.086186 | bacteria | actual | clID_2064 |
| CLOSANTEL | TBXAS1 | Thromboxane-A synthase | 5.600704 | 6.112514 | 7.19382 | mammals | actual | clID_2064 |
| CONGO RED | ache | Acetylcholinesterase | 4.8237 | 7.688367 | 6.668786 | widespread | predicted | clID_8243 |
| CONGO RED | CHAT | Choline O-acetyltransferase | 4.8237 | 4.847678 | 7 | mammals | predicted | clID_8243 |
| CONGO RED | DHFR | Dihydrofolate reductase | 4.8237 | 6.198158 | 6.39794 | widespread | predicted | clID_8243 |
| CONGO RED | ENPP2 | Ectonucleotide pyrophosphatase/phosphodiesterase family member 2 | 4.8237 | 6.580173 | 6.055487 | mammals | predicted | clID_8243 |
| CONGO RED | SUP35 | Eukaryotic peptide chain release factor GTP-binding subunit | 4.8237 | 7.855786 | 9 | fungi | predicted | clID_8243 |
| CONGO RED | SNCA | Alpha-synuclein | 4.8237 | 6.323601 | 6.373403 | mammals | predicted | clID_8243 |
| CONGO RED | MAPT | Microtubule-associated protein tau | 4.8237 | 6.818728 | 6.243805 | mammals | predicted | clID_8243 |
| CYCLOMORUSIN | ache | Acetylcholinesterase | 5.152897 | 7.688367 | 5.508638 | widespread | predicted | clID_5053 |
| CYCLOMORUSIN | BCHE | Cholinesterase | 5.152897 | 7.593404 | 5.769551 | mammals | predicted | clID_5053 |
| DIHYDROTANSHINONE I | ache | Acetylcholinesterase | 5.920819 | 7.688367 | 5.977271 | widespread | actual | clID_1151 |
| DIHYDROTANSHINONE I | ELAVL1 | ELAV-like protein 1 | 5.920819 | 4.394971 | 7.098445 | mammals | actual | clID_1151 |
| DIHYDROTANSHINONE I | EST1 | Liver carboxylesterase 1 | 5.920819 | 5.197757 | 6.400117 | mammals | actual | clID_1151 |
| DIHYDROTANSHINONE I | CES2 | Cocaine esterase | 5.920819 | 5.280596 | 6.096592 | mammals | actual | clID_1151 |
| DIHYDROTANSHINONE I | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.920819 | 7.5973 | 9 | mammals | actual | clID_1151 |
| DIHYDROTANSHINONE I | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.920819 | 7.484017 | 9 | mammals | actual | clID_1151 |
| HEXACHLOROPHENE | ADORA3 | Adenosine receptor A3 | 6.099502 | 6.389069 | 6.31847 | mammals | actual | clID_842 |
| HEXACHLOROPHENE | ache | Acetylcholinesterase | 6.099502 | 7.688367 | 5.520727 | widespread | actual | clID_842 |
| HEXACHLOROPHENE | EGFR | Epidermal growth factor receptor | 6.099502 | 8.436627 | 6.12575 | mammals | actual | clID_842 |
| HEXACHLOROPHENE | Esr2 | Estrogen receptor beta | 6.099502 | 8.418744 | 7.007579 | mammals | actual | clID_842 |
| HEXACHLOROPHENE | HSP90AA1 | Heat shock protein HSP 90-alpha | 6.099502 | 8.516022 | 6.113509 | mammals | actual | clID_842 |
| HEXACHLOROPHENE | LMNA | Prelamin-A/C [Cleaved into: Lamin-A/C | 6.099502 | 7.05737 | 6.024988 | mammals | actual | clID_842 |
| HEXACHLOROPHENE | MAPK1 | Mitogen-activated protein kinase 1 | 6.099502 | 7.159971 | 6.707744 | mammals | actual | clID_842 |
| HEXACHLOROPHENE | Mapk14 | Mitogen-activated protein kinase 14 | 6.099502 | 8.00432 | 7.057496 | mammals | actual | clID_842 |
| HEXACHLOROPHENE | PTGS2 | Prostaglandin G/H synthase 2 | 6.099502 | 6.427124 | 6.190912 | mammals | actual | clID_842 |
| HEXACHLOROPHENE | PF3D7_1443900 | Heat shock protein 90, putative | 6.099502 | 6.257667 | 6.170053 | protozoan | actual | clID_842 |
| HEXACHLOROPHENE | ska | Streptokinase A | 6.099502 | 6.6046 | 6.694649 | bacteria | actual | clID_842 |
| KUWANON E | ache | Acetylcholinesterase | 6.632607 | 7.688367 | 5.508638 | widespread | predicted | clID_3834 |
| KUWANON E | BCHE | Cholinesterase | 6.632607 | 7.593404 | 5.769551 | mammals | predicted | clID_3834 |
| KUWANONU | ache | Acetylcholinesterase | 6.696617 | 7.688367 | 5.508638 | widespread | predicted | clID_3834 |
| KUWANONU | BCHE | Cholinesterase | 6.696617 | 7.593404 | 5.769551 | mammals | predicted | clID_3834 |
| LICORICIDIN | ache | Acetylcholinesterase | 4.767951 | 7.688367 | 5.234105 | widespread | predicted | clID_10371 |
| LICORICIDIN | PTPN1 | Tyrosine-protein phosphatase non-receptor type 1 | 4.767951 | 7.108269 | 6.278754 | mammals | predicted | clID_10371 |
| LOVASTATIN | ache | Acetylcholinesterase | 5.114456 | 7.688367 | 5.334318 | widespread | actual | clID_5420 |
| LOVASTATIN | BCHE | Cholinesterase | 5.114456 | 7.593404 | 5.452869 | mammals | actual | clID_5420 |
| LOVASTATIN | GMNN | Geminin | 5.114456 | 7.313646 | 6.159255 | mammals | actual | clID_5420 |
| LOVASTATIN | HMGCR | 3-hydroxy-3-methylglutaryl-coenzyme A reductase | 5.114456 | 6.327425 | 7.012084 | mammals | actual | clID_5420 |
| LOVASTATIN | SLCO1A2 | Solute carrier organic anion transporter family member 1A2 | 5.114456 | 6.782314 | 6.278754 | mammals | actual | clID_5420 |
| LOVASTATIN | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.114456 | 7.484017 | 9 | mammals | actual | clID_5420 |
| NIFEDIPINE | ache | Acetylcholinesterase | 4.683767 | 7.688367 | 5.24897 | widespread | actual | clID_6999 |
| NIFEDIPINE | Chrm1 | Muscarinic acetylcholine receptor M1 | 4.683767 | 6.534599 | 7.344893 | mammals | actual | clID_6999 |
| NIFEDIPINE | CHRM2 | Muscarinic acetylcholine receptor M2 | 4.683767 | 6.324426 | 7.853872 | mammals | actual | clID_6999 |
| NIFEDIPINE | Chrm3 | Muscarinic acetylcholine receptor M3 | 4.683767 | 6.207045 | 7.853872 | mammals | actual | clID_6999 |
| NIFEDIPINE | CHRM4 | Muscarinic acetylcholine receptor M4 | 4.683767 | 5.866392 | 7.853872 | mammals | actual | clID_6999 |
| NIFEDIPINE | Chrm5 | Muscarinic acetylcholine receptor M5 | 4.683767 | 5.800726 | 7.853872 | mammals | actual | clID_6999 |
| NIFEDIPINE | ampC | Beta-lactamase | 4.683767 | 6.324293 | 6.450016 | bacteria | actual | clID_6999 |
| NIFEDIPINE | CACNA1C | Voltage-dependent L-type calcium channel subunit alpha-1C | 4.683767 | 8.516207 | 6.193024 | mammals | actual | clID_6999 |
| NIFEDIPINE | Cacna1d | Voltage-dependent L-type calcium channel subunit alpha-1D | 4.683767 | 8.332342 | 6.019461 | mammals | actual | clID_6999 |
| NIFEDIPINE | BCHE | Cholinesterase | 4.683767 | 7.593404 | 5.440273 | mammals | actual | clID_6999 |
| NIFEDIPINE | Cyp1a2 | Cytochrome P450 1A2 | 4.683767 | 6.507005 | 6.39223 | mammals | actual | clID_6999 |
| NIFEDIPINE | HIF1A | Hypoxia-inducible factor 1-alpha | 4.683767 | 6.498485 | 6.199993 | mammals | actual | clID_6999 |
| NIFEDIPINE | pfk | ATP-dependent 6-phosphofructokinase | 4.683767 | 6.582628 | 6.07099 | protozoan | actual | clID_6999 |
| NIFEDIPINE | RGS4 | Regulator of G-protein signaling 4 | 4.683767 | 6.451243 | 6.448927 | mammals | actual | clID_6999 |
| NIFEDIPINE | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.683767 | 7.484017 | 9 | mammals | actual | clID_6999 |
| NIFEDIPINE | TRPA1 | Transient receptor potential cation channel subfamily A member 1 | 4.683767 | 5.547422 | 6.39794 | mammals | actual | clID_6999 |
| NIFEDIPINE | TSHR | Thyrotropin receptor | 4.683767 | 6.41967 | 6.799971 | mammals | actual | clID_6999 |
