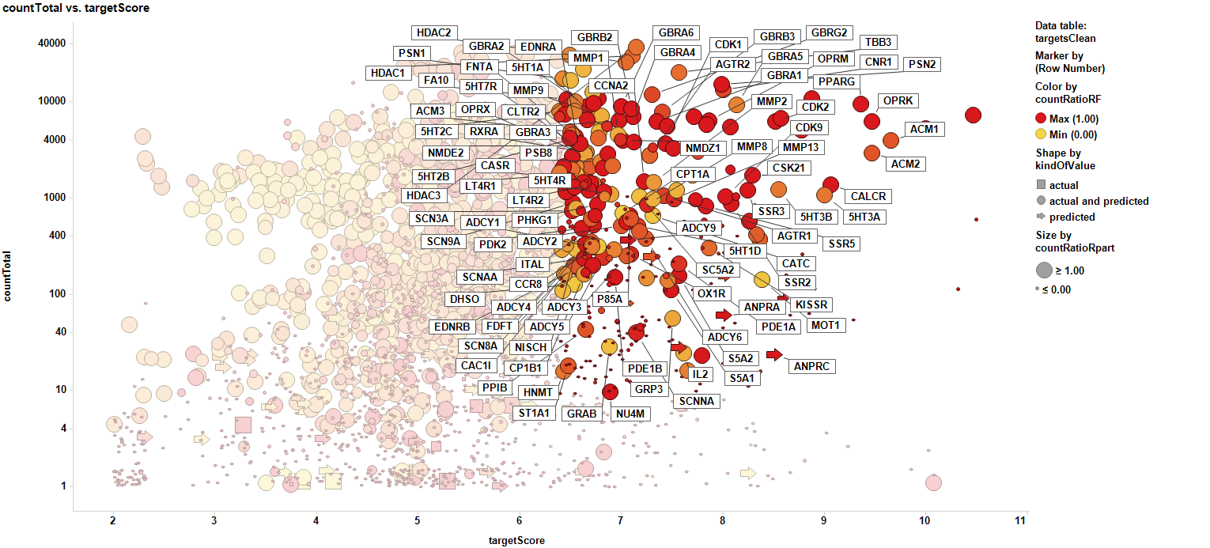
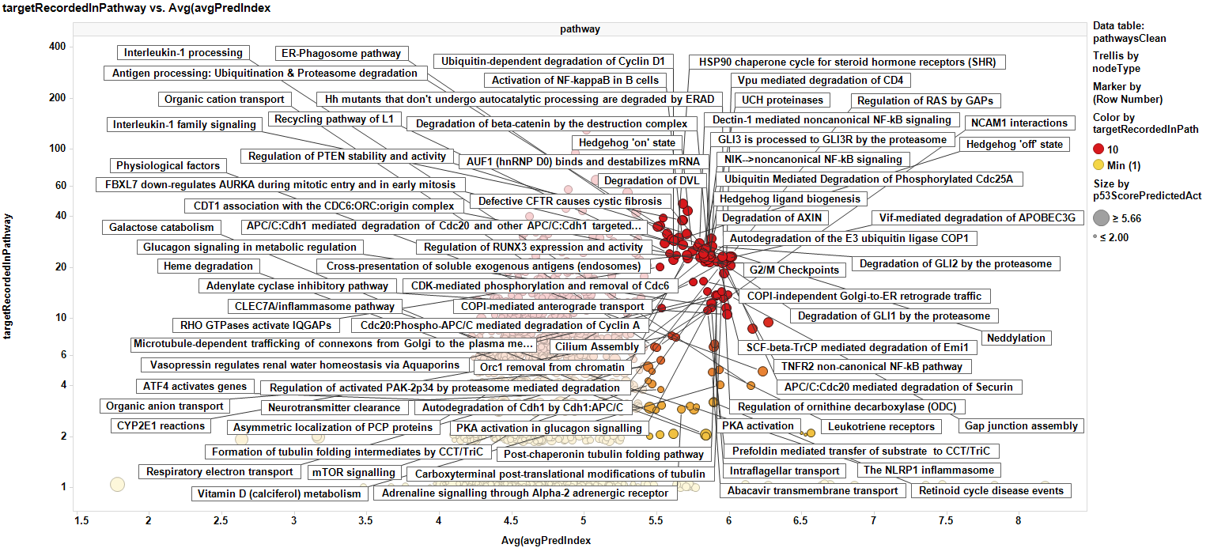
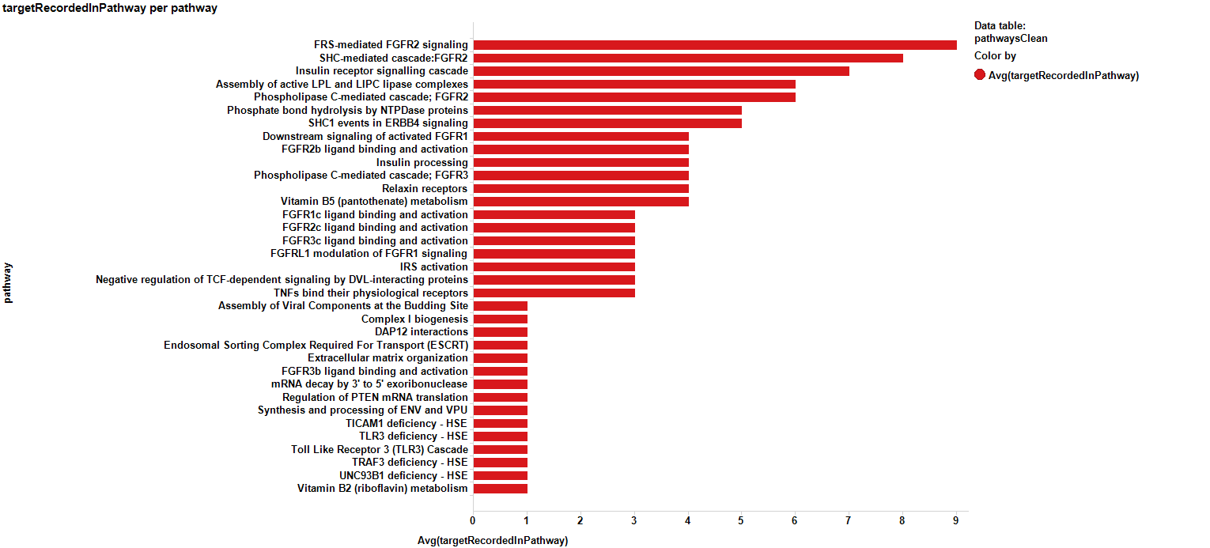
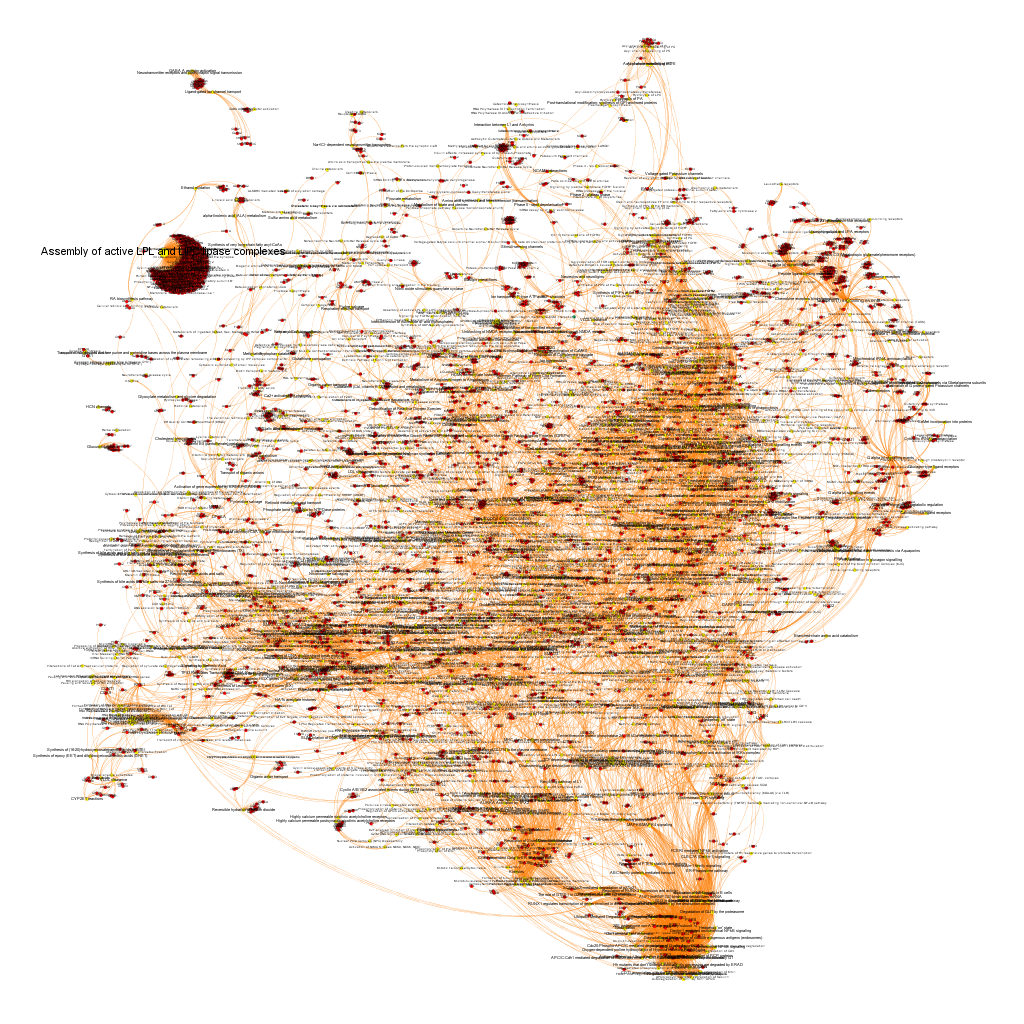
Pathways analysis with the P53 dataset.
Once targets are selected in the target ID section, we can proceed incorporating the pathways from Reactome database (Keggs and others would suit as well), in this case Reactome low level pathways.
The P53 scores are now aggregated by pathway, and there is a pathway potency score calculated too. Below a selection of best combinations of both scores.
