Histone deacetylases (EC 3.5.1.98, HDAC) are a class of enzymes that remove acetyl groups (O=C-CH3) from an ε-N-acetyl lysine amino acid on a histone, allowing the histones to wrap the DNA more tightly.[2] This is important because DNA is wrapped around histones, and DNA expression is regulated by acetylation and de-acetylation (taken from wikipedia).
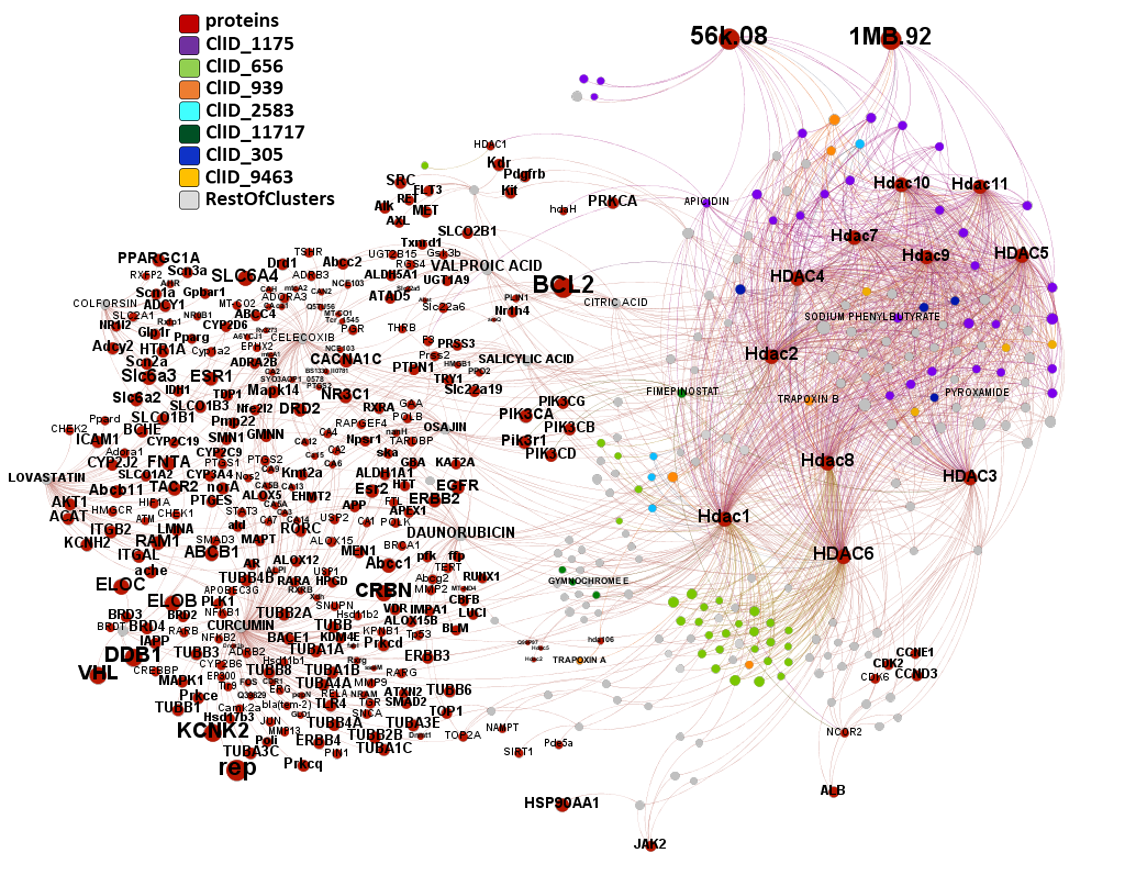
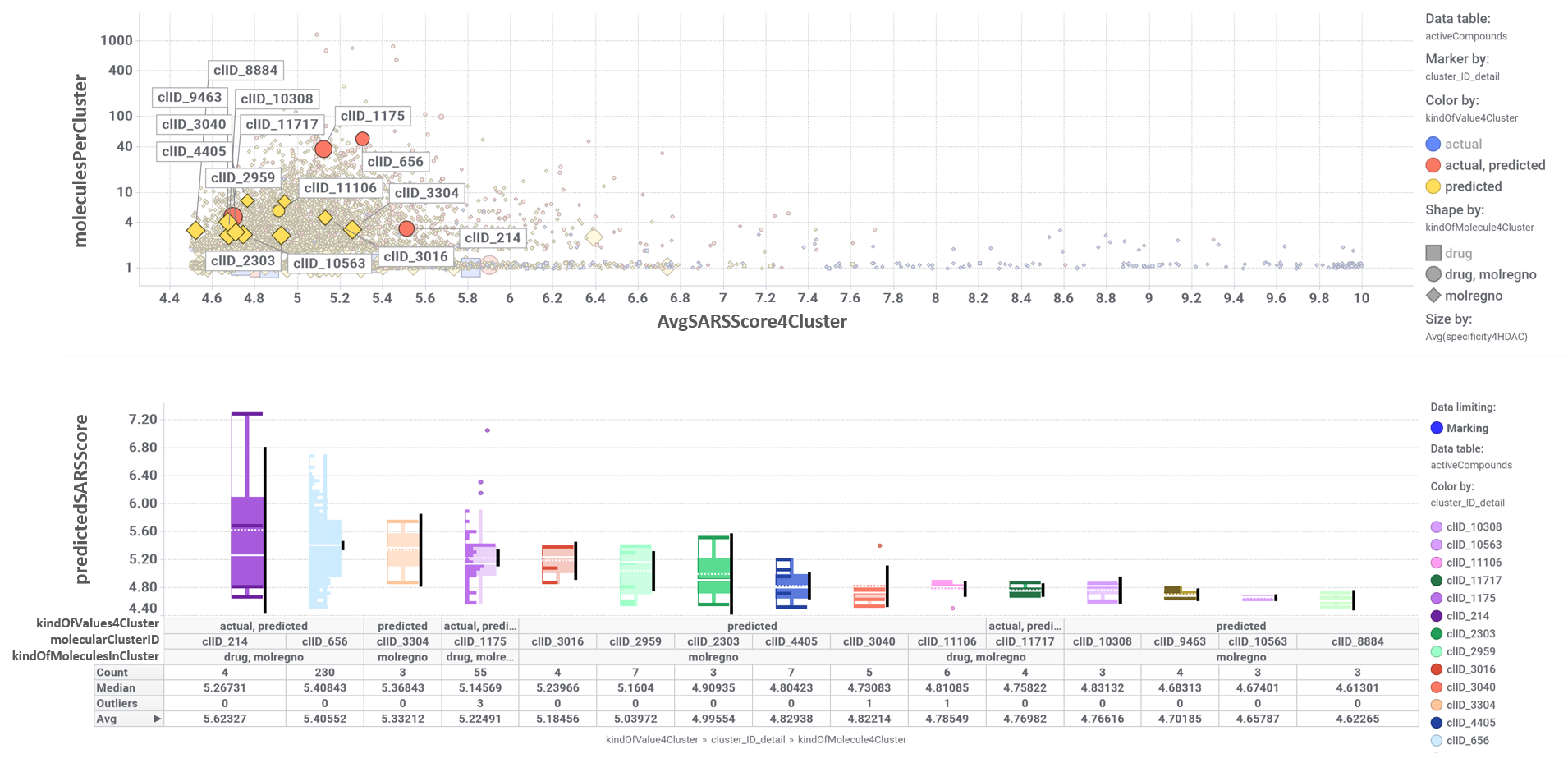
In this section we will focus on 209 molecules predicted as active against coronaviruses by machine learning models based on SARS-CoV assays in ChEMBL DB that interact with different histone deacetylases.
The network graph below displays all molecular interactions of these 209 compounds with all proteins recorded in ChEMBL. Compounds have been classified in clusters according to their structural similarity. A specific color code has been assigned to most relevant clusters. Most relevant clusters behave HDAC-specific, although there is a remarkable promiscuity of interactions for drugs like CURCUMIN, CELECOXIB, LOVASTATIN, DAUNORUBICIN, OSAJIN, VALPROIC ACID, SALICYLIC ACID, COLFORSIN and CITRIC ACID, showing activity for more than 300 non HDAC proteins. Although, among HDACs themselves the graph shows lack of selectivity of any cluster for any specific HDAC, this is most likely caused because the ChEMBL identifier for a generic non specified HDAC, is mapped in ChEMBL DB against HDAC1-11 in Uniprot.