| NYMPHAEOL A | ache | Acetylcholinesterase | 5.29243 | 7.688367 | 5.339806 | widespread | actual | clID_3834 |
| NYMPHAEOL A | ALOX5 | Arachidonate 5-lipoxygenase | 5.29243 | 6.695092 | 6.200515 | mammals | actual | clID_3834 |
| NYMPHAEOL A | BCHE | Carboxylic ester hydrolase | 5.29243 | 6.634179 | 5.887345 | mammals | actual | clID_3834 |
| OXICONAZOLE | ADORA3 | Adenosine receptor A3 | 4.795178 | 6.389069 | 6.321479 | mammals | actual | clID_2831 |
| OXICONAZOLE | ache | Acetylcholinesterase | 4.795178 | 7.688367 | 5.332874 | widespread | actual | clID_2831 |
| OXICONAZOLE | CYP2C19 | Cytochrome P450 2C19 | 4.795178 | 6.646306 | 7.39794 | mammals | actual | clID_2831 |
| OXICONAZOLE | CYP2D6 | Cytochrome P450 2D6 | 4.795178 | 6.857981 | 6 | mammals | actual | clID_2831 |
| OXICONAZOLE | CYP3A4 | Cytochrome P450 3A4 | 4.795178 | 6.852626 | 7.045757 | mammals | actual | clID_2831 |
| OXICONAZOLE | SLC6A4 | Sodium-dependent serotonin transporter | 4.795178 | 9.398949 | 6.234182 | mammals | actual | clID_2831 |
| OXICONAZOLE | TBXAS1 | Thromboxane-A synthase | 4.795178 | 6.112514 | 6.209433 | mammals | actual | clID_2831 |
| PAPAVERINE | ache | Acetylcholinesterase | 5.795233 | 7.688367 | 5.009798 | widespread | actual | clID_1423 |
| PAPAVERINE | Chrm1 | Muscarinic acetylcholine receptor M1 | 5.795233 | 6.534599 | 6.175131 | mammals | actual | clID_1423 |
| PAPAVERINE | SMPD1 | Sphingomyelin phosphodiesterase | 5.795233 | 6.037408 | 9 | mammals | actual | clID_1423 |
| PAPAVERINE | CBX1 | Chromobox protein homolog 1 | 5.795233 | 6.203048 | 8.221849 | mammals | actual | clID_1423 |
| PAPAVERINE | Pde10a | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | 5.795233 | 7.332317 | 7.304421 | mammals | actual | clID_1423 |
| PAPAVERINE | Q9XSW7 | Phosphodiesterase | 5.795233 | 6.9702 | 6.207608 | mammals | actual | clID_1423 |
| PAPAVERINE | RORC | Nuclear receptor ROR-gamma | 5.795233 | 7.548863 | 6.72642 | mammals | actual | clID_1423 |
| PAPAVERINE | Txnrd1 | Thioredoxin reductase 1, cytoplasmic | 5.795233 | 6.729015 | 6.47379 | mammals | actual | clID_1423 |
| PAZOPANIB | ache | Acetylcholinesterase | 4.969786 | 7.688367 | 6.031517 | widespread | actual | clID_7154 |
| PAZOPANIB | AURKC | Aurora kinase C | 4.969786 | 6.63628 | 6.124939 | mammals | actual | clID_7154 |
| PAZOPANIB | BMPR1B | Bone morphogenetic protein receptor type-1B | 4.969786 | 6.897843 | 6 | mammals | actual | clID_7154 |
| PAZOPANIB | BRAF | Serine/threonine-protein kinase B-raf | 4.969786 | 7.59131 | 6.251604 | mammals | actual | clID_7154 |
| PAZOPANIB | CPK1 | Calcium-dependent protein kinase 1 | 4.969786 | 4.957876 | 6.431798 | protozoan | actual | clID_7154 |
| PAZOPANIB | CSF1R | Macrophage colony-stimulating factor 1 receptor | 4.969786 | 6.594427 | 7.46054 | mammals | actual | clID_7154 |
| PAZOPANIB | DDR1 | Epithelial discoidin domain-containing receptor 1 | 4.969786 | 8.027535 | 7.122559 | mammals | actual | clID_7154 |
| PAZOPANIB | DDR2 | Discoidin domain-containing receptor 2 | 4.969786 | 7.44901 | 7.002118 | mammals | actual | clID_7154 |
| PAZOPANIB | EPHB6 | Ephrin type-B receptor 6 | 4.969786 | 7.324716 | 7.091515 | mammals | actual | clID_7154 |
| PAZOPANIB | FGFR1 | Fibroblast growth factor receptor 1 | 4.969786 | 7.460342 | 6.490082 | mammals | actual | clID_7154 |
| PAZOPANIB | FGFR2 | Fibroblast growth factor receptor 2 | 4.969786 | 7.564071 | 6.677781 | mammals | actual | clID_7154 |
| PAZOPANIB | FGFR3 | Fibroblast growth factor receptor 3 | 4.969786 | 7.237004 | 6.169188 | mammals | actual | clID_7154 |
| PAZOPANIB | FRK | Tyrosine-protein kinase FRK | 4.969786 | 6.10774 | 6.124939 | mammals | actual | clID_7154 |
| PAZOPANIB | GAK | Cyclin-G-associated kinase | 4.969786 | 6.967052 | 6.69897 | mammals | actual | clID_7154 |
| PAZOPANIB | IRAK3 | Interleukin-1 receptor-associated kinase 3 | 4.969786 | 6.910822 | 6.09691 | mammals | actual | clID_7154 |
| PAZOPANIB | Kit | Mast/stem cell growth factor receptor Kit | 4.969786 | 7.391328 | 7.592998 | mammals | actual | clID_7154 |
| PAZOPANIB | LCK | Tyrosine-protein kinase Lck | 4.969786 | 7.251968 | 6.047212 | mammals | actual | clID_7154 |
| PAZOPANIB | LYN | Tyrosine-protein kinase Lyn | 4.969786 | 6.143602 | 6.004609 | mammals | actual | clID_7154 |
| PAZOPANIB | MAP3K11 | Mitogen-activated protein kinase kinase kinase 11 | 4.969786 | 6.834073 | 6.027761 | mammals | actual | clID_7154 |
| PAZOPANIB | MAP3K19 | Mitogen-activated protein kinase kinase kinase 19 | 4.969786 | 7.360292 | 6.026872 | mammals | actual | clID_7154 |
| PAZOPANIB | MAP3K2 | Mitogen-activated protein kinase kinase kinase 2 | 4.969786 | 6.807188 | 6.537602 | mammals | actual | clID_7154 |
| PAZOPANIB | MAP3K9 | Mitogen-activated protein kinase kinase kinase 9 | 4.969786 | 6.873014 | 6.537602 | mammals | actual | clID_7154 |
| PAZOPANIB | MAP4K1 | Mitogen-activated protein kinase kinase kinase kinase 1 | 4.969786 | 6.883314 | 6.124939 | mammals | actual | clID_7154 |
| PAZOPANIB | MAP4K4 | Mitogen-activated protein kinase kinase kinase kinase 4 | 4.969786 | 5.293872 | 6.078787 | mammals | actual | clID_7154 |
| PAZOPANIB | MAP2K4 | Dual specificity mitogen-activated protein kinase kinase 4 | 4.969786 | 6.312999 | 6.229148 | mammals | actual | clID_7154 |
| PAZOPANIB | MAP2K5 | Dual specificity mitogen-activated protein kinase kinase 5 | 4.969786 | 7.421013 | 6.318759 | mammals | actual | clID_7154 |
| PAZOPANIB | NEK2 | Serine/threonine-protein kinase Nek2 | 4.969786 | 6.186541 | 6.139182 | mammals | actual | clID_7154 |
| PAZOPANIB | PDGFRA | Platelet-derived growth factor receptor alpha | 4.969786 | 6.815301 | 7.492087 | mammals | actual | clID_7154 |
| PAZOPANIB | Pdgfrb | Platelet-derived growth factor receptor beta | 4.969786 | 7.277622 | 7.720187 | mammals | actual | clID_7154 |
| PAZOPANIB | PIP4K2C | Phosphatidylinositol 5-phosphate 4-kinase type-2 gamma | 4.969786 | 6.678971 | 6.552842 | mammals | actual | clID_7154 |
| PAZOPANIB | PI4KB | Phosphatidylinositol 4-kinase beta | 4.969786 | 5.87934 | 6.017729 | mammals | actual | clID_7154 |
| PAZOPANIB | PLK4 | Serine/threonine-protein kinase PLK4 | 4.969786 | 5.412235 | 6.537602 | mammals | actual | clID_7154 |
| PAZOPANIB | RAF1 | RAF proto-oncogene serine/threonine-protein kinase | 4.969786 | 7.668057 | 6.045757 | mammals | actual | clID_7154 |
| PAZOPANIB | RET | Proto-oncogene tyrosine-protein kinase receptor Ret | 4.969786 | 6.603209 | 6.332075 | mammals | actual | clID_7154 |
| PAZOPANIB | RIOK2 | Serine/threonine-protein kinase RIO2 | 4.969786 | 6.079664 | 6.21467 | mammals | actual | clID_7154 |
| PAZOPANIB | RIPK1 | Receptor-interacting serine/threonine-protein kinase 1 | 4.969786 | 6.203598 | 6.585027 | mammals | actual | clID_7154 |
| PAZOPANIB | ROS1 | Proto-oncogene tyrosine-protein kinase ROS | 4.969786 | 6.142592 | 6.036212 | mammals | actual | clID_7154 |