And the respective barchart with the scores sorted by potency…
t may be also be interesting to consider how many proteins appear in the identified pathways…
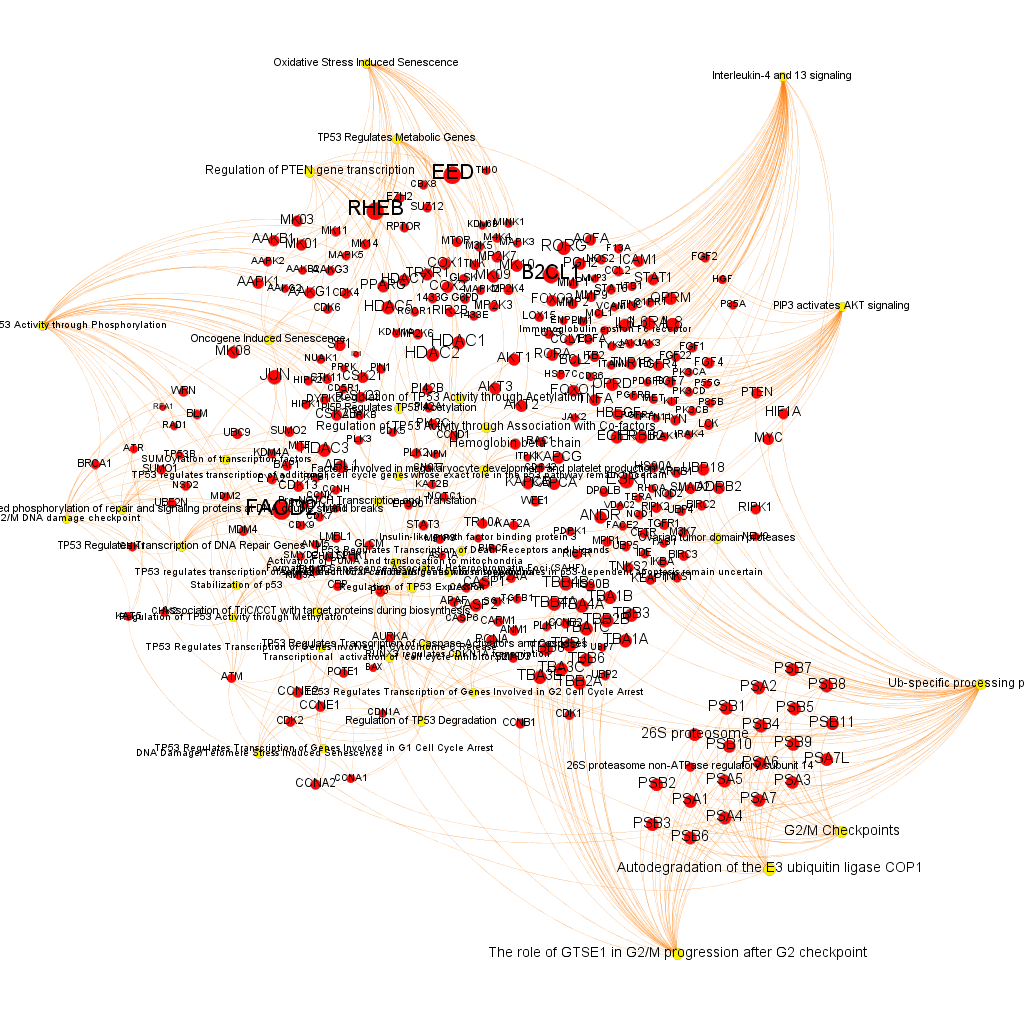
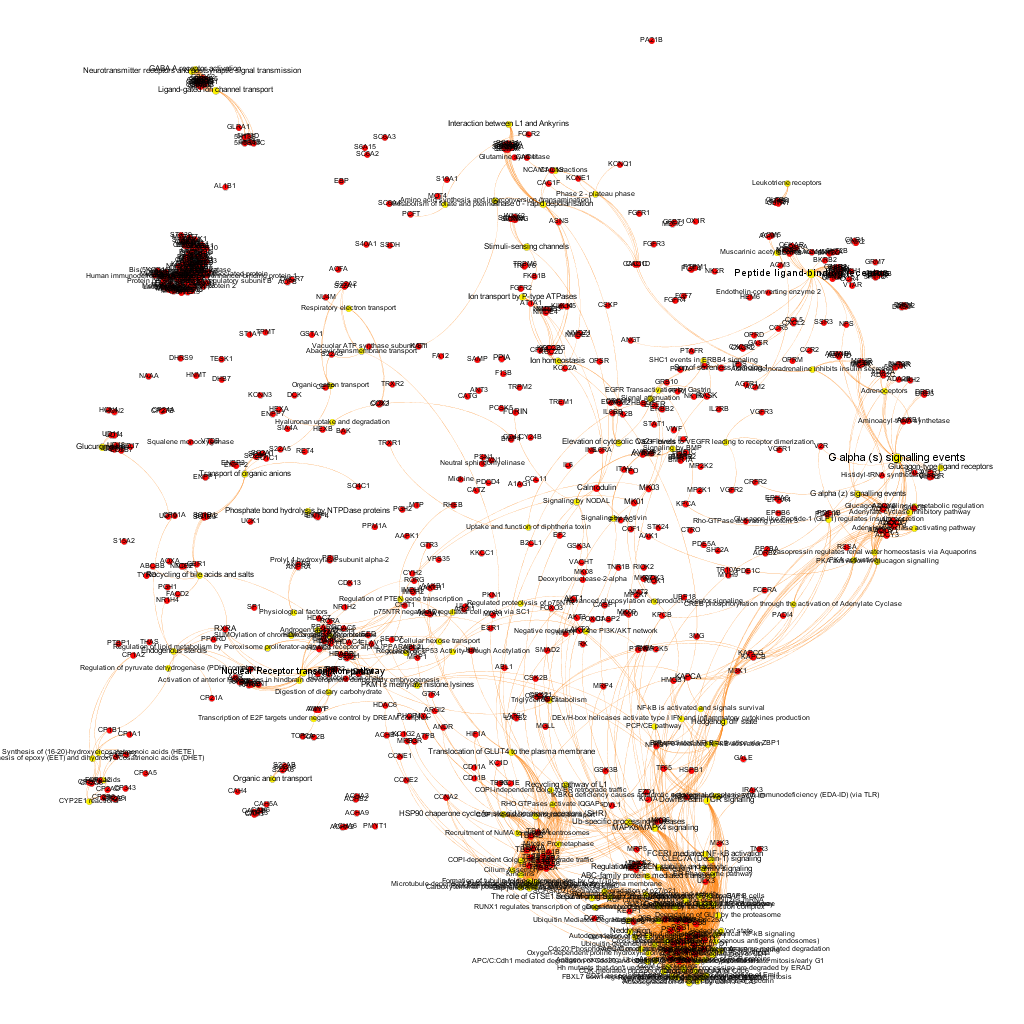
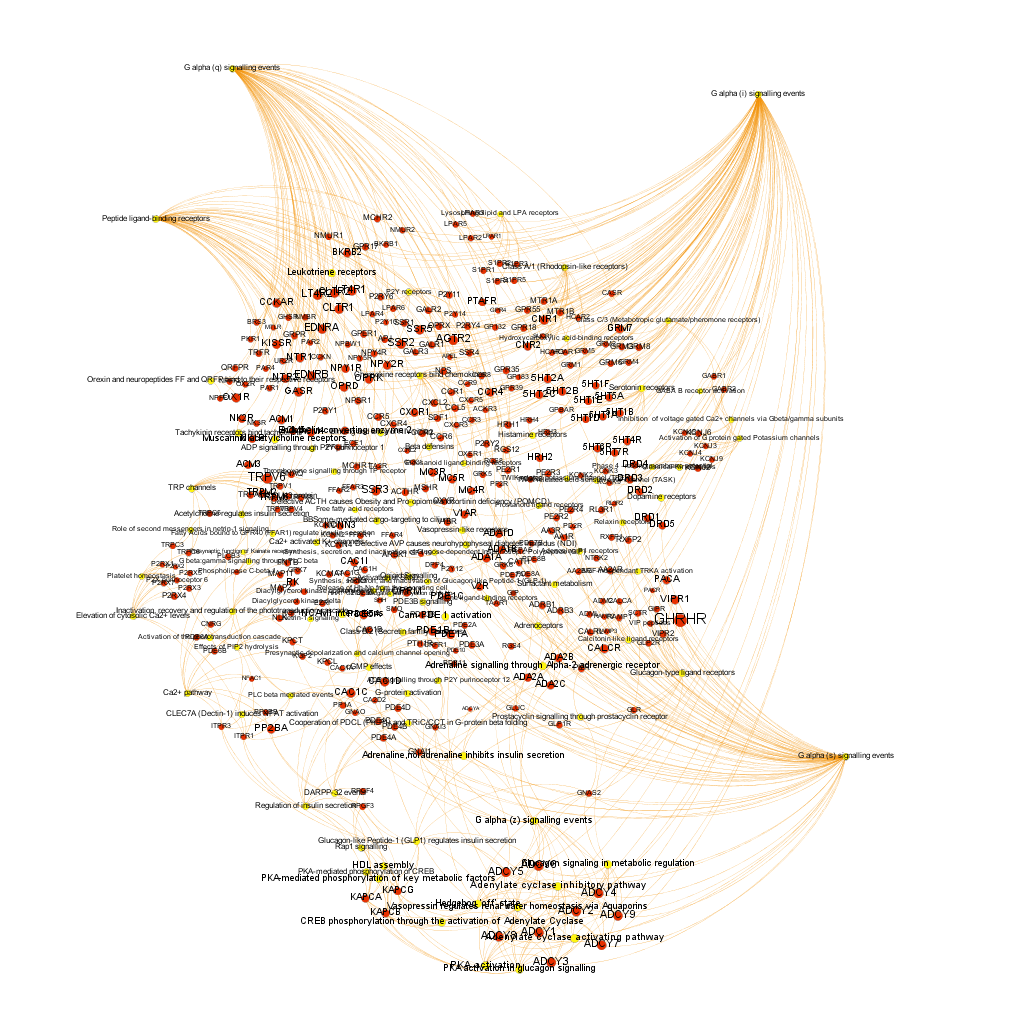
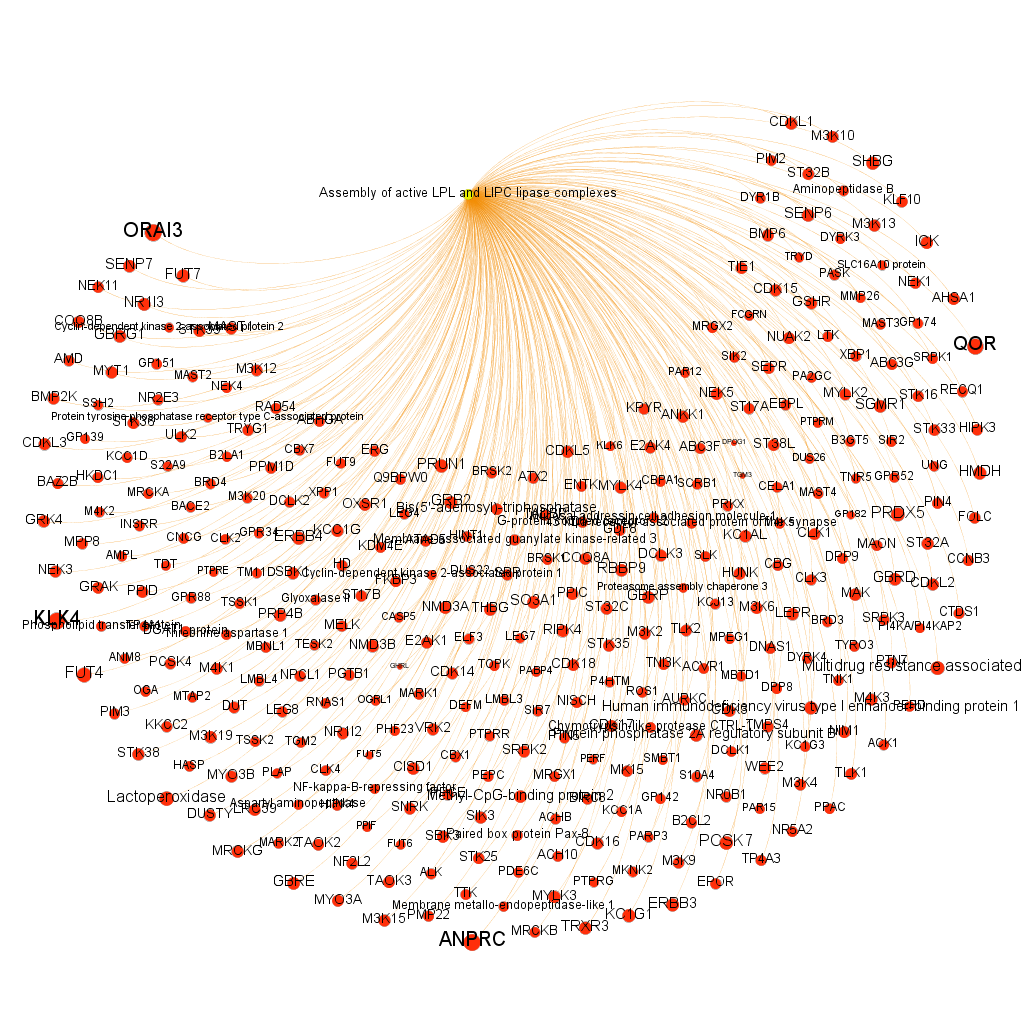
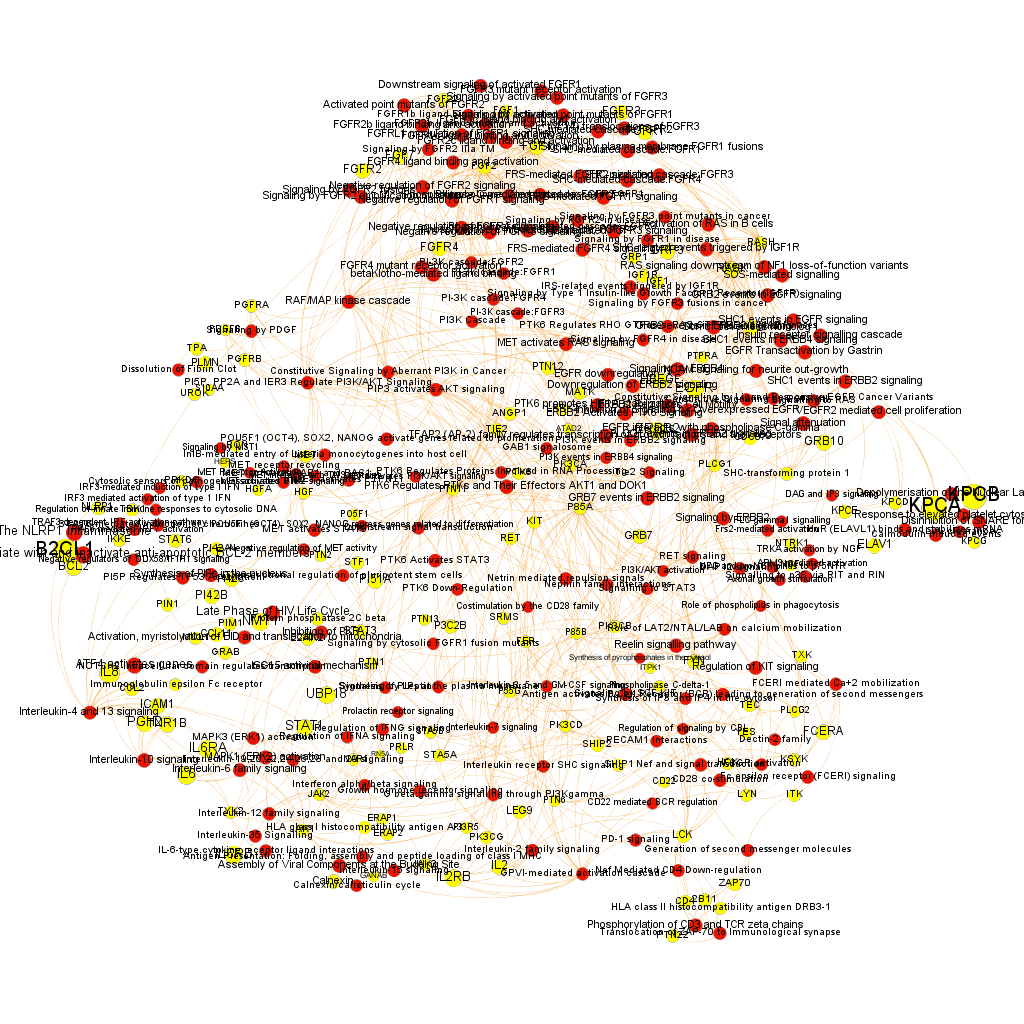
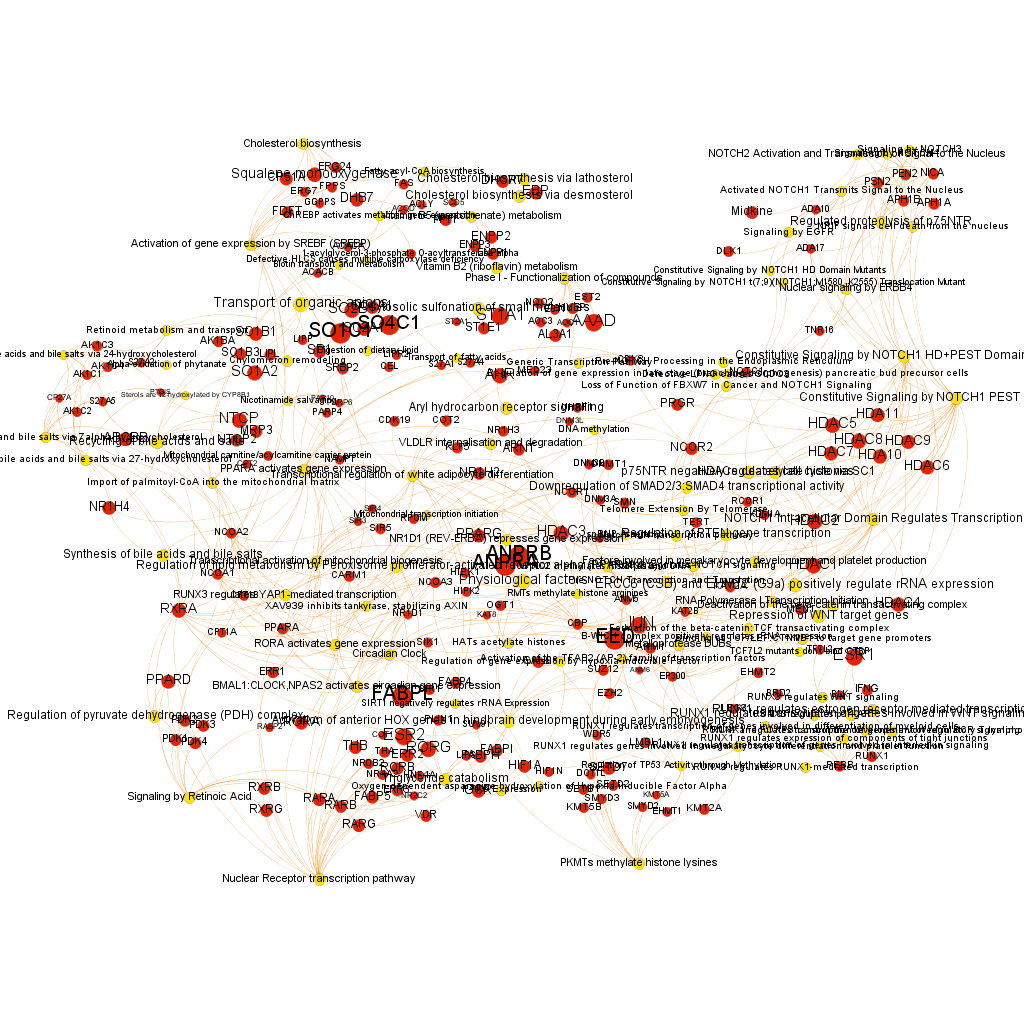
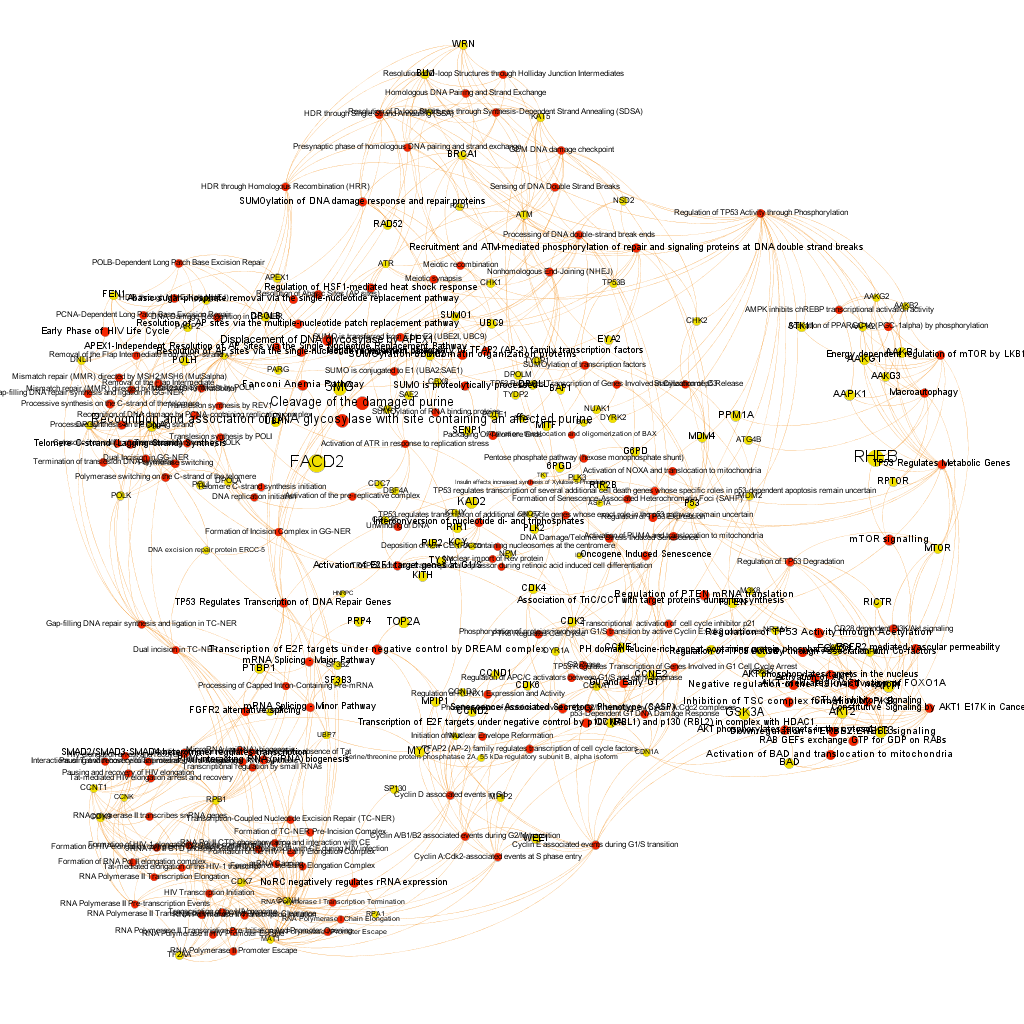
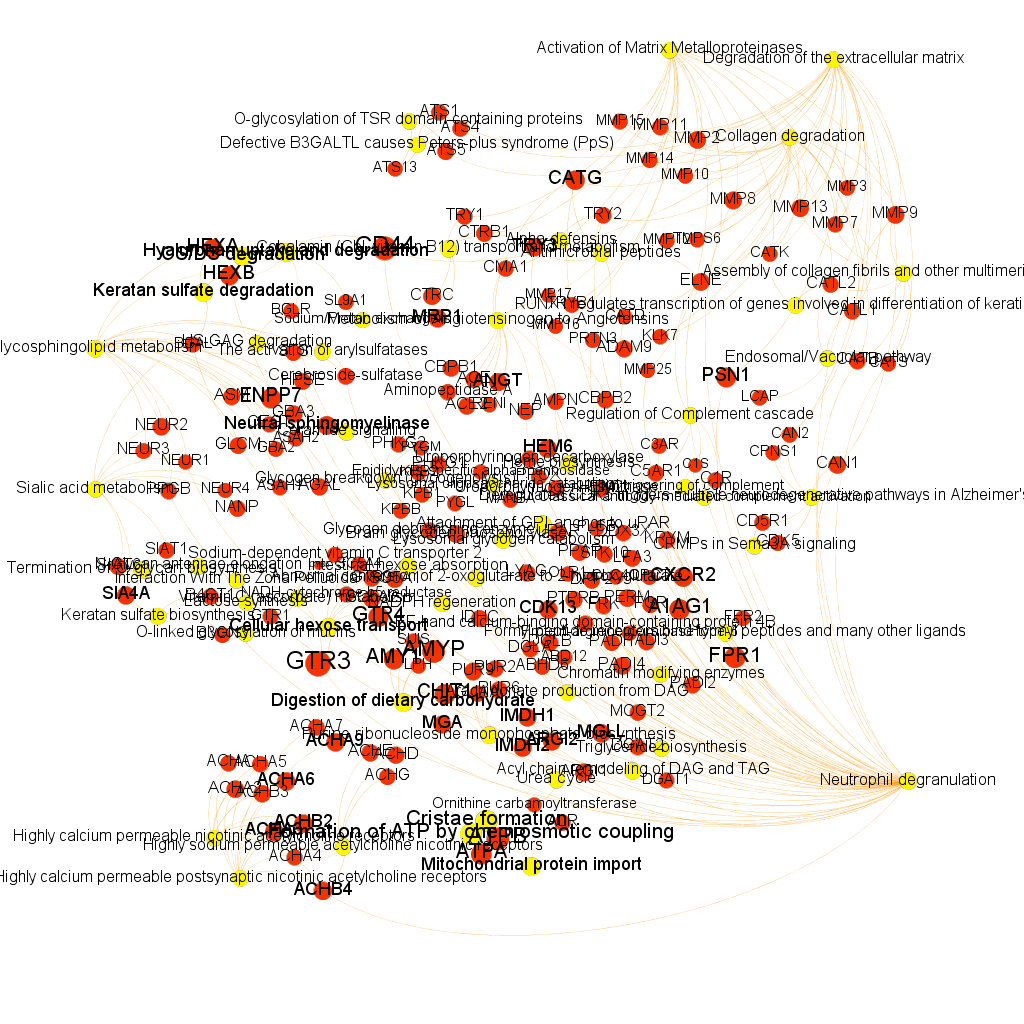
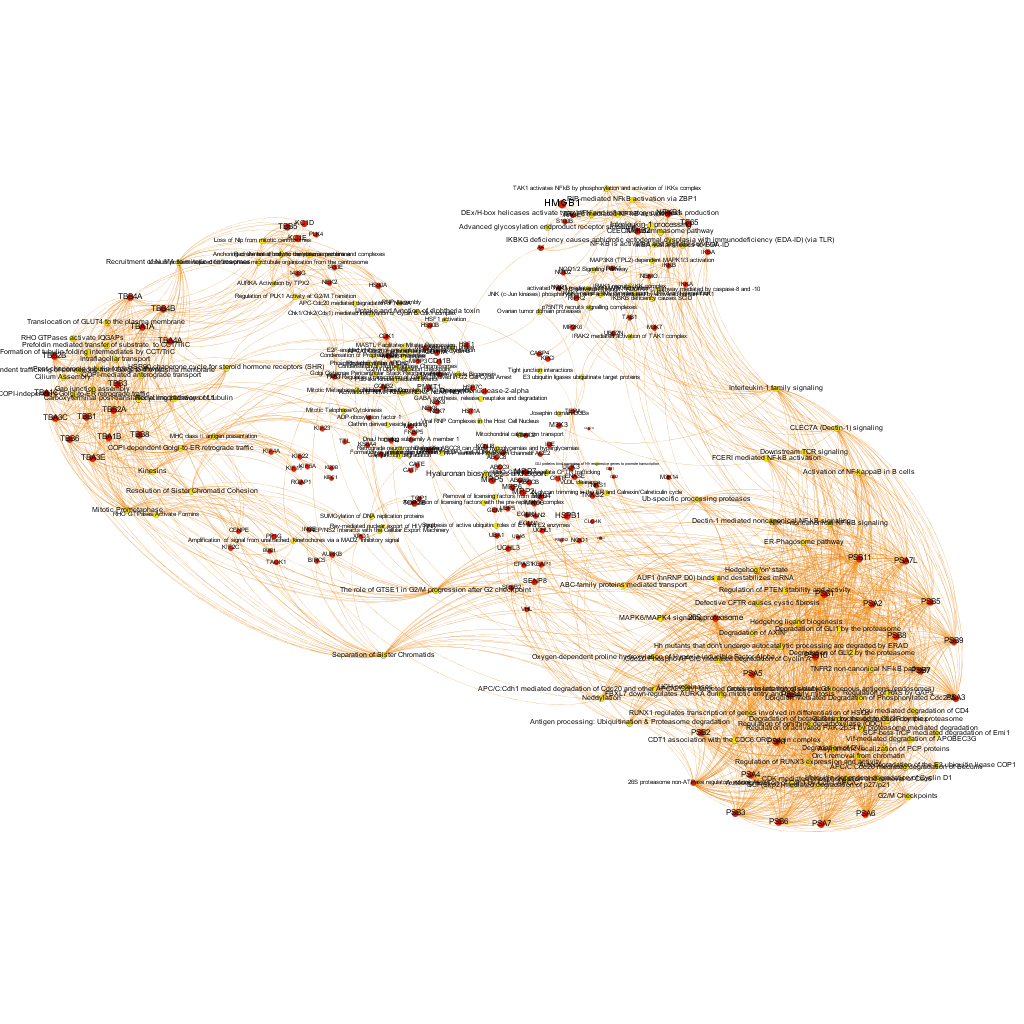
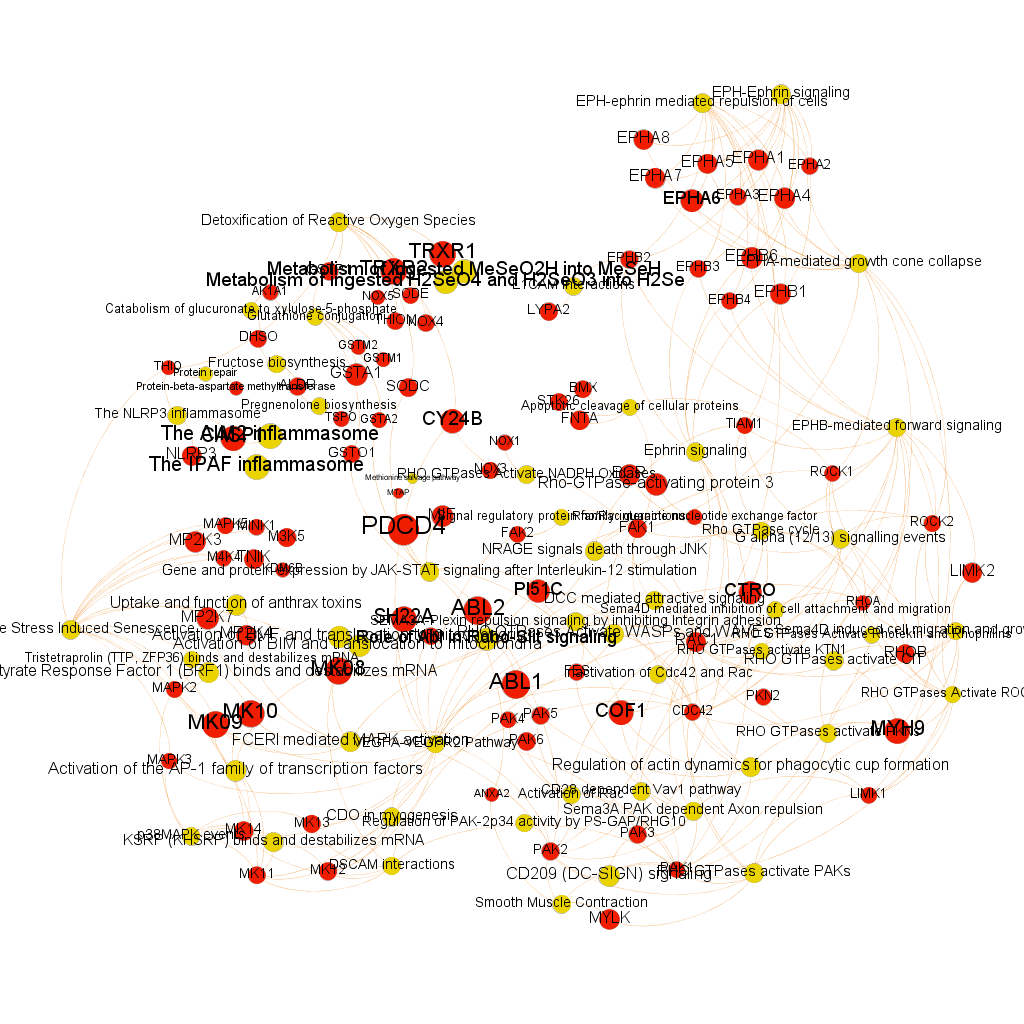
Using graph databases for pathways analysis.
What we have now is a collection of target-pathway pairs accompanied by a series of numerical and class descriptors. We can track the relationships among all pair members and quantify and categorize them using the descriptors as weights and labels. This is what graph databases do, and we can always query the database for components and properties.
This is the aspect that the whole set of relationships between targets and pathways has, but the graphs can be queried by potency of nodes, or proximity to a particular node. We can see it in the next graphs.
Once the graph is set up, it can be queried in different modes, as below, by connection to P53 or aggregated potency.
We can also try some segmentation of relations to identify the most important connection clusters
P53 Pathways Table
| pathway | pathwayID | CountActual | CountPredicted | p53ScorePredictedActual | p53ScorePredicted | targetScore | countRatioRpart | countRatioRF | countRatioAvgRpartRF | combinedPredictionScore | AvgPathwayScore | countTotal | countMolecules | targetRecordedInPathway | MostCommon(uniprot) | MostCommon(proteinName) | MostCommon(Protein names) | MostCommon(Gene names) | MostCommon(targetName) | nodeType |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Abacavir transmembrane transport | R-HSA-2161517 | 255 | 2484 | 3.382739228 | 3.336520921 | 4.96949973 | 0.869434657 | 0.541608036 | 0.705521347 | 5.103146381 | 5.831055387 | 884.2 | 547.8 | 5 | pathway | |||||
| ABC-family proteins mediated transport | R-HSA-382556 | 542 | 10836 | 3.89032962 | 3.879632905 | 5.170374999 | 0.745461163 | 0.553712816 | 0.649586989 | 5.50956448 | 5.62735647 | 1285.910448 | 169.8208955 | 36 | pathway | |||||
| Activation of anterior HOX genes in hindbrain development during early embryogenesis | R-HSA-5617472 | 1696 | 13108 | 3.49903688 | 3.437900873 | 5.315171224 | 0.738236078 | 0.473781849 | 0.606008963 | 3.988433941 | 5.206405382 | 1997.521739 | 643.6521739 | 15 | pathway | |||||
| Activation of DNA fragmentation factor | R-HSA-211227 | 532 | 1336 | 3.711786487 | 3.689311511 | 4.691398497 | 0.987948581 | 0.604091068 | 0.796019824 | 6.74369622 | 6.54406099 | 1100 | 934 | 2 | pathway | |||||
| Activation of NF-kappaB in B cells | R-HSA-1169091 | 5867 | 18506 | 3.807479011 | 3.800574839 | 5.173934813 | 0.793059984 | 0.516228963 | 0.654644474 | 5.491760709 | 5.633456161 | 1693.106061 | 369.2878788 | 29 | pathway | |||||
| Adenylate cyclase activating pathway | R-HSA-170660 | 1 | 349 | 4.003061849 | 3.937089706 | 6.355613873 | 0.833333333 | 0.644230769 | 0.738782051 | 6.289412101 | 6.259567625 | 244.8461538 | 26.92307692 | 9 | pathway | |||||
| Adenylate cyclase inhibitory pathway | R-HSA-170670 | 6 | 926 | 3.83070464 | 3.772880587 | 6.090037661 | 0.788888889 | 0.642869416 | 0.715879152 | 5.800864877 | 6.049429229 | 220.2 | 31.06666667 | 11 | pathway | |||||
| Adrenaline signalling through Alpha-2 adrenergic receptor | R-HSA-392023 | 1401 | 4073 | 3.334108363 | 3.32711232 | 4.695614958 | 0.933710747 | 0.428894347 | 0.681302547 | 4.614107328 | 5.653386435 | 1347.222222 | 608.2222222 | 3 | pathway | |||||
| Adrenaline,noradrenaline inhibits insulin secretion | R-HSA-400042 | 979 | 3664 | 3.367035392 | 3.361685989 | 5.239440826 | 0.847200465 | 0.497368541 | 0.672284503 | 4.578927926 | 5.604241814 | 662.5625 | 290.1875 | 8 | pathway | |||||
| Adrenoceptors | R-HSA-390696 | 3489 | 15013 | 3.237432177 | 3.231772577 | 5.1338662 | 0.806732796 | 0.460100052 | 0.633416424 | 4.001200713 | 5.301920216 | 1275.5 | 660.7857143 | 9 | pathway | |||||
| AKT-mediated inactivation of FOXO1A | R-HSA-211163 | 117 | 8183 | 3.162385694 | 3.161558174 | 4.871357559 | 0.997965509 | 0.276432484 | 0.637198996 | 3.86984183 | 5.302121874 | 1550.285714 | 1185.714286 | 4 | pathway | |||||
| Androgen biosynthesis | R-HSA-193048 | 1906 | 850 | 3.584178704 | 3.539117074 | 4.944344414 | 0.371542779 | 0.8450502 | 0.60829649 | 3.906066544 | 5.250036165 | 292.9 | 275.6 | 8 | pathway | |||||
| Antigen processing: Ubiquitination & Proteasome degradation | R-HSA-983168 | 351 | 7017 | 4.079214935 | 4.026941392 | 4.986451208 | 0.783663508 | 0.512688503 | 0.648176006 | 5.832287572 | 5.680911682 | 1469.611111 | 136.4444444 | 29 | pathway | |||||
| APC/C:Cdc20 mediated degradation of Securin | R-HSA-174154 | 267 | 6917 | 4.115530933 | 4.108207145 | 5.141119457 | 0.911111111 | 0.47096193 | 0.69103652 | 6.454662416 | 5.97875378 | 1758.288889 | 159.6444444 | 21 | pathway | |||||
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | R-HSA-174178 | 1937 | 16186 | 4.017526614 | 4.007103243 | 5.20999733 | 0.88461328 | 0.471156745 | 0.677885012 | 6.130406347 | 5.858408954 | 1877.816327 | 369.8571429 | 24 | pathway | |||||
| Aryl hydrocarbon receptor signalling | R-HSA-8937144 | 178 | 1354 | 3.376484989 | 3.264312671 | 4.737664705 | 0.824971924 | 0.461796779 | 0.643384351 | 4.341153414 | 5.414724005 | 760.25 | 383 | 3 | pathway | |||||