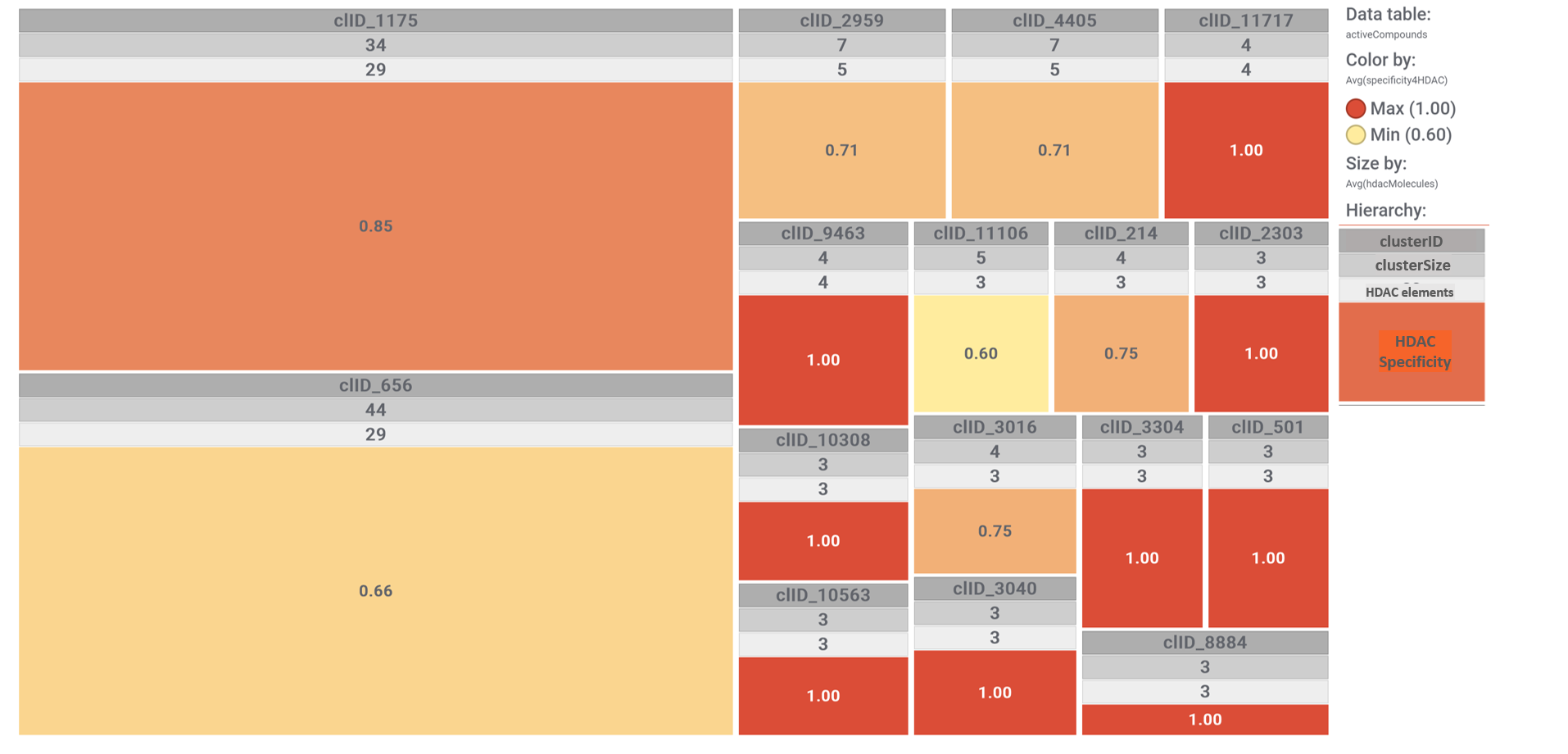
Each molecular cluster specificity for this set of molecules can be visualized with a simple tree map, where each cluster is sized according to its number of elements and its color graded by the ratio between the number of cluster elements hitting on HDACs and the cluster size. See legend.