| PAZOPANIB | Slk | STE20-like serine/threonine-protein kinase | 4.969786 | 5.863513 | 6.619789 | mammals | actual | clID_7154 |
| PAZOPANIB | STK10 | Serine/threonine-protein kinase 10 | 4.969786 | 7.104293 | 7.075721 | mammals | actual | clID_7154 |
| PAZOPANIB | STK16 | Serine/threonine-protein kinase 16 | 4.969786 | 6.533139 | 6.443697 | mammals | actual | clID_7154 |
| PAZOPANIB | STK36 | Serine/threonine-protein kinase 36 | 4.969786 | 6.695916 | 6.327902 | mammals | actual | clID_7154 |
| PAZOPANIB | TAOK1 | Serine/threonine-protein kinase TAO1 | 4.969786 | 6.323898 | 6.359894 | mammals | actual | clID_7154 |
| PAZOPANIB | TAOK3 | Serine/threonine-protein kinase TAO3 | 4.969786 | 7.019785 | 7.346787 | mammals | actual | clID_7154 |
| PAZOPANIB | TIE1 | Tyrosine-protein kinase receptor Tie-1 | 4.969786 | 6.828475 | 6.154902 | mammals | actual | clID_7154 |
| PAZOPANIB | TNIK | TRAF2 and NCK-interacting protein kinase | 4.969786 | 7.190176 | 6.508638 | mammals | actual | clID_7154 |
| PAZOPANIB | TNK1 | Non-receptor tyrosine-protein kinase TNK1 | 4.969786 | 6.490835 | 6.008774 | mammals | actual | clID_7154 |
| PAZOPANIB | TTK | Dual specificity protein kinase TTK | 4.969786 | 7.123024 | 6.823909 | mammals | actual | clID_7154 |
| PAZOPANIB | FLT1 | Vascular endothelial growth factor receptor 1 | 4.969786 | 7.159396 | 7.703298 | mammals | actual | clID_7154 |
| PAZOPANIB | Kdr | Vascular endothelial growth factor receptor 2 | 4.969786 | 7.82472 | 7.15109 | mammals | actual | clID_7154 |
| PAZOPANIB | FLT4 | Vascular endothelial growth factor receptor 3 | 4.969786 | 6.814388 | 7.450604 | mammals | actual | clID_7154 |
| PROPOXUR | ache | Acetylcholinesterase | 5.197005 | 7.688367 | 6.190098 | widespread | actual | clID_4636 |
| PROPOXUR | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 5.197005 | 7.5973 | 9 | mammals | actual | clID_4636 |
| PROPOXUR | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 5.197005 | 7.484017 | 9 | mammals | actual | clID_4636 |
| QUINIDINE | BCHE | Cholinesterase | 4.856749 | 7.593404 | 5.521314 | mammals | actual | clID_530 |
| QUINIDINE | CYP2D6 | Cytochrome P450 2D6 | 4.856749 | 6.857981 | 7.02473 | mammals | actual | clID_530 |
| QUINIDINE | KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 4.856749 | 7.946483 | 6.364291 | mammals | actual | clID_530 |
| QUINIDINE | SLCO1A2 | Solute carrier organic anion transporter family member 1A2 | 4.856749 | 6.782314 | 6.004799 | mammals | actual | clID_530 |
| RUTIN | Adora1 | Adenosine receptor A1 | 4.838698 | 6.453453 | 9 | mammals | actual | clID_1263 |
| RUTIN | ache | Acetylcholinesterase | 4.838698 | 7.688367 | 7.920819 | widespread | actual | clID_1263 |
| RUTIN | ESR1 | Estrogen receptor | 4.838698 | 8.667211 | 7.49793 | mammals | actual | clID_1263 |
| RUTIN | Acp5 | Tartrate-resistant acid phosphatase type 5 | 4.838698 | 8.083753 | 7.238371 | mammals | actual | clID_1263 |
| SACCHARIN | A6YCJ1 | Astrosclerin-3 | 4.783147 | 4.659198 | 6.12436 | notRecorded | actual | clID_8142 |
| SACCHARIN | SYO3AOP1_0578 | Carbonate dehydratase | 4.783147 | 4.640052 | 6.057496 | bacteria | actual | clID_8142 |
| SACCHARIN | CA | Alpha carbonic anhydrase | 4.783147 | 4.73442 | 7.394695 | notRecorded | actual | clID_8142 |
| SACCHARIN | CA2 | Carbonic anhydrase | 4.783147 | 4.844608 | 6.982967 | notRecorded | actual | clID_8142 |
| SACCHARIN | CA12 | Carbonic anhydrase 12 | 4.783147 | 5.439747 | 6.196678 | mammals | actual | clID_8142 |
| SACCHARIN | CA14 | Carbonic anhydrase 14 | 4.783147 | 5.297097 | 6.111821 | mammals | actual | clID_8142 |
| SACCHARIN | CA6 | Carbonic anhydrase 6 | 4.783147 | 5.235941 | 6.029188 | mammals | actual | clID_8142 |
| SACCHARIN | CA7 | Carbonic anhydrase 7 | 4.783147 | 5.589434 | 8 | mammals | actual | clID_8142 |
| SACCHARIN | CA9 | Carbonic anhydrase 9 | 4.783147 | 5.53441 | 6.990372 | mammals | actual | clID_8142 |
| SACCHARIN | BCHE | Cholinesterase | 4.783147 | 7.593404 | 5.437828 | mammals | actual | clID_8142 |
| SACCHARIN | GMNN | Geminin | 4.783147 | 7.313646 | 9.09691 | mammals | actual | clID_8142 |
| SACCHARIN | LMNA | Prelamin-A/C [Cleaved into: Lamin-A/C | 4.783147 | 7.05737 | 8.251812 | mammals | actual | clID_8142 |
| SACCHARIN | mtcA2 | Carbonic anhydrase 2 | 4.783147 | 4.619486 | 6.101275 | bacteria | actual | clID_8142 |
| SACCHARIN | Tcr_1545 | Carbonic anhydrase, alpha family | 4.783147 | 4.602024 | 6.030118 | bacteria | actual | clID_8142 |
| SACCHARIN | 1272966 | Carbonic anhydrase | 4.783147 | 4.784702 | 7.020907 | arthropoda | actual | clID_8142 |
| SACCHARIN | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 4.783147 | 7.5973 | 9 | mammals | actual | clID_8142 |
| SACCHARIN | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 4.783147 | 7.484017 | 9 | mammals | actual | clID_8142 |
| SACCHARIN | THRB | Thyroid hormone receptor beta | 4.783147 | 5.722362 | 8.60206 | mammals | actual | clID_8142 |
| SACCHARIN | VDR | Vitamin D3 receptor | 4.783147 | 7.025036 | 8.251812 | mammals | actual | clID_8142 |
| TRIFLUPROMAZINE | Chrm1 | Muscarinic acetylcholine receptor M1 | 5.780435 | 6.534599 | 6.399985 | mammals | actual | clID_1458 |
| TRIFLUPROMAZINE | BCHE | Cholinesterase | 5.780435 | 7.593404 | 5.130768 | mammals | actual | clID_1458 |
| TRIFLUPROMAZINE | CYP2D6 | Cytochrome P450 2D6 | 5.780435 | 6.857981 | 6.250005 | mammals | actual | clID_1458 |
| TRIFLUPROMAZINE | Drd1 | D(1A) dopamine receptor | 5.780435 | 7.436039 | 6.143136 | mammals | actual | clID_1458 |
Use search function in table to view values for specific interactions
| molName | geneName | protname | sarsScoreMolecule | geneScore | interactionScore | organismType | kindOfValue | molecularCluster |
|---|---|---|---|---|---|---|---|---|
| KETOCONAZOLE | CYP11B1 | Cytochrome P450 11B1, mitochondrial | 5.777396 | 5.119144 | 6.535429 | mammals | actual | clID_6688 |
| KETOCONAZOLE | Cyp11b2 | Cytochrome P450 11B2, mitochondrial | 5.777396 | 4.682681 | 6.481372 | mammals | actual | clID_6688 |
| KETOCONAZOLE | CYP17A1 | Steroid 17-alpha-hydroxylase/17,20 lyase | 5.777396 | 6.366169 | 6.011085 | mammals | actual | clID_6688 |
| KETOCONAZOLE | CYP24A1 | 1,25-dihydroxyvitamin D(3) 24-hydroxylase, mitochondrial | 5.777396 | 8.196691 | 6.313272 | mammals | actual | clID_6688 |
| KETOCONAZOLE | CYP27B1 | 25-hydroxyvitamin D-1 alpha hydroxylase, mitochondrial | 5.777396 | 8.453908 | 6.765812 | mammals | actual | clID_6688 |
| KETOCONAZOLE | Cyp2a2 | Cytochrome P450 2A2 | 5.777396 | 9.532294 | 6.349087 | mammals | actual | clID_6688 |
| KETOCONAZOLE | CYP3A4 | Cytochrome P450 3A4 | 5.777396 | 6.852626 | 6.763857 | mammals | actual | clID_6688 |
| KETOCONAZOLE | CYP3A5 | Cytochrome P450 3A5 | 5.777396 | 6.541939 | 6.275275 | mammals | actual | clID_6688 |