| Asymmetric localization of PCP proteins | R-HSA-4608870 | 271 | 6936 | 4.117324396 | 4.108543857 | 5.108221876 | 0.893617021 | 0.485635442 | 0.689626232 | 6.393599776 | 5.969949677 | 1683.957447 | 153.3404255 | 23 | pathway | |||||
| ATF4 activates genes | R-HSA-380994 | 229 | 156 | 4.410650143 | 4.381922191 | 5.881126847 | 0.323476703 | 0.869175627 | 0.596326165 | 4.63647165 | 5.44572448 | 136.3333333 | 128.3333333 | 3 | pathway | |||||
| AUF1 (hnRNP D0) binds and destabilizes mRNA | R-HSA-450408 | 1422 | 7302 | 4.113753283 | 4.070386328 | 5.060548299 | 0.857084838 | 0.506570692 | 0.681827765 | 6.267219061 | 5.91676953 | 1652.734694 | 178.0408163 | 25 | pathway | |||||
| Autodegradation of Cdh1 by Cdh1:APC/C | R-HSA-174084 | 255 | 6581 | 4.114052555 | 4.10640202 | 5.137884039 | 0.906976744 | 0.472837012 | 0.689906878 | 6.430816533 | 5.970730039 | 1741.976744 | 158.9767442 | 19 | pathway | |||||
| Autodegradation of the E3 ubiquitin ligase COP1 | R-HSA-349425 | 11612 | 8006 | 4.123544643 | 4.0872092 | 5.148560925 | 0.836734694 | 0.510539702 | 0.673637198 | 6.17626059 | 5.865429535 | 1921.755102 | 400.3673469 | 23 | pathway | |||||
| BH3-only proteins associate with and inactivate anti-apoptotic BCL-2 members | R-HSA-111453 | 24 | 1725 | 3.052926874 | 2.965997496 | 6.083099028 | 0.795638117 | 0.6445893 | 0.720113708 | 4.459987335 | 5.818400347 | 541.75 | 437.25 | 3 | pathway | |||||
| Budding and maturation of HIV virion | R-HSA-162588 | 207 | 409 | 3.980601094 | 3.856274839 | 4.944834165 | 0.522139303 | 0.656033002 | 0.589086153 | 4.257322298 | 5.25410805 | 414 | 308 | 2 | pathway | |||||
| Cam-PDE 1 activation | R-HSA-111957 | 32 | 553 | 3.392294948 | 3.370794053 | 5.299936654 | 0.595238095 | 0.671393785 | 0.63331594 | 4.147680261 | 5.35287125 | 97.71428571 | 83.57142857 | 4 | pathway | |||||
| Carboxyterminal post-translational modifications of tubulin | R-HSA-8955332 | 368 | 32170 | 3.558969454 | 3.561395653 | 5.396218782 | 0.865602248 | 0.556070162 | 0.710836205 | 5.160549409 | 5.925231184 | 22598.62857 | 929.6571429 | 15 | pathway | |||||
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | R-HSA-174184 | 369 | 12384 | 3.887138744 | 3.881000895 | 5.346770602 | 0.814351815 | 0.504766993 | 0.659559404 | 5.68079224 | 5.692775609 | 1568.214286 | 227.7321429 | 24 | pathway | |||||
| CDK-mediated phosphorylation and removal of Cdc6 | R-HSA-69017 | 340 | 11948 | 3.9643409 | 3.957399416 | 5.264239145 | 0.868183247 | 0.477503641 | 0.672843444 | 5.989231611 | 5.80706993 | 1692.647059 | 240.9411765 | 22 | pathway | |||||
| CDT1 association with the CDC6:ORC:origin complex | R-HSA-68827 | 6773 | 8090 | 4.120089972 | 4.103498543 | 5.139495333 | 0.891894153 | 0.48154171 | 0.686717932 | 6.384723086 | 5.951482868 | 1954.456522 | 323.1086957 | 22 | pathway | |||||
| Cellular hexose transport | R-HSA-189200 | 30 | 3460 | 3.329764731 | 3.328436061 | 5.284108241 | 0.557987369 | 0.688793505 | 0.623390437 | 3.882519935 | 5.265857826 | 559.4285714 | 498.5714286 | 7 | pathway | |||||
| Cholesterol biosynthesis via desmosterol | R-HSA-6807047 | 14 | 97 | 3.452481089 | 3.442157089 | 4.805865094 | 0.75 | 0.469652993 | 0.609826496 | 3.870132743 | 5.216337006 | 68.5 | 27.75 | 2 | pathway | |||||
| Cilium Assembly | R-HSA-5617833 | 250 | 26943 | 3.527916923 | 3.530083012 | 5.432915193 | 0.90225918 | 0.514945666 | 0.708602423 | 5.089121221 | 5.89998846 | 22912.10526 | 1431.210526 | 15 | pathway | |||||
| CLEC7A (Dectin-1) signaling | R-HSA-5607764 | 5960 | 22237 | 3.904635975 | 3.878815947 | 5.069010546 | 0.763979142 | 0.488027295 | 0.626003219 | 5.299025829 | 5.474900116 | 1695.661765 | 414.6617647 | 35 | pathway | |||||
| CLEC7A/inflammasome pathway | R-HSA-5660668 | 5441 | 3725 | 3.745721387 | 3.715783489 | 4.96401314 | 0.762516184 | 0.602257721 | 0.682386952 | 5.347419791 | 5.797820145 | 3971.8 | 1833.2 | 3 | pathway | |||||
| COPI-dependent Golgi-to-ER retrograde traffic | R-HSA-6811434 | 354 | 17247 | 3.240276124 | 3.227073337 | 4.845668055 | 0.576446656 | 0.692280976 | 0.634363816 | 4.011356778 | 5.309184149 | 14182.60714 | 628.6071429 | 25 | pathway | |||||
| COPI-independent Golgi-to-ER retrograde traffic | R-HSA-6811436 | 185 | 16265 | 3.564986763 | 3.566907305 | 5.405340917 | 0.807569041 | 0.580611845 | 0.694090443 | 4.973043036 | 5.815598542 | 20824.36842 | 865.7894737 | 16 | pathway | |||||
| COPI-mediated anterograde transport | R-HSA-6807878 | 187 | 16129 | 3.63996045 | 3.642026753 | 5.293361228 | 0.807401967 | 0.564822261 | 0.686112114 | 4.941358313 | 5.78740091 | 19777.8 | 815.8 | 17 | pathway | |||||
| CREB phosphorylation through the activation of Adenylate Cyclase | R-HSA-442720 | 62 | 3531 | 3.746680555 | 3.66011776 | 5.641442355 | 0.851588838 | 0.520155179 | 0.685872008 | 5.264575685 | 5.821373573 | 871.3 | 359.3 | 7 | pathway | |||||
| Cristae formation | R-HSA-8949613 | 0 | 4 | 2 | 2 | 5 | 1 | 0.75 | 0.875 | 3.75 | 6.5 | 6 | 2 | 2 | pathway | |||||
| Cross-presentation of soluble exogenous antigens (endosomes) | R-HSA-1236978 | 267 | 6927 | 3.955596096 | 3.948870169 | 5.101995663 | 0.836734694 | 0.514148711 | 0.675441702 | 6.059771352 | 5.821476714 | 1614.959184 | 146.8163265 | 23 | pathway | |||||
| CS/DS degradation | R-HSA-2024101 | 4 | 92 | 3.143298808 | 3.13297168 | 4.88696119 | 1 | 0.329188988 | 0.664594494 | 4.198482127 | 5.47839623 | 57.5 | 24 | 2 | pathway | |||||
| CYP2E1 reactions | R-HSA-211999 | 2932 | 7918 | 3.559875043 | 3.531009146 | 3.805595868 | 0.656934402 | 0.666688876 | 0.661811639 | 4.756790554 | 5.598702609 | 2106 | 1356.25 | 8 | pathway | |||||
| Cytosolic sulfonation of small molecules | R-HSA-156584 | 3 | 14 | 3.053355729 | 3.053355729 | 4.876441084 | 1 | 0.324786325 | 0.662393162 | 4.409315313 | 5.433739659 | 9 | 5.666666667 | 3 | pathway | |||||
