Have a look on what Machine Learning can do for the P53 community:
- Design of compound libraries by prediction of activity.
- Identify new targets.
- Pathways analysis.
- Repurpose drugs.
Make your own P53 ad hoc compound set
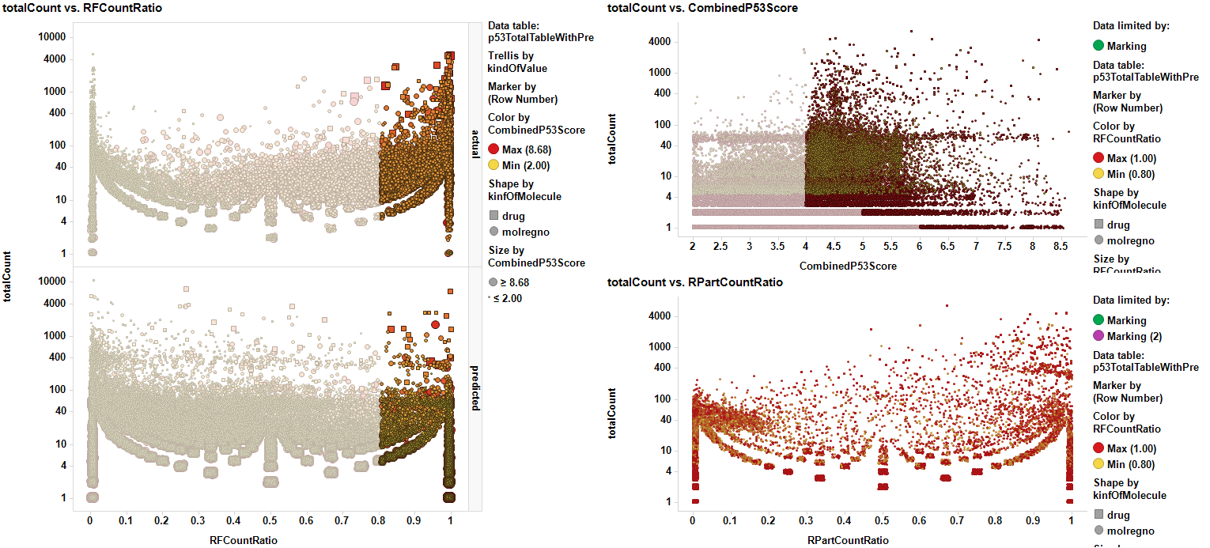
Select from tables or downloads section compounds predicted to be active enhancing P53 expression. Predictions have been carried out by combination of machine learning algorithms applied to experimental data stored in the ChEMBL DB.
Go to prediction of activity of compounds on P53 enhancement
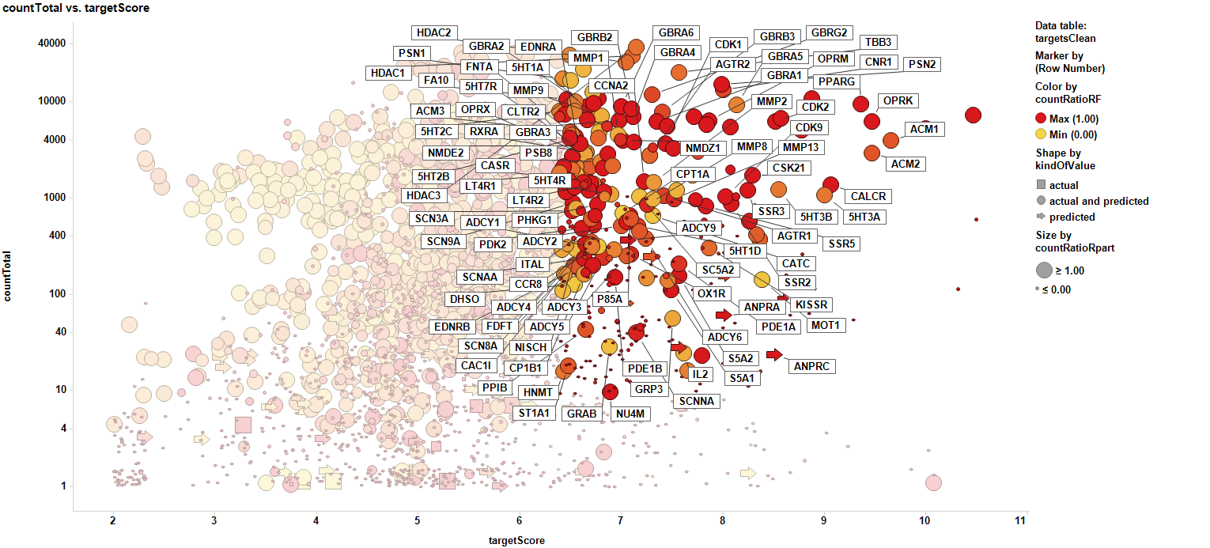
Test known and unforeseen protein targets involved in P53 expression
