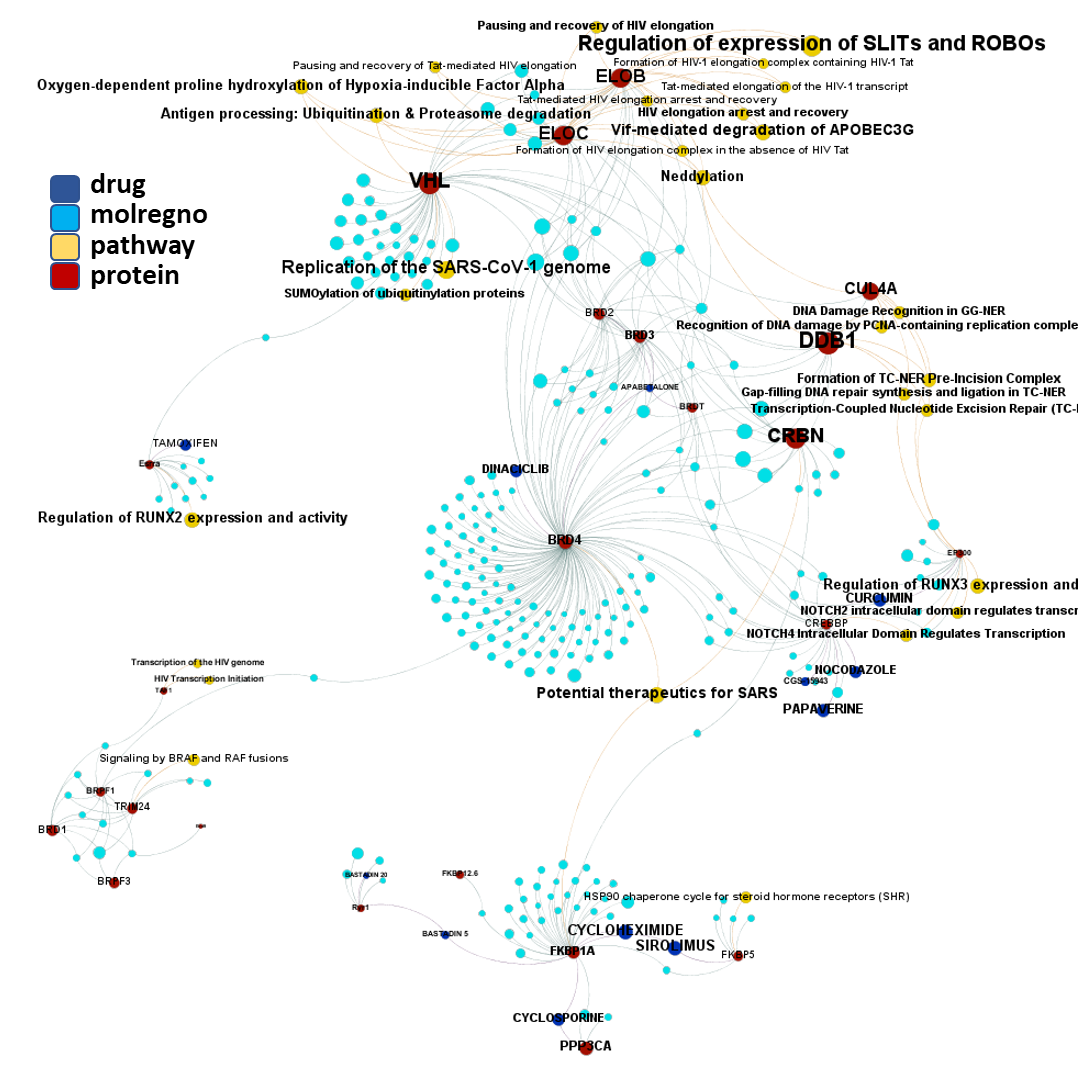
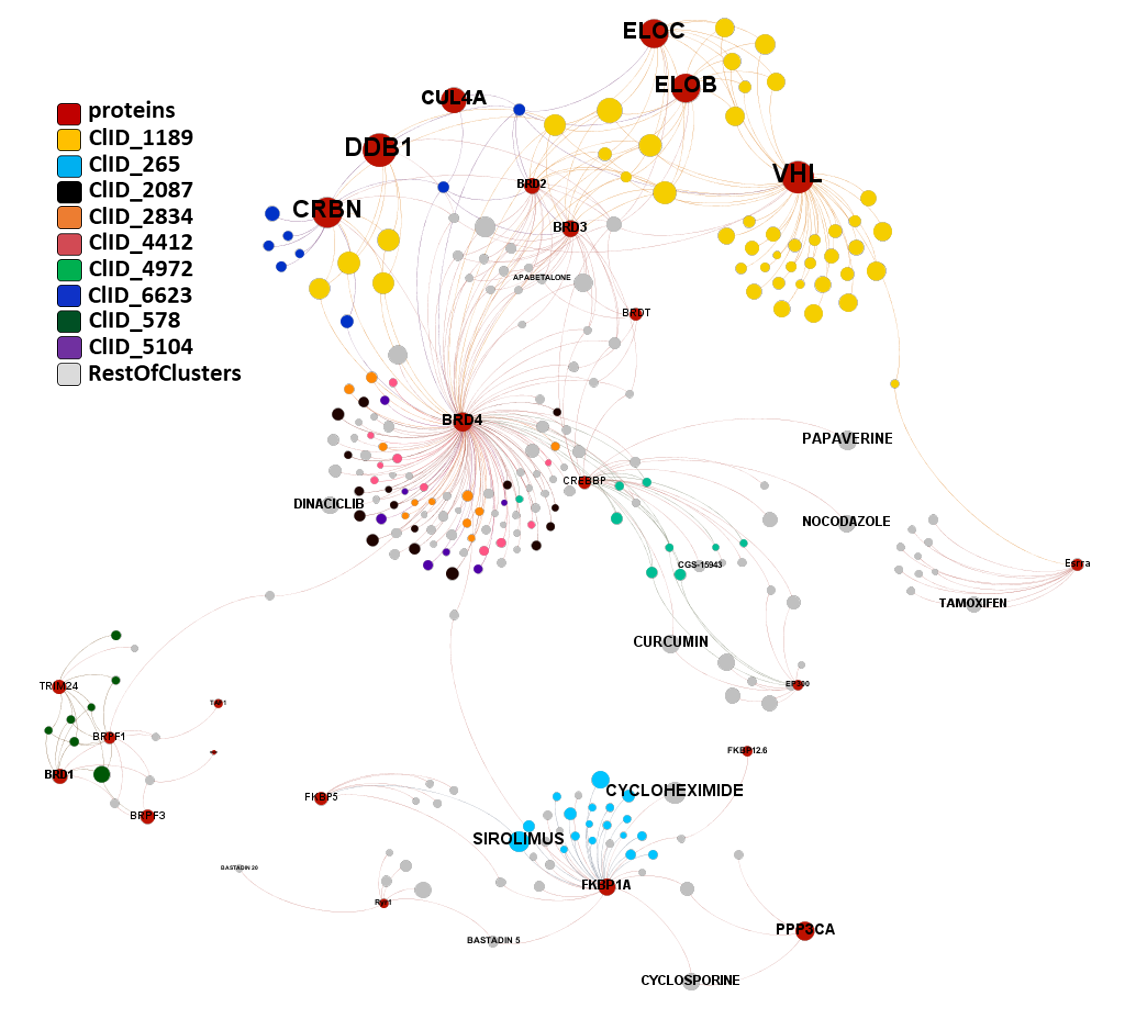
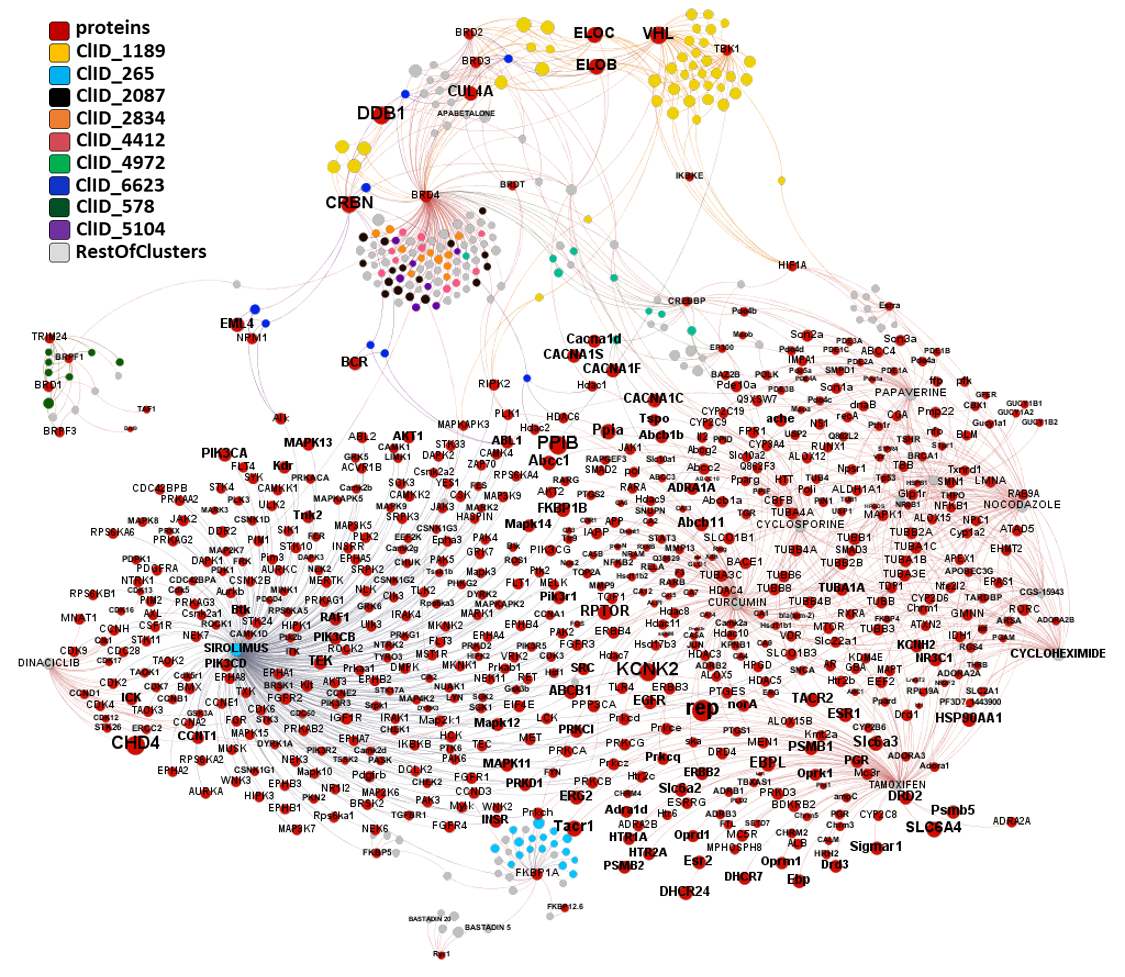