| Dectin-1 mediated noncanonical NF-kB signaling | R-HSA-5607761 | 3590 | 10641 | 4.020356106 | 4.013355929 | 5.182342049 | 0.862951924 | 0.476680667 | 0.669816296 | 6.060093769 | 5.805560674 | 1857.480769 | 273.6730769 | 26 | pathway | |||||
| Defective CFTR causes cystic fibrosis | R-HSA-5678895 | 280 | 7381 | 4.063835033 | 4.055316794 | 5.13439375 | 0.893562185 | 0.47505142 | 0.684306803 | 6.285286738 | 5.916657028 | 1697.06383 | 163 | 23 | pathway | |||||
| Degradation of AXIN | R-HSA-4641257 | 276 | 6991 | 4.081292517 | 4.075061387 | 5.259498777 | 0.848292634 | 0.508927581 | 0.678610107 | 6.194944736 | 5.884498221 | 1617.122449 | 148.3061224 | 23 | pathway | |||||
| Degradation of beta-catenin by the destruction complex | R-HSA-195253 | 353 | 10585 | 4.008188995 | 4.001721254 | 5.260659138 | 0.85753843 | 0.487883873 | 0.672711152 | 5.981789255 | 5.820804009 | 1586.698113 | 206.3773585 | 26 | pathway | |||||
| Degradation of DVL | R-HSA-4641258 | 267 | 6924 | 4.166170229 | 4.159304178 | 5.121164387 | 0.854166667 | 0.504026809 | 0.679096738 | 6.282141 | 5.916034995 | 1648.5625 | 149.8125 | 24 | pathway | |||||
| Degradation of GLI1 by the proteasome | R-HSA-5610780 | 328 | 10343 | 4.060265044 | 4.053773495 | 5.129349933 | 0.878510674 | 0.449900743 | 0.664205708 | 6.033853433 | 5.781459737 | 1668.942308 | 205.2115385 | 25 | pathway | |||||
| Degradation of GLI2 by the proteasome | R-HSA-5610783 | 408 | 13971 | 3.971843273 | 3.965585159 | 5.10068832 | 0.899610601 | 0.44686942 | 0.673240011 | 5.95757099 | 5.812214496 | 1666.981818 | 261.4363636 | 27 | pathway | |||||
| DEx/H-box helicases activate type I IFN and inflammatory cytokines production | R-HSA-3134963 | 7108 | 5312 | 3.563884321 | 3.546851934 | 4.794801493 | 0.706020133 | 0.707392109 | 0.706706121 | 5.38968845 | 5.899335579 | 3115.444444 | 1380 | 7 | pathway | |||||
| Digestion of dietary carbohydrate | R-HSA-189085 | 16 | 334 | 3.074058296 | 3.06667375 | 5.281938905 | 0.663650075 | 0.650369064 | 0.65700957 | 4.123539751 | 5.404749896 | 66.33333333 | 58.33333333 | 6 | pathway | |||||
| Displacement of DNA glycosylase by APEX1 | R-HSA-110357 | 1856 | 505 | 3.986170457 | 3.661314321 | 5.347838232 | 0.504674883 | 0.793623389 | 0.649149136 | 5.20498631 | 5.656384391 | 1244.5 | 1180.5 | 2 | pathway | |||||
| Downregulation of ERBB2:ERBB3 signaling | R-HSA-1358803 | 593 | 11786 | 3.244083057 | 3.243647186 | 5.053830529 | 0.887221408 | 0.338858855 | 0.613040131 | 3.725085024 | 5.168295228 | 1915.777778 | 1375.444444 | 4 | pathway | |||||
| Downregulation of SMAD2/3:SMAD4 transcriptional activity | R-HSA-2173795 | 7232 | 15248 | 3.392277551 | 3.264985555 | 5.669705681 | 0.660864419 | 0.553440876 | 0.607152647 | 3.68780169 | 5.1784435 | 3309.125 | 1405 | 7 | pathway | |||||
| Downstream TCR signaling | R-HSA-202424 | 6466 | 30469 | 3.714828361 | 3.705670404 | 5.375198663 | 0.690084194 | 0.554457912 | 0.622271053 | 4.88483384 | 5.386749807 | 1592.580247 | 455.9876543 | 42 | pathway | |||||
| ERCC6 (CSB) and EHMT2 (G9a) positively regulate rRNA expression | R-HSA-427389 | 5575 | 23432 | 3.430432007 | 3.377146209 | 5.761884452 | 0.873555574 | 0.463333251 | 0.668444413 | 4.417942838 | 5.599773422 | 10921.875 | 3625.875 | 3 | pathway | |||||
| ER-Phagosome pathway | R-HSA-1236974 | 359 | 12505 | 3.858171332 | 3.852596292 | 5.177510395 | 0.762603951 | 0.536270427 | 0.649437189 | 5.50348706 | 5.615638368 | 1474.644068 | 218.0338983 | 31 | pathway | |||||
| Fanconi Anemia Pathway | R-HSA-6783310 | 1273 | 1800 | 4.280496216 | 4.007691643 | 5.529703643 | 0.403333886 | 0.779036571 | 0.591185228 | 5.344938859 | 5.368066926 | 803.25 | 768.25 | 4 | pathway | |||||
| Fatty acids | R-HSA-211935 | 726 | 2682 | 3.978607374 | 3.958455542 | 4.304525124 | 0.420817728 | 0.757117464 | 0.588967596 | 4.230006862 | 5.252653098 | 615.375 | 426 | 8 | pathway | |||||
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | R-HSA-8854050 | 307 | 11364 | 4.084332357 | 4.077159362 | 5.151235401 | 0.912844837 | 0.463452409 | 0.688148623 | 6.370729125 | 5.949101606 | 1836.021739 | 253.7173913 | 22 | pathway | |||||
| FCERI mediated NF-kB activation | R-HSA-2871837 | 5979 | 22742 | 3.843684258 | 3.833001978 | 5.193536948 | 0.770007992 | 0.508263513 | 0.639135752 | 5.302878619 | 5.542133101 | 1714.347826 | 416.2463768 | 35 | pathway | |||||
| Formation of ATP by chemiosmotic coupling | R-HSA-163210 | 0 | 4 | 2 | 2 | 5 | 1 | 0.75 | 0.875 | 3.75 | 6.5 | 6 | 2 | 2 | pathway | |||||
| Formation of tubulin folding intermediates by CCT/TriC | R-HSA-389960 | 171 | 14934 | 3.579955872 | 3.582609527 | 5.385181577 | 0.884361273 | 0.542459321 | 0.713410297 | 5.217456649 | 5.949387271 | 22951.625 | 944.0625 | 13 | pathway | |||||
| G alpha (z) signalling events | R-HSA-418597 | 7777 | 20316 | 3.780162929 | 3.736435146 | 5.437832055 | 0.656580492 | 0.593419021 | 0.624999756 | 4.698069989 | 5.426719353 | 857.9193548 | 453.1129032 | 23 | pathway | |||||
| G2/M Checkpoints | R-HSA-69481 | 11376 | 7983 | 4.117888304 | 4.087677913 | 5.161130481 | 0.854166667 | 0.501227426 | 0.677697046 | 6.238583686 | 5.890609744 | 1955.833333 | 403.3125 | 22 | pathway | |||||
| GABA A receptor activation | R-HSA-977441 | 272 | 7037 | 3.370539017 | 3.367597224 | 6.079932096 | 0.736230504 | 0.516583876 | 0.62640719 | 3.932334179 | 5.299560939 | 2410.6875 | 152.2708333 | 12 | pathway | |||||
| Galactose catabolism | R-HSA-70370 | 14 | 5 | 4.189742164 | 4.22531655 | 5.326312083 | 0.5 | 0.833333333 | 0.666666667 | 5.493756594 | 5.841025166 | 9.5 | 9.5 | 2 | pathway | |||||