| KETOCONAZOLE | CYP51A1 | Lanosterol 14-alpha demethylase | 5.777396 | 7.713871 | 6.886986 | mammals | actual | clID_6688 |
| KETOCONAZOLE | Cyp7a1 | Cytochrome P450 7A1 | 5.777396 | 7.643246 | 6.164877 | mammals | actual | clID_6688 |
| KETOCONAZOLE | camC | Camphor 5-monooxygenase | 5.777396 | 4.581474 | 6.261439 | bacteria | actual | clID_6688 |
| KETOCONAZOLE | GNRHR | Gonadotropin-releasing hormone receptor | 5.777396 | 9.328642 | 7.717881 | mammals | actual | clID_6688 |
| KETOCONAZOLE | HSP90AA1 | Heat shock protein HSP 90-alpha | 5.777396 | 8.516022 | 6.361511 | mammals | actual | clID_6688 |
| KETOCONAZOLE | Kcnma1 | Calcium-activated potassium channel subunit alpha-1 | 5.777396 | 5.67589 | 7.818086 | mammals | actual | clID_6688 |
| KETOCONAZOLE | KCNMB1 | Calcium-activated potassium channel subunit beta-1 | 5.777396 | 6.648685 | 7.818086 | mammals | actual | clID_6688 |
| KETOCONAZOLE | LMNA | Prelamin-A/C [Cleaved into: Lamin-A/C | 5.777396 | 7.05737 | 6.42539 | mammals | actual | clID_6688 |
| KETOCONAZOLE | SLC6A4 | Sodium-dependent serotonin transporter | 5.777396 | 9.398949 | 6.412328 | mammals | actual | clID_6688 |
| KETOCONAZOLE | SLCO1A2 | Solute carrier organic anion transporter family member 1A2 | 5.777396 | 6.782314 | 9 | mammals | actual | clID_6688 |
| SUFUGOLIX | GNRHR | Gonadotropin-releasing hormone receptor | 5.148198 | 9.328642 | 8.303246 | mammals | predicted | clID_5096 |
Use search function in table to view values for specific interactions
| molName | geneName | protname | sarsScoreMolecule | geneScore | interactionScore | organismType | kindOfValue | molecularCluster |
|---|---|---|---|---|---|---|---|---|
| AMENTOFLAVONE | GABRA1 | Gamma-aminobutyric acid receptor subunit alpha-1 | 5.080922 | 6.595702 | 7.934496 | mammals | actual | clID_459 |
| AMENTOFLAVONE | GABRA2 | Gamma-aminobutyric acid receptor subunit alpha-2 | 5.080922 | 6.56009 | 8.09017 | mammals | actual | clID_459 |
| AMENTOFLAVONE | GABRA3 | Gamma-aminobutyric acid receptor subunit alpha-3 | 5.080922 | 6.540145 | 8.09017 | mammals | actual | clID_459 |
| AMENTOFLAVONE | GABRA4 | Gamma-aminobutyric acid receptor subunit alpha-4 | 5.080922 | 6.455052 | 8.09017 | mammals | actual | clID_459 |
| AMENTOFLAVONE | Gabra5 | Gamma-aminobutyric acid receptor subunit alpha-5 | 5.080922 | 6.549437 | 8.09017 | mammals | actual | clID_459 |
| AMENTOFLAVONE | Gabra6 | Gamma-aminobutyric acid receptor subunit alpha-6 | 5.080922 | 6.496227 | 8.09017 | mammals | actual | clID_459 |
| AMENTOFLAVONE | GABRB1 | Gamma-aminobutyric acid receptor subunit beta-1 | 5.080922 | 6.448271 | 8.09017 | mammals | actual | clID_459 |
| AMENTOFLAVONE | GABRB2 | Gamma-aminobutyric acid receptor subunit beta-2 | 5.080922 | 6.507898 | 7.934496 | mammals | actual | clID_459 |
| AMENTOFLAVONE | GABRB3 | Gamma-aminobutyric acid receptor subunit beta-3 | 5.080922 | 6.609073 | 8.09017 | mammals | actual | clID_459 |
| AMENTOFLAVONE | GABRD | Gamma-aminobutyric acid receptor subunit delta | 5.080922 | 6.475717 | 8.09017 | mammals | actual | clID_459 |
| AMENTOFLAVONE | GABRE | Gamma-aminobutyric acid receptor subunit epsilon | 5.080922 | 6.43215 | 8.09017 | mammals | actual | clID_459 |
| AMENTOFLAVONE | Gabrg1 | Gamma-aminobutyric acid receptor subunit gamma-1 | 5.080922 | 6.445115 | 8.09017 | mammals | actual | clID_459 |
| AMENTOFLAVONE | GABRG2 | Gamma-aminobutyric acid receptor subunit gamma-2 | 5.080922 | 6.599199 | 7.934496 | mammals | actual | clID_459 |
| AMENTOFLAVONE | Gabrg3 | Gamma-aminobutyric acid receptor subunit gamma-3 | 5.080922 | 6.440188 | 8.09017 | mammals | actual | clID_459 |
| AMENTOFLAVONE | GABRP | Gamma-aminobutyric acid receptor subunit pi | 5.080922 | 6.440268 | 8.09017 | mammals | actual | clID_459 |
| AMENTOFLAVONE | Gabrq | Gamma-aminobutyric acid receptor subunit theta | 5.080922 | 5.60614 | 8.221849 | mammals | actual | clID_459 |
| AMENTOFLAVONE | Oprd1 | Delta-type opioid receptor | 5.080922 | 7.770012 | 6.88524 | mammals | actual | clID_459 |
| AMENTOFLAVONE | Oprk1 | Kappa-type opioid receptor | 5.080922 | 7.699064 | 6.309804 | mammals | actual | clID_459 |
| AMENTOFLAVONE | PGF | Placenta growth factor | 5.080922 | 5.038878 | 8.086186 | mammals | actual | clID_459 |
| AMENTOFLAVONE | Gabrq | GABA theta subunit | 5.080922 | 6.382318 | 7.826814 | mammals | actual | clID_459 |
| AMENTOFLAVONE | reverse | Reverse transcriptase | 5.080922 | 7.682466 | 6.556749 | virus | actual | clID_459 |
| AMENTOFLAVONE | VCP | Transitional endoplasmic reticulum ATPase | 5.080922 | 7.802163 | 5.04653 | mammals | actual | clID_459 |
| AMENTOFLAVONE | VEGFA | Vascular endothelial growth factor A | 5.080922 | 6.3527 | 7.782516 | mammals | actual | clID_459 |
| AMENTOFLAVONE | Xdh | Xanthine dehydrogenase/oxidase [Includes: Xanthine dehydrogenase | 5.080922 | 4.956934 | 9 | mammals | actual | clID_459 |
| CB-5083 | PRKDC | DNA-dependent protein kinase catalytic subunit | 5.495867 | 5.887635 | 6.30103 | mammals | actual | clID_2541 |
| CB-5083 | VCP | Transitional endoplasmic reticulum ATPase | 5.495867 | 7.802163 | 6.327254 | mammals | actual | clID_2541 |
| DIAZIRINYL ARCHAZOLID A | VATC | V-type proton ATPase subunit C | 6.182461 | 12.665952 | 6.221849 | arthropoda | predicted | clID_722 |
| DIGITOXIGENIN | ATP12A | Potassium-transporting ATPase alpha chain 2 | 5.176634 | 8.798384 | 7.09691 | mammals | actual | clID_364 |
| DIGITOXIGENIN | ATP1A1 | Sodium/potassium-transporting ATPase subunit alpha-1 | 5.176634 | 7.258587 | 7.11674 | mammals | actual | clID_364 |
| DIGITOXIGENIN | Atp1a2 | Sodium/potassium-transporting ATPase subunit alpha-2 | 5.176634 | 7.204732 | 7.145873 | mammals | actual | clID_364 |
| DIGITOXIGENIN | Atp1a3 | Sodium/potassium-transporting ATPase subunit alpha-3 | 5.176634 | 7.206256 | 7.145873 | mammals | actual | clID_364 |
| DIGITOXIGENIN | ATP1A4 | Sodium/potassium-transporting ATPase subunit alpha-4 | 5.176634 | 7.166569 | 7.145873 | mammals | actual | clID_364 |
| DIGITOXIGENIN | ATP1B1 | Sodium/potassium-transporting ATPase subunit beta-1 | 5.176634 | 7.169881 | 7.145873 | mammals | actual | clID_364 |
| DIGITOXIGENIN | ATP1B2 | Sodium/potassium-transporting ATPase subunit beta-2 | 5.176634 | 7.167865 | 7.145873 | mammals | actual | clID_364 |
| DIGITOXIGENIN | ATP1B3 | Sodium/potassium-transporting ATPase subunit beta-3 | 5.176634 | 7.165775 | 7.145873 | mammals | actual | clID_364 |
| DIGITOXIGENIN | ATAD5 | ATPase family AAA domain-containing protein 5 | 5.176634 | 7.207699 | 6.118216 | mammals | actual | clID_364 |
| DIGITOXIGENIN | FXYD2 | Sodium/potassium-transporting ATPase subunit gamma | 5.176634 | 7.167865 | 7.145873 | mammals | actual | clID_364 |
| DIGITOXIGENIN | Glp1r | Glucagon-like peptide 1 receptor | 5.176634 | 7.163203 | 6.299989 | mammals | actual | clID_364 |
| DIGITOXIGENIN | IDH1 | Isocitrate dehydrogenase [NADP] cytoplasmic | 5.176634 | 6.995416 | 6.286174 | mammals | actual | clID_364 |