You can use the information stored in the P53 target identification section to select targets with different properties activating P53. In this section, the positive molecule-P53 pairs are aggregated by protein name while several prediction scores ara calculated and compared with aggregated Target scores.
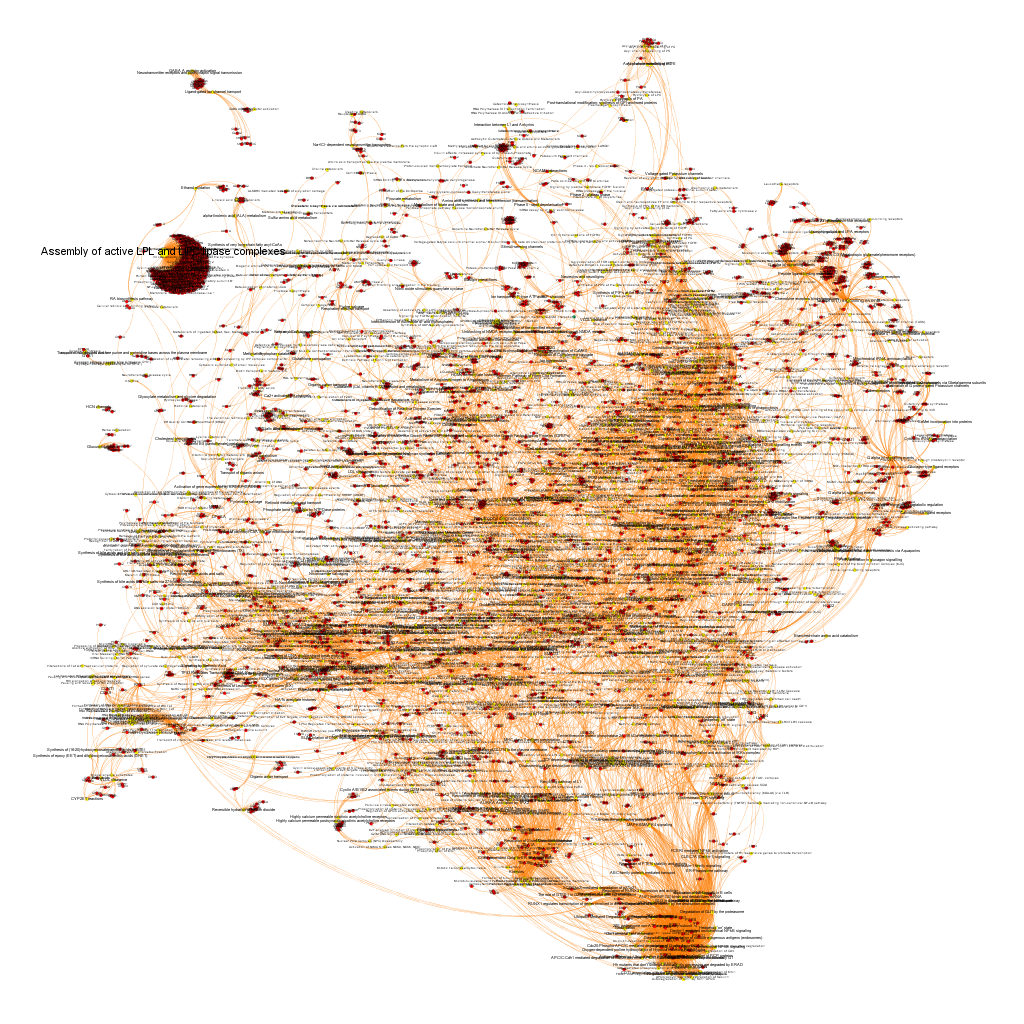
Perform P53 pathways analysis
In this section scored targets are associated to pathways which are equally scored for their role in P53 enhancement, the average potency of the positive interactions target-molecule, and the number of protein targets in the pathway having positive records in the ChEMBL DB. Pathways and targets are transformed in graphs so that their relations can be explored and queried.