| Gap junction assembly | R-HSA-190861 | 184 | 16080 | 3.581033645 | 3.583531203 | 5.370695805 | 0.891061137 | 0.543013402 | 0.71703727 | 5.250371738 | 5.973926346 | 23263 | 956.7058824 | 14 | pathway | |||||
| GLI3 is processed to GLI3R by the proteasome | R-HSA-5610785 | 408 | 13966 | 3.95618758 | 3.949813574 | 5.090256623 | 0.916270057 | 0.447737373 | 0.682003715 | 6.032395252 | 5.865420624 | 1697.759259 | 266.1851852 | 26 | pathway | |||||
| Glucagon signaling in metabolic regulation | R-HSA-163359 | 4820 | 1348 | 3.924226713 | 3.833377082 | 6.341653701 | 0.677083333 | 0.707066143 | 0.692074738 | 5.603783725 | 5.921907158 | 586.125 | 385.5 | 12 | pathway | |||||
| HCN channels | R-HSA-1296061 | 2 | 49 | 3.133772117 | 3.133772117 | 5.615318691 | 0.666666667 | 0.625925926 | 0.646296296 | 4.175422987 | 5.353232681 | 19.66666667 | 17 | 3 | pathway | |||||
| HDACs deacetylate histones | R-HSA-3214815 | 379 | 51957 | 3.205908927 | 3.178472481 | 5.698606473 | 0.826448429 | 0.454543402 | 0.640495916 | 3.900111474 | 5.338609081 | 8516.434783 | 2275.478261 | 9 | pathway | |||||
| HDL assembly | R-HSA-8963896 | 61 | 3418 | 3.291100856 | 3.289502838 | 4.735164191 | 0.936511008 | 0.340310358 | 0.638410683 | 4.086144659 | 5.353104839 | 1530.8 | 695.8 | 3 | pathway | |||||
| Hedgehog ligand biogenesis | R-HSA-5358346 | 290 | 7010 | 4.116452803 | 4.102804084 | 5.13158916 | 0.854166667 | 0.496848282 | 0.675507474 | 6.225270236 | 5.875534097 | 1651.229167 | 152.0833333 | 24 | pathway | |||||
| Hedgehog 'off' state | R-HSA-5610787 | 5040 | 18924 | 3.766149137 | 3.728791394 | 5.679313569 | 0.780423716 | 0.537442721 | 0.658933219 | 5.020087928 | 5.648271169 | 9383.184211 | 630.6315789 | 29 | pathway | |||||
| Hedgehog 'on' state | R-HSA-5632684 | 290 | 7609 | 4.129924836 | 4.123324057 | 5.185374734 | 0.85572621 | 0.461506286 | 0.658616248 | 6.079174585 | 5.767416597 | 1602.06 | 157.98 | 26 | pathway | |||||
| Heme degradation | R-HSA-189483 | 21 | 55 | 3.530543349 | 3.440411709 | 3.993716078 | 1 | 0.409232955 | 0.704616477 | 5.16063957 | 5.874290965 | 64.5 | 38 | 2 | pathway | |||||
| Hh mutants that don't undergo autocatalytic processing are degraded by ERAD | R-HSA-5362768 | 277 | 6999 | 4.120934633 | 4.112397961 | 5.133465914 | 0.872340426 | 0.486142926 | 0.679241676 | 6.29539609 | 5.901922718 | 1685.744681 | 154.8085106 | 23 | pathway | |||||
| HSP90 chaperone cycle for steroid hormone receptors (SHR) | R-HSA-3371497 | 3334 | 27652 | 3.525242267 | 3.485424656 | 5.426869967 | 0.771867136 | 0.545382731 | 0.658624934 | 4.503613131 | 5.565913647 | 14285.46667 | 1032.866667 | 25 | pathway | |||||
| Hyaluronan uptake and degradation | R-HSA-2160916 | 7 | 137 | 3.121192848 | 3.105439062 | 4.94313968 | 0.714285714 | 0.596271259 | 0.655278486 | 3.995514384 | 5.408920859 | 41 | 20.57142857 | 5 | pathway | |||||
| IkBA variant leads to EDA-ID | R-HSA-5603029 | 7149 | 8190 | 3.135112945 | 3.119245345 | 5.157491608 | 0.708460267 | 0.559602202 | 0.634031234 | 4.06005358 | 5.271912544 | 2956.090909 | 1394.454545 | 7 | pathway | |||||
| Interaction between L1 and Ankyrins | R-HSA-445095 | 284 | 2240 | 3.376446799 | 3.37504474 | 5.494758854 | 0.603615234 | 0.701649765 | 0.652632499 | 4.578894473 | 5.476365595 | 289.8695652 | 109.7391304 | 11 | pathway | |||||
| Interleukin-1 family signaling | R-HSA-446652 | 6050 | 17240 | 3.796748821 | 3.780263283 | 5.152795315 | 0.815968354 | 0.493861427 | 0.654914891 | 5.441854297 | 5.631682211 | 1615.940299 | 347.6119403 | 38 | pathway | |||||
| Interleukin-1 processing | R-HSA-448706 | 7782 | 5366 | 3.676526526 | 3.644810078 | 4.691200303 | 0.917226645 | 0.575535819 | 0.746381232 | 6.011459499 | 6.201383722 | 4925.166667 | 2191.333333 | 4 | pathway | |||||
| Intraflagellar transport | R-HSA-5620924 | 158 | 13788 | 3.578734395 | 3.581564961 | 5.401598785 | 0.876768093 | 0.541831363 | 0.709299728 | 5.180152882 | 5.92157632 | 22598.73333 | 929.7333333 | 12 | pathway | |||||
| Keratan sulfate degradation | R-HSA-2022857 | 6 | 122 | 3.172655983 | 3.159719696 | 4.571209862 | 0.875 | 0.38210119 | 0.628550595 | 3.917750622 | 5.247889296 | 53 | 25.6 | 3 | pathway | |||||
| Kinesins | R-HSA-983189 | 354 | 17248 | 3.217284753 | 3.204081966 | 4.822989484 | 0.576446656 | 0.692280976 | 0.634363816 | 3.99411325 | 5.301520358 | 14182.64286 | 628.6428571 | 24 | pathway | |||||
| Leukotriene receptors | R-HSA-391906 | 466 | 3207 | 3.505856023 | 3.495351839 | 5.936687619 | 0.996298549 | 0.425582335 | 0.710940442 | 5.212074234 | 5.908221619 | 1131.75 | 459.125 | 5 | pathway | |||||
| Ligand-gated ion channel transport | R-HSA-975298 | 353 | 9232 | 3.332848558 | 3.327240228 | 6.051097066 | 0.724048508 | 0.531042112 | 0.62754531 | 3.916611347 | 5.294584919 | 1864.382353 | 140.9558824 | 22 | pathway | |||||
| MAPK6/MAPK4 signaling | R-HSA-5687128 | 2799 | 18843 | 3.695716858 | 3.673987863 | 4.953773522 | 0.800213662 | 0.497020781 | 0.648617222 | 5.192501939 | 5.556020431 | 1344.526316 | 284.7631579 | 41 | pathway | |||||
| Metabolism of folate and pterines | R-HSA-196757 | 20 | 1096 | 3.420606139 | 3.414810062 | 5.313860062 | 0.799030068 | 0.444213942 | 0.621622005 | 3.925539132 | 5.284348745 | 365.4 | 223.2 | 5 | pathway | |||||
| Microtubule-dependent trafficking of connexons from Golgi to the plasma membrane | R-HSA-190840 | 184 | 16080 | 3.581033645 | 3.583531203 | 5.370695805 | 0.891061137 | 0.543013402 | 0.71703727 | 5.250371738 | 5.973926346 | 23263 | 956.7058824 | 14 | pathway | |||||
| Mitochondrial protein import | R-HSA-1268020 | 148 | 31 | 2.546910922 | 2.283554329 | 4.492930011 | 0.5 | 0.856321839 | 0.67816092 | 2.985721998 | 5.370043104 | 47.5 | 44.75 | 4 | pathway | |||||