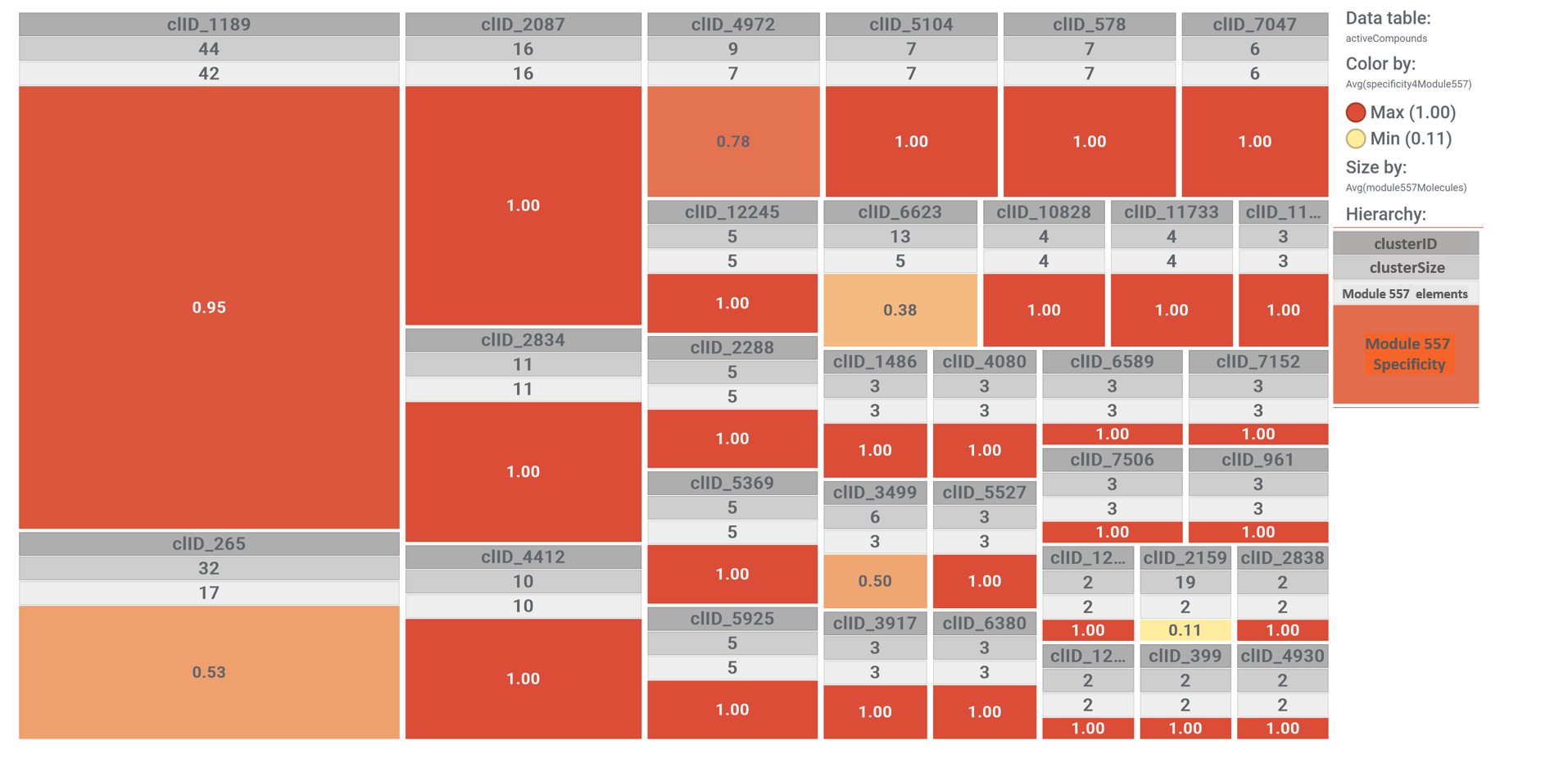
In this section we will focus on molecules predicted as active against coronaviruses by machine learning models based on SARS-CoV assays in ChEMBL DB that interact with E3-ubiquitin ligase components, bromodomain proteins and FK501 binding proteins.