| DIGITOXIGENIN | Nfe2l2 | Nuclear factor erythroid 2-related factor 2 | 5.176634 | 6.942004 | 6.636165 | mammals | actual | clID_364 |
| DIGITOXIGENIN | TSHR | Thyrotropin receptor | 5.176634 | 6.41967 | 6.321027 | mammals | actual | clID_364 |
| DIGITOXIN | ALB | Serum albumin | 6.638272 | 7.03501 | 6.453498 | mammals | actual | clID_364 |
| DIGITOXIN | AR | Androgen receptor | 6.638272 | 6.822547 | 7.300162 | mammals | actual | clID_364 |
| DIGITOXIN | ATP1A1 | Sodium/potassium-transporting ATPase subunit alpha-1 | 6.638272 | 7.258587 | 8.09691 | mammals | actual | clID_364 |
| DIGITOXIN | Atp1a2 | Sodium/potassium-transporting ATPase subunit alpha-2 | 6.638272 | 7.204732 | 8.09691 | mammals | actual | clID_364 |
| DIGITOXIN | Atp1a3 | Sodium/potassium-transporting ATPase subunit alpha-3 | 6.638272 | 7.206256 | 8.09691 | mammals | actual | clID_364 |
| DIGITOXIN | ATP1A4 | Sodium/potassium-transporting ATPase subunit alpha-4 | 6.638272 | 7.166569 | 8.09691 | mammals | actual | clID_364 |
| DIGITOXIN | ATP1B1 | Sodium/potassium-transporting ATPase subunit beta-1 | 6.638272 | 7.169881 | 8.09691 | mammals | actual | clID_364 |
| DIGITOXIN | ATP1B2 | Sodium/potassium-transporting ATPase subunit beta-2 | 6.638272 | 7.167865 | 8.09691 | mammals | actual | clID_364 |
| DIGITOXIN | ATP1B3 | Sodium/potassium-transporting ATPase subunit beta-3 | 6.638272 | 7.165775 | 8.09691 | mammals | actual | clID_364 |
| DIGITOXIN | ATAD5 | ATPase family AAA domain-containing protein 5 | 6.638272 | 7.207699 | 6.836242 | mammals | actual | clID_364 |
| DIGITOXIN | FXYD2 | Sodium/potassium-transporting ATPase subunit gamma | 6.638272 | 7.167865 | 8.09691 | mammals | actual | clID_364 |
| DIGITOXIN | ESR1 | Estrogen receptor | 6.638272 | 8.667211 | 6.300006 | mammals | actual | clID_364 |
| DIGITOXIN | NR3C1 | Glucocorticoid receptor | 6.638272 | 8.072674 | 6.450016 | mammals | actual | clID_364 |
| DIGITOXIN | GMNN | Geminin | 6.638272 | 7.313646 | 6.755689 | mammals | actual | clID_364 |
| DIGITOXIN | IDH1 | Isocitrate dehydrogenase [NADP] cytoplasmic | 6.638272 | 6.995416 | 6.686133 | mammals | actual | clID_364 |
| DIGITOXIN | LMNA | Prelamin-A/C [Cleaved into: Lamin-A/C | 6.638272 | 7.05737 | 6.050018 | mammals | actual | clID_364 |
| DIGITOXIN | Nfe2l2 | Nuclear factor erythroid 2-related factor 2 | 6.638272 | 6.942004 | 7.02446 | mammals | actual | clID_364 |
| DIGITOXIN | NPC1 | NPC intracellular cholesterol transporter 1 | 6.638272 | 6.59825 | 6.59998 | mammals | actual | clID_364 |
| DIGITOXIN | Nr1h4 | Bile acid receptor | 6.638272 | 6.87787 | 6.349984 | mammals | actual | clID_364 |
| DIGITOXIN | Tp53 | Cellular tumor antigen p53 | 6.638272 | 6.353776 | 7.500313 | mammals | actual | clID_364 |
| DIGITOXIN | Ppard | Peroxisome proliferator-activated receptor delta | 6.638272 | 6.116712 | 6.550009 | mammals | actual | clID_364 |
| DIGITOXIN | Pparg | Peroxisome proliferator-activated receptor gamma | 6.638272 | 6.626045 | 6.500038 | mammals | actual | clID_364 |
| DIGITOXIN | RAB9A | Ras-related protein Rab-9A | 6.638272 | 6.645667 | 6.649946 | mammals | actual | clID_364 |
| DIGITOXIN | RXRA | Retinoic acid receptor RXR-alpha | 6.638272 | 6.799335 | 7.625039 | mammals | actual | clID_364 |
| DIGITOXIN | SLCO4C1 | Solute carrier organic anion transporter family member 4C1 | 6.638272 | 9.861004 | 6.920819 | mammals | actual | clID_364 |
| DIGITOXIN | STAT3 | Signal transducer and activator of transcription 3 | 6.638272 | 6.290226 | 6.154902 | mammals | actual | clID_364 |
| DIGITOXIN | THRB | Thyroid hormone receptor beta | 6.638272 | 5.722362 | 7.149967 | mammals | actual | clID_364 |
| DIGITOXIN | TDP1 | Tyrosyl-DNA phosphodiesterase 1 | 6.638272 | 7.013656 | 6.528682 | mammals | actual | clID_364 |
| DIGITOXIN | VDR | Vitamin D3 receptor | 6.638272 | 7.025036 | 6.250032 | mammals | actual | clID_364 |
| DIGOXIN | AR | Androgen receptor | 6.721246 | 6.822547 | 6.333324 | mammals | actual | clID_364 |
| DIGOXIN | ATP1A1 | Sodium/potassium-transporting ATPase subunit alpha-1 | 6.721246 | 7.258587 | 6.061936 | mammals | actual | clID_364 |
| DIGOXIN | Atp1a2 | Sodium/potassium-transporting ATPase subunit alpha-2 | 6.721246 | 7.204732 | 6.071068 | mammals | actual | clID_364 |
| DIGOXIN | Atp1a3 | Sodium/potassium-transporting ATPase subunit alpha-3 | 6.721246 | 7.206256 | 6.071068 | mammals | actual | clID_364 |
| DIGOXIN | ATP1A4 | Sodium/potassium-transporting ATPase subunit alpha-4 | 6.721246 | 7.166569 | 6.194303 | mammals | actual | clID_364 |
| DIGOXIN | ATP1B1 | Sodium/potassium-transporting ATPase subunit beta-1 | 6.721246 | 7.169881 | 6.194303 | mammals | actual | clID_364 |
| DIGOXIN | ATP1B2 | Sodium/potassium-transporting ATPase subunit beta-2 | 6.721246 | 7.167865 | 6.022192 | mammals | actual | clID_364 |
| DIGOXIN | ATP1B3 | Sodium/potassium-transporting ATPase subunit beta-3 | 6.721246 | 7.165775 | 6.022192 | mammals | actual | clID_364 |
| DIGOXIN | ATAD5 | ATPase family AAA domain-containing protein 5 | 6.721246 | 7.207699 | 6.68634 | mammals | actual | clID_364 |
| DIGOXIN | FXYD2 | Sodium/potassium-transporting ATPase subunit gamma | 6.721246 | 7.167865 | 6.022192 | mammals | actual | clID_364 |
| DIGOXIN | ESR1 | Estrogen receptor | 6.721246 | 8.667211 | 6.216672 | mammals | actual | clID_364 |
| DIGOXIN | NR3C1 | Glucocorticoid receptor | 6.721246 | 8.072674 | 6.125016 | mammals | actual | clID_364 |
| DIGOXIN | GMNN | Geminin | 6.721246 | 7.313646 | 6.824155 | mammals | actual | clID_364 |
| DIGOXIN | IDH1 | Isocitrate dehydrogenase [NADP] cytoplasmic | 6.721246 | 6.995416 | 6.6362 | mammals | actual | clID_364 |
| DIGOXIN | CXCL8 | Interleukin-8 | 6.721246 | 4.812332 | 6.525056 | mammals | actual | clID_364 |
| DIGOXIN | lef | Lethal factor | 6.721246 | 6.363322 | 7.500313 | bacteria | actual | clID_364 |
| DIGOXIN | Abcc2 | Canalicular multispecific organic anion transporter 1 | 6.721246 | 7.3938 | 6.995635 | mammals | actual | clID_364 |
| DIGOXIN | Nfe2l2 | Nuclear factor erythroid 2-related factor 2 | 6.721246 | 6.942004 | 6.367298 | mammals | actual | clID_364 |
| DIGOXIN | NPC1 | NPC intracellular cholesterol transporter 1 | 6.721246 | 6.59825 | 6.649946 | mammals | actual | clID_364 |
| DIGOXIN | Nr1h4 | Bile acid receptor | 6.721246 | 6.87787 | 6.383195 | mammals | actual | clID_364 |
| DIGOXIN | Tp53 | Cellular tumor antigen p53 | 6.721246 | 6.353776 | 6.733362 | mammals | actual | clID_364 |
| DIGOXIN | PAX8 | Paired box protein Pax-8 | 6.721246 | 5.085583 | 6.383 | mammals | actual | clID_364 |
| DIGOXIN | Ppard | Peroxisome proliferator-activated receptor delta | 6.721246 | 6.116712 | 6.249965 | mammals | actual | clID_364 |
| DIGOXIN | RAB9A | Ras-related protein Rab-9A | 6.721246 | 6.645667 | 6.600002 | mammals | actual | clID_364 |