| mTOR signalling | R-HSA-165159 | 352 | 10862 | 3.384714666 | 3.327263825 | 4.75597385 | 0.828575734 | 0.470437932 | 0.649506833 | 4.379099193 | 5.458283773 | 2447.5 | 1869 | 4 | pathway | |||||
| Muscarinic acetylcholine receptors | R-HSA-390648 | 2296 | 10906 | 3.103274816 | 3.101904147 | 5.989798131 | 0.883211731 | 0.427643197 | 0.655427464 | 3.991410969 | 5.403941364 | 1658.642857 | 943 | 5 | pathway | |||||
| NCAM1 interactions | R-HSA-419037 | 111 | 861 | 3.434706967 | 3.394636919 | 5.691729953 | 0.717822298 | 0.582255093 | 0.650038696 | 4.289873617 | 5.478493628 | 224.8 | 97.2 | 7 | pathway | |||||
| Neddylation | R-HSA-8951664 | 1054 | 7640 | 4.014663902 | 3.967857272 | 5.112012297 | 0.751767486 | 0.53627167 | 0.644019578 | 5.639698597 | 5.631685153 | 1377.40678 | 147.3559322 | 32 | pathway | |||||
| Negative regulation of the PI3K/AKT network | R-HSA-199418 | 118 | 8208 | 3.460114301 | 3.45985695 | 4.972344948 | 0.87321982 | 0.341532801 | 0.607376311 | 3.833153016 | 5.202546839 | 1359.875 | 1040.75 | 5 | pathway | |||||
| Neurophilin interactions with VEGF and VEGFR | R-HSA-194306 | 522 | 11865 | 3.334084697 | 3.334547003 | 5.4543995 | 0.758480549 | 0.489665383 | 0.624072966 | 4.01336523 | 5.271848006 | 3531.8 | 2477.4 | 3 | pathway | |||||
| Neurotransmitter clearance | R-HSA-112311 | 609 | 6706 | 3.190603962 | 3.152934924 | 4.944768657 | 0.967082053 | 0.389303309 | 0.678192681 | 4.543359553 | 5.584819192 | 2697 | 1828.75 | 4 | pathway | |||||
| Neurotransmitter receptors and postsynaptic signal transmission | R-HSA-112314 | 272 | 7037 | 3.370539017 | 3.367597224 | 6.079932096 | 0.736230504 | 0.516583876 | 0.62640719 | 3.932334179 | 5.299560939 | 2410.6875 | 152.2708333 | 12 | pathway | |||||
| NIK-->noncanonical NF-kB signaling | R-HSA-5676590 | 1936 | 9288 | 4.046268725 | 4.039352778 | 5.187258129 | 0.859614313 | 0.47791137 | 0.668762842 | 6.102221657 | 5.807175186 | 1773.78 | 224.48 | 25 | pathway | |||||
| Norepinephrine Neurotransmitter Release Cycle | R-HSA-181430 | 590 | 2632 | 3.231269152 | 3.198687623 | 4.859763806 | 0.776292092 | 0.537690693 | 0.656991392 | 4.361536362 | 5.457032333 | 951.8 | 644.4 | 4 | pathway | |||||
| NOTCH1 Intracellular Domain Regulates Transcription | R-HSA-2122947 | 4637 | 110316 | 3.31115049 | 3.268722983 | 5.357093759 | 0.70691444 | 0.518146212 | 0.612530326 | 3.70516342 | 5.187252339 | 8957.4375 | 2394.854167 | 21 | pathway | |||||
| Orc1 removal from chromatin | R-HSA-68949 | 386 | 13959 | 3.868389266 | 3.86223947 | 5.403099908 | 0.794372749 | 0.514294289 | 0.654333519 | 5.578024658 | 5.651686546 | 1540.741379 | 247.3275862 | 25 | pathway | |||||
| Organic anion transport | R-HSA-561048 | 32 | 393 | 3.838933511 | 3.845104208 | 4.794910838 | 0.595867183 | 0.650916691 | 0.623391937 | 4.570192781 | 5.435590751 | 95.8 | 85 | 5 | pathway | |||||
| Organic cation transport | R-HSA-549127 | 856 | 309 | 3.60560085 | 3.3879826 | 4.7669483 | 0.614663913 | 0.680968747 | 0.64781633 | 4.5412708 | 5.520642481 | 326 | 166.4285714 | 7 | pathway | |||||
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | R-HSA-1234176 | 521 | 7620 | 3.898398994 | 3.889482124 | 5.32896104 | 0.706774187 | 0.591274812 | 0.6490245 | 5.537378341 | 5.62629633 | 1366.644068 | 137.9830508 | 27 | pathway | |||||
| p75NTR negatively regulates cell cycle via SC1 | R-HSA-193670 | 211 | 32490 | 3.119978831 | 3.119328237 | 5.310940059 | 0.855906075 | 0.452623173 | 0.654264624 | 3.995186549 | 5.401757102 | 8524.285714 | 2335.785714 | 5 | pathway | |||||
| Phase 0 - rapid depolarisation | R-HSA-5576892 | 458 | 7112 | 3.389891763 | 3.384044188 | 5.375341596 | 0.743449046 | 0.561745425 | 0.652597235 | 4.489935683 | 5.480612157 | 401 | 189.25 | 21 | pathway | |||||
| Phase 2 - plateau phase | R-HSA-5576893 | 76 | 1106 | 3.126506861 | 3.120812435 | 5.613673307 | 0.685879594 | 0.614683306 | 0.65028145 | 4.042491743 | 5.37737862 | 215.2307692 | 90.92307692 | 8 | pathway | |||||
| Physiological factors | R-HSA-5578768 | 55 | 1577 | 4.16682572 | 4.161292913 | 5.868916068 | 0.77054188 | 0.6701431 | 0.72034249 | 6.803206851 | 6.191225173 | 339.1666667 | 272 | 5 | pathway | |||||
| PKA activation | R-HSA-163615 | 69 | 3800 | 3.771857851 | 3.718377102 | 5.842840239 | 0.869257283 | 0.546134305 | 0.707695794 | 5.606298017 | 5.975257909 | 573.0526316 | 203.6315789 | 13 | pathway | |||||
| PKA activation in glucagon signalling | R-HSA-164378 | 4880 | 4508 | 3.831491424 | 3.754565941 | 5.854376708 | 0.816625704 | 0.580964499 | 0.698795102 | 5.543086246 | 5.935797819 | 878.5789474 | 494.1052632 | 13 | pathway | |||||
| PKA-mediated phosphorylation of key metabolic factors | R-HSA-163358 | 61 | 3418 | 3.291100856 | 3.289502838 | 4.735164191 | 0.936511008 | 0.340310358 | 0.638410683 | 4.086144659 | 5.353104839 | 1530.8 | 695.8 | 3 | pathway | |||||
| Post-chaperonin tubulin folding pathway | R-HSA-389977 | 171 | 14934 | 3.579955872 | 3.582609527 | 5.385181577 | 0.884361273 | 0.542459321 | 0.713410297 | 5.217456649 | 5.949387271 | 22951.625 | 944.0625 | 13 | pathway | |||||
| Prefoldin mediated transfer of substrate to CCT/TriC | R-HSA-389957 | 145 | 12642 | 3.577338422 | 3.580371171 | 5.420361309 | 0.868090174 | 0.541113697 | 0.704601935 | 5.137520005 | 5.889792375 | 22195.42857 | 913.3571429 | 11 | pathway | |||||
| Recruitment of NuMA to mitotic centrosomes | R-HSA-380320 | 2180 | 28952 | 3.345578615 | 3.332006619 | 5.061025814 | 0.713500496 | 0.551378402 | 0.632439449 | 4.052630159 | 5.331455867 | 11374.89744 | 798.2564103 | 25 | pathway | |||||