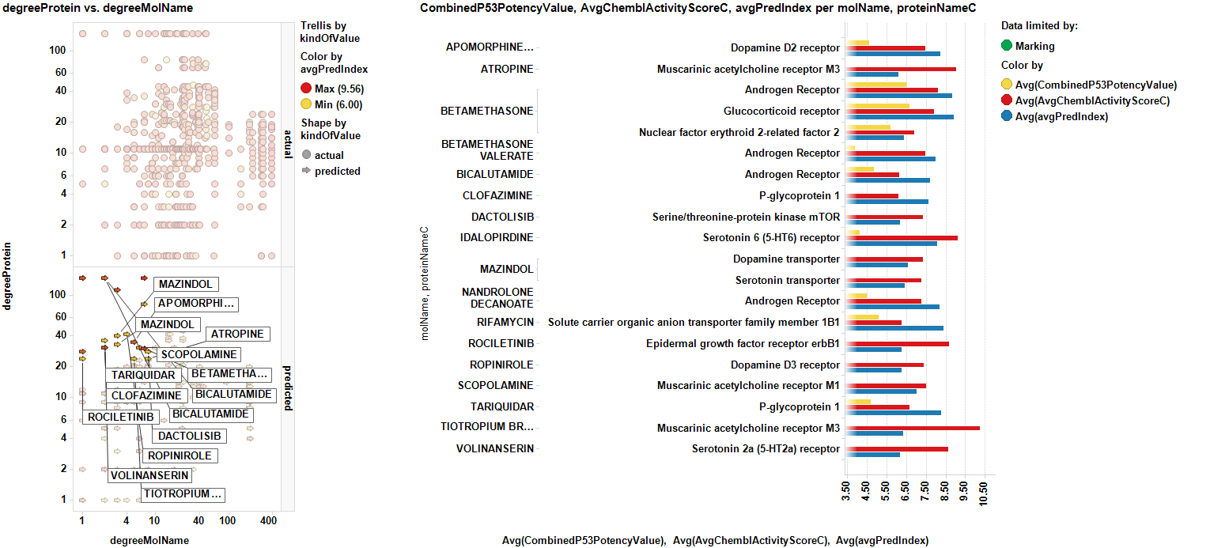
Repurpose drugs to increase P53 expression
ChEMBL DB has about 14M experimental records for 1.5M molecules. About 3k out of them are prescription drugs and there is a prediction based on machine learning algorithms for P53 enhancing activity. Wouldn’t it be easy to test a few of them at t5he bench? In this section you can go to plots and tables and decide what drugs to test. There is drug-target(s) interaction information for each one and a smart analysis based on graphs to remove most promiscuous drugs from selection. Extended information may be obtained at the download section.