| DIGOXIN | RXRA | Retinoic acid receptor RXR-alpha | 6.721246 | 6.799335 | 6.887602 | mammals | actual | clID_364 |
| DIGOXIN | Slco1a4 | Solute carrier organic anion transporter family member 1A4 | 6.721246 | 6.998048 | 6.342883 | mammals | actual | clID_364 |
| DIGOXIN | THRB | Thyroid hormone receptor beta | 6.721246 | 5.722362 | 6.633358 | mammals | actual | clID_364 |
| DIGOXIN | TNF | Tumor necrosis factor | 6.721246 | 5.878526 | 6.425043 | mammals | actual | clID_364 |
| DIGOXIN | TDP1 | Tyrosyl-DNA phosphodiesterase 1 | 6.721246 | 7.013656 | 6.203708 | mammals | actual | clID_364 |
| DIGOXIN | VDR | Vitamin D3 receptor | 6.721246 | 7.025036 | 6.125016 | mammals | actual | clID_364 |
| HEXACHLOROPHENE | ADORA3 | Adenosine receptor A3 | 6.099502 | 6.389069 | 6.31847 | mammals | actual | clID_842 |
| HEXACHLOROPHENE | EGFR | Epidermal growth factor receptor | 6.099502 | 8.436627 | 6.12575 | mammals | actual | clID_842 |
| HEXACHLOROPHENE | Esr2 | Estrogen receptor beta | 6.099502 | 8.418744 | 7.007579 | mammals | actual | clID_842 |
| HEXACHLOROPHENE | HSP90AA1 | Heat shock protein HSP 90-alpha | 6.099502 | 8.516022 | 6.113509 | mammals | actual | clID_842 |
| HEXACHLOROPHENE | LMNA | Prelamin-A/C [Cleaved into: Lamin-A/C | 6.099502 | 7.05737 | 6.024988 | mammals | actual | clID_842 |
| HEXACHLOROPHENE | MAPK1 | Mitogen-activated protein kinase 1 | 6.099502 | 7.159971 | 6.707744 | mammals | actual | clID_842 |
| HEXACHLOROPHENE | Mapk14 | Mitogen-activated protein kinase 14 | 6.099502 | 8.00432 | 7.057496 | mammals | actual | clID_842 |
| HEXACHLOROPHENE | PTGS2 | Prostaglandin G/H synthase 2 | 6.099502 | 6.427124 | 6.190912 | mammals | actual | clID_842 |
| HEXACHLOROPHENE | PF3D7_1443900 | Heat shock protein 90, putative | 6.099502 | 6.257667 | 6.170053 | protozoan | actual | clID_842 |
| HEXACHLOROPHENE | ska | Streptokinase A | 6.099502 | 6.6046 | 6.694649 | bacteria | actual | clID_842 |
| HEXACHLOROPHENE | VCP | Transitional endoplasmic reticulum ATPase | 6.099502 | 7.802163 | 5.260586 | mammals | actual | clID_842 |
| IVERMECTIN | ADORA3 | Adenosine receptor A3 | 5.52 | 6.389069 | 6.414573 | mammals | actual | clID_2585 |
| IVERMECTIN | ATP1A1 | Sodium/potassium-transporting ATPase subunit alpha-1 | 5.52 | 7.258587 | 5.0846 | mammals | actual | clID_2585 |
| IVERMECTIN | Atp1a2 | Sodium/potassium-transporting ATPase subunit alpha-2 | 5.52 | 7.204732 | 5.188425 | mammals | actual | clID_2585 |
| IVERMECTIN | Atp1a3 | Sodium/potassium-transporting ATPase subunit alpha-3 | 5.52 | 7.206256 | 5.188425 | mammals | actual | clID_2585 |
| IVERMECTIN | ATP2A1 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | 5.52 | 6.762896 | 4.772113 | mammals | actual | clID_2585 |
| IVERMECTIN | Atp2a2 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | 5.52 | 9.76373 | 5.172631 | mammals | actual | clID_2585 |
| IVERMECTIN | Glra1 | Glycine receptor subunit alpha-1 | 5.52 | 5.677586 | 6.550195 | mammals | actual | clID_2585 |
| IVERMECTIN | Glra2 | Glycine receptor subunit alpha-2 | 5.52 | 5.850529 | 6.550195 | mammals | actual | clID_2585 |
| IVERMECTIN | GLRA3 | Glycine receptor subunit alpha-3 | 5.52 | 5.216047 | 6.550195 | mammals | actual | clID_2585 |
| IVERMECTIN | Glrb | Glycine receptor subunit beta | 5.52 | 4.995783 | 6.550195 | mammals | actual | clID_2585 |
| IVERMECTIN | ABCB1 | ATP-dependent translocase ABCB1 | 5.52 | 8.720249 | 6.349589 | widespread | actual | clID_2585 |
| IVERMECTIN | Abcb1a | ATP-dependent translocase ABCB1 | 5.52 | 7.382312 | 6.30103 | mammals | actual | clID_2585 |
| IVERMECTIN | Abcb1b | ATP-dependent translocase ABCB1 | 5.52 | 7.980944 | 6.30103 | mammals | actual | clID_2585 |
| IVERMECTIN | Nr1h4 | Bile acid receptor | 5.52 | 6.87787 | 6.05983 | mammals | actual | clID_2585 |
| OMEPRAZOLE | ATP12A | Potassium-transporting ATPase alpha chain 2 | 4.681041 | 8.798384 | 5.481486 | mammals | actual | clID_3072 |
| OMEPRAZOLE | Atp4a | Potassium-transporting ATPase alpha chain 1 | 4.681041 | 8.233493 | 5.733525 | mammals | actual | clID_3072 |
| OMEPRAZOLE | ATP4B | Potassium-transporting ATPase subunit beta | 4.681041 | 8.289388 | 5.733525 | mammals | actual | clID_3072 |
| OMEPRAZOLE | Abcc2 | Canalicular multispecific organic anion transporter 1 | 4.681041 | 7.3938 | 9 | mammals | actual | clID_3072 |
| OMEPRAZOLE | VDR | Vitamin D3 receptor | 4.681041 | 7.025036 | 6.002598 | mammals | actual | clID_3072 |
| OUABAIN | AR | Androgen receptor | 7.013228 | 6.822547 | 7.525049 | mammals | actual | clID_364 |
| OUABAIN | ATP1A1 | Sodium/potassium-transporting ATPase subunit alpha-1 | 7.013228 | 7.258587 | 5.177937 | mammals | actual | clID_364 |
| OUABAIN | Atp1a2 | Sodium/potassium-transporting ATPase subunit alpha-2 | 7.013228 | 7.204732 | 5.260468 | mammals | actual | clID_364 |
| OUABAIN | Atp1a3 | Sodium/potassium-transporting ATPase subunit alpha-3 | 7.013228 | 7.206256 | 5.303944 | mammals | actual | clID_364 |
| OUABAIN | ATP1A4 | Sodium/potassium-transporting ATPase subunit alpha-4 | 7.013228 | 7.166569 | 5.542759 | mammals | actual | clID_364 |
| OUABAIN | ATP1B1 | Sodium/potassium-transporting ATPase subunit beta-1 | 7.013228 | 7.169881 | 5.533483 | mammals | actual | clID_364 |
| OUABAIN | ATP1B2 | Sodium/potassium-transporting ATPase subunit beta-2 | 7.013228 | 7.167865 | 5.16615 | mammals | actual | clID_364 |
| OUABAIN | ATP1B3 | Sodium/potassium-transporting ATPase subunit beta-3 | 7.013228 | 7.165775 | 5.376655 | mammals | actual | clID_364 |
| OUABAIN | ATAD5 | ATPase family AAA domain-containing protein 5 | 7.013228 | 7.207699 | 7.002949 | mammals | actual | clID_364 |
| OUABAIN | ATXN2 | Ataxin-2 | 7.013228 | 6.573244 | 7.287879 | mammals | actual | clID_364 |
| OUABAIN | ESR1 | Estrogen receptor | 7.013228 | 8.667211 | 6.749975 | mammals | actual | clID_364 |
| OUABAIN | NR3C1 | Glucocorticoid receptor | 7.013228 | 8.072674 | 6.550059 | mammals | actual | clID_364 |
| OUABAIN | GMNN | Geminin | 7.013228 | 7.313646 | 6.318968 | mammals | actual | clID_364 |
| OUABAIN | IDH1 | Isocitrate dehydrogenase [NADP] cytoplasmic | 7.013228 | 6.995416 | 6.886057 | mammals | actual | clID_364 |
| OUABAIN | KLF5 | Krueppel-like factor 5 | 7.013228 | 4.870683 | 6.783702 | mammals | actual | clID_364 |
| OUABAIN | LMNA | Prelamin-A/C [Cleaved into: Lamin-A/C | 7.013228 | 7.05737 | 6.540064 | mammals | actual | clID_364 |
| OUABAIN | NPC1 | NPC intracellular cholesterol transporter 1 | 7.013228 | 6.59825 | 6.799971 | mammals | actual | clID_364 |
| OUABAIN | Nr1h4 | Bile acid receptor | 7.013228 | 6.87787 | 6.550059 | mammals | actual | clID_364 |
| OUABAIN | Tp53 | Cellular tumor antigen p53 | 7.013228 | 6.353776 | 7.300162 | mammals | actual | clID_364 |
| OUABAIN | Ppard | Peroxisome proliferator-activated receptor delta | 7.013228 | 6.116712 | 6.674995 | mammals | actual | clID_364 |