| Recycling of bile acids and salts | R-HSA-159418 | 1818 | 3756 | 3.58602971 | 3.474117181 | 4.737760614 | 0.617962831 | 0.641899252 | 0.629931041 | 4.299966471 | 5.394883512 | 578.9333333 | 371.6 | 13 | pathway | |||||
| Recycling pathway of L1 | R-HSA-437239 | 2349 | 45494 | 3.379700641 | 3.37822314 | 4.939090518 | 0.847474607 | 0.488592276 | 0.668033441 | 4.509305818 | 5.580123156 | 16953.54167 | 996.7291667 | 25 | pathway | |||||
| Regulated proteolysis of p75NTR | R-HSA-193692 | 5545 | 26785 | 3.449733323 | 3.419818481 | 5.531069322 | 0.69300073 | 0.528019247 | 0.610509989 | 3.992320828 | 5.219977698 | 7249.153846 | 2486.923077 | 9 | pathway | |||||
| Regulation of activated PAK-2p34 by proteasome mediated degradation | R-HSA-211733 | 291 | 7510 | 4.08617844 | 4.079018613 | 5.119888145 | 0.912955394 | 0.463357338 | 0.688156366 | 6.373348287 | 5.949768588 | 1740.282609 | 169.5869565 | 22 | pathway | |||||
| Regulation of gene expression in beta cells | R-HSA-210745 | 3 | 927 | 3.887200619 | 3.885320117 | 4.117514573 | 0.666666667 | 0.502237438 | 0.584452052 | 3.697195988 | 5.192080556 | 454.3333333 | 310 | 3 | pathway | |||||
| Regulation of lipid metabolism by Peroxisome proliferator-activated receptor alpha (PPARalpha) | R-HSA-400206 | 753 | 14190 | 3.483243984 | 3.362196277 | 5.688042558 | 0.665304159 | 0.566369557 | 0.615836858 | 3.890419418 | 5.266660382 | 2577.722222 | 830.1666667 | 10 | pathway | |||||
| Regulation of ornithine decarboxylase (ODC) | R-HSA-350562 | 273 | 7252 | 4.068181113 | 4.061158105 | 5.137850939 | 0.914822458 | 0.461126692 | 0.687974575 | 6.33313764 | 5.942557536 | 1692.595745 | 160.106383 | 23 | pathway | |||||
| Regulation of PTEN stability and activity | R-HSA-8948751 | 564 | 20121 | 3.878277806 | 3.87179509 | 5.325043064 | 0.818415472 | 0.493614538 | 0.656015005 | 5.502285799 | 5.666192635 | 1507.430769 | 318.2307692 | 32 | pathway | |||||
| Regulation of pyruvate dehydrogenase (PDH) complex | R-HSA-204174 | 1600 | 4721 | 3.449405106 | 3.442347385 | 5.549421423 | 0.799753599 | 0.435992975 | 0.617873287 | 3.825148115 | 5.268956948 | 1185.3 | 632.1 | 6 | pathway | |||||
| Regulation of RAS by GAPs | R-HSA-5658442 | 269 | 6942 | 4.085308675 | 4.078314571 | 5.106185033 | 0.893617021 | 0.478744237 | 0.686180629 | 6.334219494 | 5.936307086 | 1684.12766 | 153.4255319 | 23 | pathway | |||||
| Regulation of RUNX3 expression and activity | R-HSA-8941858 | 1058 | 12494 | 4.012742533 | 3.989400475 | 5.177212955 | 0.800641733 | 0.527118045 | 0.663879889 | 5.897931152 | 5.763446768 | 1682.72 | 271.04 | 24 | pathway | |||||
| Regulation of TP53 Activity through Acetylation | R-HSA-6804758 | 11372 | 31763 | 3.414505315 | 3.343655426 | 5.430646155 | 0.765862218 | 0.474660697 | 0.620261457 | 3.920616323 | 5.273244819 | 6255.588235 | 2537.352941 | 7 | pathway | |||||
| Repression of WNT target genes | R-HSA-4641265 | 144 | 25140 | 3.334880797 | 3.334485014 | 5.726347537 | 0.79900742 | 0.52441713 | 0.661712275 | 4.281212859 | 5.523042099 | 8405.6 | 2528.4 | 2 | pathway | |||||
| Respiratory electron transport | R-HSA-611105 | 5 | 106 | 3.392988688 | 3.390741972 | 5.188951008 | 0.666666667 | 0.657602339 | 0.662134503 | 4.436567375 | 5.545226249 | 21.5 | 18.5 | 6 | pathway | |||||
| Retinoid cycle disease events | R-HSA-2453864 | 34 | 402 | 3.297791423 | 3.22947319 | 5.048347474 | 0.897555272 | 0.550964066 | 0.724259669 | 4.914294508 | 5.927661602 | 220.3333333 | 145.3333333 | 3 | pathway | |||||
| RHO GTPases activate IQGAPs | R-HSA-5626467 | 14784 | 19182 | 3.525218144 | 3.506435967 | 5.218275549 | 0.733311924 | 0.575730454 | 0.654521189 | 4.505412822 | 5.53854731 | 16624.8 | 1358.64 | 19 | pathway | |||||
| RIP-mediated NFkB activation via ZBP1 | R-HSA-1810476 | 7188 | 8844 | 3.302810238 | 3.291679688 | 4.854012559 | 0.608907444 | 0.64090016 | 0.624903802 | 4.084218177 | 5.266962093 | 1967.588235 | 943.0588235 | 13 | pathway | |||||
| Role of Abl in Robo-Slit signaling | R-HSA-428890 | 134 | 11022 | 3.041414491 | 3.041147112 | 5.452634469 | 0.989083218 | 0.29067716 | 0.639880189 | 3.78144553 | 5.279672757 | 2668.6 | 1115.6 | 2 | pathway | |||||
| RUNX1 regulates estrogen receptor mediated transcription | R-HSA-8931987 | 2341 | 5258 | 3.869028211 | 3.569689225 | 5.205214358 | 0.395735866 | 0.798565073 | 0.597150469 | 4.142072776 | 5.270679199 | 2928 | 1519.8 | 3 | pathway | |||||
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | R-HSA-8939236 | 20234 | 16229 | 3.959275997 | 3.91855434 | 5.214117009 | 0.749401701 | 0.534501781 | 0.641951741 | 5.51549306 | 5.599436937 | 2000.483333 | 607.7166667 | 29 | pathway | |||||
| RUNX1 regulates transcription of genes involved in WNT signaling | R-HSA-8939256 | 2341 | 5258 | 3.869028211 | 3.569689225 | 5.205214358 | 0.395735866 | 0.798565073 | 0.597150469 | 4.142072776 | 5.270679199 | 2928 | 1519.8 | 3 | pathway | |||||
| SCF(Skp2)-mediated degradation of p27/p21 | R-HSA-187577 | 474 | 21592 | 3.647267719 | 3.641759565 | 5.442852299 | 0.680867996 | 0.567285085 | 0.62407654 | 4.820785232 | 5.376266176 | 1293.918919 | 298.1891892 | 28 | pathway | |||||
| SCF-beta-TrCP mediated degradation of Emi1 | R-HSA-174113 | 267 | 6917 | 4.115530933 | 4.108207145 | 5.141119457 | 0.911111111 | 0.47096193 | 0.69103652 | 6.454662416 | 5.97875378 | 1758.288889 | 159.6444444 | 21 | pathway | |||||
| Separation of Sister Chromatids | R-HSA-2467813 | 2110 | 28838 | 3.796177917 | 3.789566869 | 5.380333837 | 0.782811507 | 0.543491579 | 0.663151543 | 5.430394132 | 5.686402923 | 6359.315789 | 407.2105263 | 48 | pathway | |||||
| Signaling by BMP | R-HSA-201451 | 91 | 628 | 3.444120028 | 3.444702732 | 5.199996702 | 0.995488722 | 0.278848827 | 0.637168774 | 4.417849828 | 5.395831839 | 135.7142857 | 102.7142857 | 7 | pathway | |||||
| Stimuli-sensing channels | R-HSA-2672351 | 111 | 3661 | 3.204074122 | 3.159018646 | 4.874614381 | 0.669551517 | 0.623295991 | 0.646423754 | 4.174745328 | 5.3775164 | 264.8947368 | 198.5263158 | 17 | pathway | |||||
| Synthesis of bile acids and bile salts | R-HSA-192105 | 907 | 1937 | 3.837563372 | 3.601760569 | 4.667139783 | 0.499154285 | 0.693474933 | 0.596314609 | 4.035982205 | 5.254618516 | 878.6666667 | 474 | 4 | pathway | |||||
| Synthesis of epoxy (EET) and dihydroxyeicosatrienoic acids (DHET) | R-HSA-2142670 | 2080 | 6545 | 3.342034003 | 3.30835251 | 5.054499645 | 0.612781595 | 0.625469983 | 0.619125789 | 3.928346862 | 5.241516594 | 1576.75 | 1078.125 | 8 | pathway | |||||
| Tachykinin receptors bind tachykinins | R-HSA-380095 | 895 | 4604 | 3.603614146 | 3.603560506 | 6.204880073 | 0.660832364 | 0.559372602 | 0.610102483 | 4.119543879 | 5.268554604 | 2717.666667 | 1833 | 3 | pathway | |||||
| The NLRP1 inflammasome | R-HSA-844455 | 31 | 1614 | 3.444769745 | 3.255164903 | 5.809388476 | 0.545638117 | 0.823160729 | 0.684399423 | 4.414631737 | 5.7109194 | 514.25 | 411.25 | 3 | pathway | |||||
| The role of GTSE1 in G2/M progression after G2 checkpoint | R-HSA-8852276 | 13499 | 34685 | 3.803801721 | 3.774033903 | 5.25808962 | 0.803493874 | 0.520395157 | 0.661944516 | 5.382723933 | 5.680897347 | 6384.8375 | 602.3 | 43 | pathway | |||||
| TNFR1-mediated ceramide production | R-HSA-5626978 | 53 | 52 | 3.380175312 | 3.334182293 | 4.907337269 | 0.398611111 | 0.814637681 | 0.606624396 | 3.80979345 | 5.170887745 | 44 | 35 | 3 | pathway | |||||
| TNFR2 non-canonical NF-kB pathway | R-HSA-5668541 | 320 | 7294 | 4.154220536 | 4.1452292 | 5.154012325 | 0.796073718 | 0.540233308 | 0.668153513 | 6.07364348 | 5.839096933 | 1532.826923 | 146.4230769 | 27 | pathway | |||||
| TRAF6 mediated NF-kB activation | R-HSA-933542 | 7203 | 8884 | 3.196140737 | 3.185010186 | 5.004852148 | 0.667730973 | 0.596712719 | 0.632221846 | 4.098735843 | 5.280192554 | 1965.352941 | 946.2941176 | 12 | pathway | |||||
| Translocation of GLUT4 to the plasma membrane | R-HSA-1445148 | 435 | 24358 | 3.492088485 | 3.485154628 | 5.081142732 | 0.520295468 | 0.690122046 | 0.605208757 | 3.871977405 | 5.198754544 | 8876.847826 | 538.9782609 | 28 | pathway | |||||
| Transport of bile salts and organic acids, metal ions and amine compounds | R-HSA-425366 | 117 | 70 | 3.717643521 | 3.564999442 | 4.790819237 | 0.666666667 | 0.712349931 | 0.689508299 | 5.289736129 | 5.8359365 | 95 | 62.33333333 | 3 | pathway | |||||
| Transport of organic anions | R-HSA-879518 | 948 | 1518 | 3.896152259 | 3.867755765 | 5.363097789 | 0.872549792 | 0.607544707 | 0.74004725 | 6.338725458 | 6.232365751 | 343.2222222 | 274 | 9 | pathway | |||||
| Triglyceride catabolism | R-HSA-163560 | 123 | 4018 | 3.369785307 | 3.302066851 | 5.465511409 | 0.697143373 | 0.552083978 | 0.624613675 | 3.947027035 | 5.287352937 | 531.0625 | 258.8125 | 13 | pathway | |||||
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | R-HSA-69601 | 614 | 15170 | 4.018967544 | 4.004926581 | 5.097583229 | 0.884279539 | 0.466252174 | 0.675265857 | 6.085093467 | 5.841428226 | 1796.6 | 315.68 | 25 | pathway | |||||
| Ubiquitin-dependent degradation of Cyclin D1 | R-HSA-69229 | 313 | 18331 | 3.857254911 | 3.852024144 | 5.23440367 | 0.755912944 | 0.535174813 | 0.645543878 | 5.38168726 | 5.589377494 | 1472.84127 | 295.9365079 | 23 | pathway | |||||
| Ub-specific processing proteases | R-HSA-5689880 | 21610 | 18786 | 3.836169317 | 3.772923192 | 5.368535344 | 0.550126022 | 0.656495046 | 0.603310534 | 4.677022044 | 5.300793334 | 1291.383838 | 408.040404 | 57 | pathway | |||||
| UCH proteinases | R-HSA-5689603 | 827 | 7594 | 3.996753442 | 3.987811911 | 5.09370225 | 0.857404808 | 0.490212969 | 0.673808889 | 6.002698953 | 5.824310405 | 1527.87037 | 155.9444444 | 29 | pathway | |||||
| Uptake and function of diphtheria toxin | R-HSA-5336415 | 224 | 2015 | 3.853091195 | 3.719674301 | 5.076820093 | 0.627364999 | 0.552709544 | 0.590037271 | 4.06182002 | 5.217945541 | 632.75 | 279.875 | 5 | pathway | |||||
| Vasopressin regulates renal water homeostasis via Aquaporins | R-HSA-432040 | 4883 | 5342 | 3.795952128 | 3.722875075 | 5.872467754 | 0.825794419 | 0.567233308 | 0.696513863 | 5.471365108 | 5.908743132 | 898.5 | 511.25 | 14 | pathway | |||||
| Vif-mediated degradation of APOBEC3G | R-HSA-180585 | 267 | 6917 | 4.115530933 | 4.108207145 | 5.141119457 | 0.911111111 | 0.47096193 | 0.69103652 | 6.454662416 | 5.97875378 | 1758.288889 | 159.6444444 | 21 | pathway | |||||
| Vitamin D (calciferol) metabolism | R-HSA-196791 | 2 | 100 | 3.350809632 | 3.349471868 | 4.664983287 | 0.75 | 0.694977169 | 0.722488584 | 5.036397261 | 5.933527107 | 39.75 | 25.5 | 4 | pathway | |||||
| Vitamins | R-HSA-211916 | 1 | 96 | 3.186199386 | 3.18595113 | 5.952186498 | 0.666666667 | 0.615525114 | 0.64109589 | 4.002834163 | 5.336039065 | 49.33333333 | 32.33333333 | 3 | pathway | |||||
| VLDL assembly | R-HSA-8866423 | 13 | 129 | 3.751741081 | 3.667102768 | 7.023795566 | 0.233009709 | 0.949838188 | 0.591423948 | 3.835212212 | 5.193406682 | 52.33333333 | 47.33333333 | 3 | pathway | |||||
| Vpu mediated degradation of CD4 | R-HSA-180534 | 267 | 6928 | 4.116595979 | 4.109431405 | 5.135962782 | 0.891304348 | 0.482462757 | 0.686883553 | 6.3822435 | 5.951422343 | 1720.586957 | 156.4130435 | 22 | pathway | |||||
| Xenobiotics | R-HSA-211981 | 3964 | 12846 | 3.405351795 | 3.365014169 | 4.361208198 | 0.597841093 | 0.695232847 | 0.64653697 | 4.304697109 | 5.445363729 | 1355.736842 | 884.7368421 | 17 | pathway |