| OUABAIN | Pparg | Peroxisome proliferator-activated receptor gamma | 7.013228 | 6.626045 | 6.724919 | mammals | actual | clID_364 |
| OUABAIN | RAB9A | Ras-related protein Rab-9A | 7.013228 | 6.645667 | 6.899974 | mammals | actual | clID_364 |
| OUABAIN | RXRA | Retinoic acid receptor RXR-alpha | 7.013228 | 6.799335 | 6.949987 | mammals | actual | clID_364 |
| OUABAIN | SMAD3 | Mothers against decapentaplegic homolog 3 | 7.013228 | 6.518109 | 6.151709 | mammals | actual | clID_364 |
| OUABAIN | SMN1 | Survival motor neuron protein | 7.013228 | 7.034367 | 6.662505 | mammals | actual | clID_364 |
| OUABAIN | SLCO1B1 | Solute carrier organic anion transporter family member 1B1 | 7.013228 | 7.5973 | 7.004343 | mammals | actual | clID_364 |
| OUABAIN | SLCO1B3 | Solute carrier organic anion transporter family member 1B3 | 7.013228 | 7.484017 | 6.71996 | mammals | actual | clID_364 |
| OUABAIN | SLCO4C1 | Solute carrier organic anion transporter family member 4C1 | 7.013228 | 9.861004 | 6.420216 | mammals | actual | clID_364 |
| OUABAIN | THRB | Thyroid hormone receptor beta | 7.013228 | 5.722362 | 6.899974 | mammals | actual | clID_364 |
| OUABAIN | VDR | Vitamin D3 receptor | 7.013228 | 7.025036 | 7.025117 | mammals | actual | clID_364 |
Use search function in table to view values for specific interactions
| molName | geneName | protname | sarsScoreMolecule | geneScore | interactionScore | organismType | kindOfValue | molecularCluster |
|---|---|---|---|---|---|---|---|---|
| APICIDIN | Hdac10 | Polyamine deacetylase HDAC10 | 4.559786 | 6.714438 | 7.896842 | mammals | actual | clID_1175 |
| APICIDIN | Hdac11 | Histone deacetylase 11 | 4.559786 | 6.69745 | 7.896842 | mammals | actual | clID_1175 |
| APICIDIN | Hdac1 | Histone deacetylase 1 | 4.559786 | 6.964004 | 7.898024 | mammals | actual | clID_1175 |
| APICIDIN | Hdac2 | Histone deacetylase 2 | 4.559786 | 6.79746 | 7.914313 | mammals | actual | clID_1175 |
| APICIDIN | HDAC3 | Histone deacetylase 3 | 4.559786 | 6.743628 | 7.907742 | mammals | actual | clID_1175 |
| APICIDIN | HDAC4 | Histone deacetylase 4 | 4.559786 | 6.710525 | 7.746167 | mammals | actual | clID_1175 |
| APICIDIN | HDAC5 | Histone deacetylase 5 | 4.559786 | 6.706175 | 7.896842 | mammals | actual | clID_1175 |
| APICIDIN | HDAC6 | Histone deacetylase 6 | 4.559786 | 6.928568 | 7.818034 | mammals | actual | clID_1175 |
| APICIDIN | Hdac7 | Histone deacetylase 7 | 4.559786 | 6.68009 | 7.868874 | mammals | actual | clID_1175 |
| APICIDIN | Hdac8 | Histone deacetylase 8 | 4.559786 | 6.869963 | 7.810528 | mammals | actual | clID_1175 |
| APICIDIN | Hdac9 | Histone deacetylase 9 | 4.559786 | 6.70873 | 7.896842 | mammals | actual | clID_1175 |
| APICIDIN | NCOR2 | Nuclear receptor corepressor 2 | 4.559786 | 5.990339 | 8.012228 | mammals | actual | clID_1175 |
| APICIDIN | 1MB.92 | Histone deacetylase | 4.559786 | 10.092065 | 8.329622 | protozoan | actual | clID_1175 |
| APICIDIN | 56k.08 | Histone deacetylase | 4.559786 | 10.372722 | 9 | protozoan | actual | clID_1175 |
| APICIDIN | HDAC1 | Histone deacetylase | 4.559786 | 5.852775 | 9 | protozoan | actual | clID_1175 |
| APICIDIN | F3 | Tissue factor | 4.559786 | 6.262633 | 6.219567 | mammals | actual | clID_1175 |
| CITRIC ACID | Hdac10 | Polyamine deacetylase HDAC10 | 4.931063 | 6.714438 | 9 | mammals | actual | clID_8565 |
| CITRIC ACID | Hdac11 | Histone deacetylase 11 | 4.931063 | 6.69745 | 9 | mammals | actual | clID_8565 |
| CITRIC ACID | Hdac1 | Histone deacetylase 1 | 4.931063 | 6.964004 | 9 | mammals | actual | clID_8565 |
| CITRIC ACID | Hdac2 | Histone deacetylase 2 | 4.931063 | 6.79746 | 9 | mammals | actual | clID_8565 |
| CITRIC ACID | HDAC3 | Histone deacetylase 3 | 4.931063 | 6.743628 | 9 | mammals | actual | clID_8565 |
| CITRIC ACID | HDAC4 | Histone deacetylase 4 | 4.931063 | 6.710525 | 9 | mammals | actual | clID_8565 |
| CITRIC ACID | HDAC5 | Histone deacetylase 5 | 4.931063 | 6.706175 | 9 | mammals | actual | clID_8565 |
| CITRIC ACID | HDAC6 | Histone deacetylase 6 | 4.931063 | 6.928568 | 9 | mammals | actual | clID_8565 |
| CITRIC ACID | Hdac7 | Histone deacetylase 7 | 4.931063 | 6.68009 | 9 | mammals | actual | clID_8565 |
| CITRIC ACID | Hdac8 | Histone deacetylase 8 | 4.931063 | 6.869963 | 9 | mammals | actual | clID_8565 |
| CITRIC ACID | Hdac9 | Histone deacetylase 9 | 4.931063 | 6.70873 | 9 | mammals | actual | clID_8565 |
| CITRIC ACID | Nr1h4 | Bile acid receptor | 4.931063 | 6.87787 | 8.346787 | mammals | actual | clID_8565 |
| CITRIC ACID | SLCO2B1 | Solute carrier organic anion transporter family member 2B1 | 4.931063 | 7.313814 | 6.628167 | mammals | actual | clID_8565 |
| CITRIC ACID | THRB | Thyroid hormone receptor beta | 4.931063 | 5.722362 | 8.69897 | mammals | actual | clID_8565 |
| DAUNORUBICIN | KAT2A | Histone acetyltransferase KAT2A | 6.177141 | 6.643569 | 6.500038 | mammals | actual | clID_732 |
| DAUNORUBICIN | Abcc1 | Multidrug resistance-associated protein 1 | 6.177141 | 9.003589 | 6.03199 | mammals | actual | clID_732 |
| DAUNORUBICIN | Pmp22 | Peripheral myelin protein 22 | 6.177141 | 7.357957 | 6.900319 | mammals | actual | clID_732 |
| DAUNORUBICIN | TDP1 | Tyrosyl-DNA phosphodiesterase 1 | 6.177141 | 7.013656 | 6.62515 | mammals | actual | clID_732 |
| FIMEPINOSTAT | Hdac10 | Polyamine deacetylase HDAC10 | 4.771095 | 6.714438 | 8.226776 | mammals | actual | clID_11717 |
| FIMEPINOSTAT | Hdac11 | Histone deacetylase 11 | 4.771095 | 6.69745 | 8.175834 | mammals | actual | clID_11717 |
| FIMEPINOSTAT | Hdac1 | Histone deacetylase 1 | 4.771095 | 6.964004 | 7.953884 | mammals | actual | clID_11717 |
| FIMEPINOSTAT | Hdac2 | Histone deacetylase 2 | 4.771095 | 6.79746 | 7.946298 | mammals | actual | clID_11717 |
| FIMEPINOSTAT | HDAC3 | Histone deacetylase 3 | 4.771095 | 6.743628 | 8.00176 | mammals | actual | clID_11717 |
| FIMEPINOSTAT | HDAC4 | Histone deacetylase 4 | 4.771095 | 6.710525 | 8.019223 | mammals | actual | clID_11717 |
| FIMEPINOSTAT | HDAC5 | Histone deacetylase 5 | 4.771095 | 6.706175 | 8.001145 | mammals | actual | clID_11717 |
| FIMEPINOSTAT | HDAC6 | Histone deacetylase 6 | 4.771095 | 6.928568 | 7.973844 | mammals | actual | clID_11717 |
| FIMEPINOSTAT | Hdac7 | Histone deacetylase 7 | 4.771095 | 6.68009 | 8.017749 | mammals | actual | clID_11717 |
| FIMEPINOSTAT | Hdac8 | Histone deacetylase 8 | 4.771095 | 6.869963 | 8.046781 | mammals | actual | clID_11717 |
| FIMEPINOSTAT | Hdac9 | Histone deacetylase 9 | 4.771095 | 6.70873 | 8.008241 | mammals | actual | clID_11717 |
| FIMEPINOSTAT | NCOR2 | Nuclear receptor corepressor 2 | 4.771095 | 5.990339 | 8.744727 | mammals | actual | clID_11717 |
| FIMEPINOSTAT | Pik3r1 | Phosphatidylinositol 3-kinase regulatory subunit alpha | 4.771095 | 8.165486 | 7.505594 | mammals | actual | clID_11717 |
| FIMEPINOSTAT | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | 4.771095 | 8.414227 | 7.721246 | mammals | actual | clID_11717 |
| FIMEPINOSTAT | PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform | 4.771095 | 7.674333 | 7.267606 | mammals | actual | clID_11717 |
| FIMEPINOSTAT | PIK3CD | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | 4.771095 | 7.906179 | 7.408935 | mammals | actual | clID_11717 |
| FIMEPINOSTAT | PIK3CG | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | 4.771095 | 7.447055 | 6.50724 | mammals | actual | clID_11717 |
| PYROXAMIDE | Hdac1 | Histone deacetylase 1 | 4.83654 | 6.964004 | 6.01667 | mammals | actual | clID_214 |
| PYROXAMIDE | HDAC6 | Histone deacetylase 6 | 4.83654 | 6.928568 | 6.005806 | mammals | actual | clID_214 |
| SALICYLIC ACID | Hdac10 | Polyamine deacetylase HDAC10 | 5.39542 | 6.714438 | 6.060698 | mammals | actual | clID_3107 |
| SALICYLIC ACID | Hdac11 | Histone deacetylase 11 | 5.39542 | 6.69745 | 6.060698 | mammals | actual | clID_3107 |
| SALICYLIC ACID | Hdac1 | Histone deacetylase 1 | 5.39542 | 6.964004 | 6.060698 | mammals | actual | clID_3107 |
| SALICYLIC ACID | Hdac2 | Histone deacetylase 2 | 5.39542 | 6.79746 | 6.060698 | mammals | actual | clID_3107 |
| SALICYLIC ACID | HDAC3 | Histone deacetylase 3 | 5.39542 | 6.743628 | 6.060698 | mammals | actual | clID_3107 |
| SALICYLIC ACID | HDAC4 | Histone deacetylase 4 | 5.39542 | 6.710525 | 6.060698 | mammals | actual | clID_3107 |
| SALICYLIC ACID | HDAC5 | Histone deacetylase 5 | 5.39542 | 6.706175 | 6.060698 | mammals | actual | clID_3107 |
| SALICYLIC ACID | HDAC6 | Histone deacetylase 6 | 5.39542 | 6.928568 | 6.060698 | mammals | actual | clID_3107 |
| SALICYLIC ACID | Hdac7 | Histone deacetylase 7 | 5.39542 | 6.68009 | 6.060698 | mammals | actual | clID_3107 |
| SALICYLIC ACID | Hdac8 | Histone deacetylase 8 | 5.39542 | 6.869963 | 6.060698 | mammals | actual | clID_3107 |
| SALICYLIC ACID | Hdac9 | Histone deacetylase 9 | 5.39542 | 6.70873 | 6.060698 | mammals | actual | clID_3107 |
| SALICYLIC ACID | HMGB1 | High mobility group protein B1 | 5.39542 | 4.956066 | 6 | mammals | actual | clID_3107 |
| SALICYLIC ACID | PPO2 | Polyphenol oxidase 2 | 5.39542 | 5.497037 | 6.50965 | fungi | actual | clID_3107 |
| TRAPOXIN A | Hdac1 | Histone deacetylase 1 | 4.575447 | 6.964004 | 6.086186 | mammals | predicted | clID_939 |
| TRAPOXIN A | hda106 | HD2 type histone deacetylase HDA106 | 4.575447 | 4.923718 | 8 | plants | predicted | clID_939 |
| TRAPOXIN B | Hdac10 | Polyamine deacetylase HDAC10 | 4.785863 | 6.714438 | 8.25 | mammals | predicted | clID_939 |
| TRAPOXIN B | Hdac11 | Histone deacetylase 11 | 4.785863 | 6.69745 | 8.25 | mammals | predicted | clID_939 |
| TRAPOXIN B | Hdac1 | Histone deacetylase 1 | 4.785863 | 6.964004 | 8.25997 | mammals | predicted | clID_939 |
| TRAPOXIN B | Hdac2 | Histone deacetylase 2 | 4.785863 | 6.79746 | 8.25 | mammals | predicted | clID_939 |
| TRAPOXIN B | HDAC3 | Histone deacetylase 3 | 4.785863 | 6.743628 | 8.25 | mammals | predicted | clID_939 |
| TRAPOXIN B | HDAC4 | Histone deacetylase 4 | 4.785863 | 6.710525 | 8.25 | mammals | predicted | clID_939 |
| TRAPOXIN B | HDAC5 | Histone deacetylase 5 | 4.785863 | 6.706175 | 8.25 | mammals | predicted | clID_939 |
| TRAPOXIN B | HDAC6 | Histone deacetylase 6 | 4.785863 | 6.928568 | 8.25 | mammals | predicted | clID_939 |
| TRAPOXIN B | Hdac7 | Histone deacetylase 7 | 4.785863 | 6.68009 | 8.25 | mammals | predicted | clID_939 |
| TRAPOXIN B | Hdac8 | Histone deacetylase 8 | 4.785863 | 6.869963 | 8.25 | mammals | predicted | clID_939 |
| TRAPOXIN B | Hdac9 | Histone deacetylase 9 | 4.785863 | 6.70873 | 8.25 | mammals | predicted | clID_939 |
| TRAPOXIN B | hda106 | HD2 type histone deacetylase HDA106 | 4.785863 | 4.923718 | 8 | plants | predicted | clID_939 |
Use search function in table to view values for specific interactions
| molName | geneName | protname | sarsScoreMolecule | geneScore | interactionScore | organismType | kindOfValue | molecularCluster |
|---|---|---|---|---|---|---|---|---|
| ACULEACIN A | CaFKS1 | 1,3-beta-D-glucan synthase catalytic subunit | 4.724035 | 5.759173 | 5.09691 | fungi | predicted | clID_939 |
| ANIDULAFUNGIN | CYP2C8 | Cytochrome P450 2C8 | 5.333482 | 6.575757 | 4.819274 | mammals | actual | clID_3316 |
| ANIDULAFUNGIN | CaFKS1 | 1,3-beta-D-glucan synthase catalytic subunit | 5.333482 | 5.759173 | 8.794549 | fungi | actual | clID_3316 |
| ANIDULAFUNGIN | GSL2 | Glucan synthase | 5.333482 | 11.256083 | 7.890738 | fungi | actual | clID_3316 |
| ANIDULAFUNGIN | GSC1 | Beta-1,3-glucan synthase catalytic subunit 1 | 5.333482 | 10.055226 | 5.477121 | fungi | actual | clID_3316 |
| CILOFUNGIN | GSC1 | Beta-1,3-glucan synthase catalytic subunit 1 | 6.073807 | 10.055226 | 5.500602 | fungi | predicted | clID_3316 |
| FR-901469 | GSC1 | Beta-1,3-glucan synthase catalytic subunit 1 | 4.538801 | 10.055226 | 8.070581 | fungi | predicted | clID_939 |
| MICAFUNGIN | ALB | Serum albumin | 7.087835 | 7.03501 | 7.521574 | mammals | predicted | clID_3316 |
| MICAFUNGIN | CaFKS1 | 1,3-beta-D-glucan synthase catalytic subunit | 7.087835 | 5.759173 | 7.829745 | fungi | predicted | clID_3316 |
| MICAFUNGIN | GSL2 | Glucan synthase | 7.087835 | 11.256083 | 7.551859 | fungi | predicted | clID_3316 |
Use search function in table to view values for specific interactions
| molName | geneName | protname | sarsScoreMolecule | geneScore | interactionScore | organismType | kindOfValue | molecularCluster |
|---|---|---|---|---|---|---|---|---|
| CE-224535 | P2rx7 | P2X purinoceptor 7 | 4.848405 | 6.846055 | 8.239312 | mammals | predicted | clID_10341 |
| NIPHATOXIN C | P2rx7 | P2X purinoceptor 7 | 5.027325 | 6.846055 | 4.841932 | mammals | predicted | clID_6424 |
| PSAMMAPLYSENE D | P2rx7 | P2X purinoceptor 7 | 4.891308 | 6.846055 | 5.410174 | mammals | predicted | clID_9299 |
Use search function in table to view values for specific interactions
| molName | geneName | protname | sarsScoreMolecule | geneScore | interactionScore | organismType | kindOfValue | molecularCluster |
|---|---|---|---|---|---|---|---|---|
| ISPINESIB | CENPE | Centromere-associated protein E | 6.052502 | 9.243621 | 6.690196 | mammals | actual | clID_6821 |
| ISPINESIB | KIF11 | Kinesin-like protein KIF11 | 6.052502 | 6.848695 | 8.053093 | mammals | actual | clID_6821 |
| ISPINESIB | KIF2C | Kinesin-like protein KIF2C | 6.052502 | 12.017143 | 6.466913 | mammals | actual | clID_6821 |
| ISPINESIB | KIFC3 | Kinesin-like protein KIFC3 | 6.052502 | 11.293771 | 6.556749 | mammals | actual | clID_6821 |
| ISPINESIB | KIF5B | Kinesin-1 heavy chain | 6.052502 | 6.047941 | 9 | mammals | actual | clID_6821 |
| ISPINESIB MESYLATE | KIF11 | Kinesin-like protein KIF11 | 7.358363 | 6.848695 | 8.920819 | mammals | predicted | clID_6821 |
| SB-743921 | KIF11 | Kinesin-like protein KIF11 | 5.572534 | 6.848695 | 6.788229 | mammals | actual | clID_2183 |