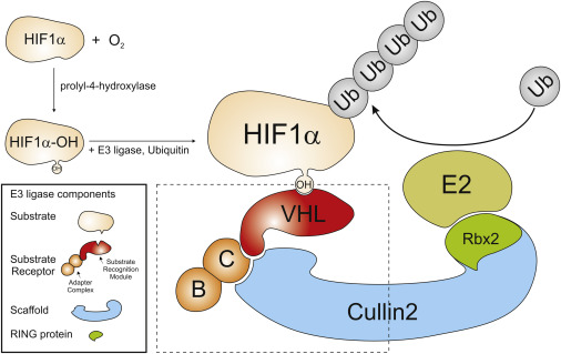
- Ubiquitin ligases are protein complexes that conduct proteins ubiquitination thus targeting them to proteasomal degradation. The standard complex is based on the assembly of a receptor for protein to be degraded (here represented by CRBN and VHL) bound to a main scaffold (cullin protein, here represented by CULA4) by an adaptor (ELOB, ELOC and DDB1 in this section). Additional proteins to enable binding and function of the E2 Ub-conjugating enzymes do not appear in this ChEMBL module. Ub-ligases show specificity among the scaffold, the adaptor, the protein receptor and the proteins to be degraded.
- Bromodomain proteins (BRDs), also known as BET (bromodomain and extra terminal domain) recognize acetylated histones to modulate gene expression either as scaffolds that facilitate the assembly of larger protein complexes or acting as transcription factors or regulators of transcription.
- FK501 binding proteins (FKBPs) are involved in diverse cellular functions including protein folding, cellular signaling, apoptosis and transcription. They elicit their function through direct binding and altering conformation of their target proteins, hence acting as molecular switches.
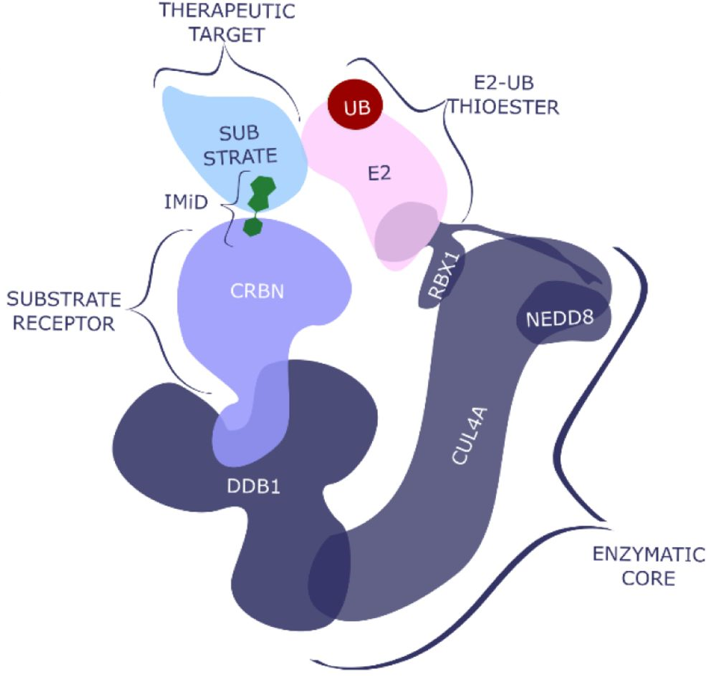
The three categories of proteins seem to be connected in this interaction module by a class of molecules called PROTACs (proteolysis targeting chimeras). PROTACs consist of two covalently linked protein-binding molecules: one capable of engaging an E3 ubiquitin ligase, and another that binds to a target protein meant for degradation. in this case, connecting Ub-ligases to BRDs or FFKBPs.
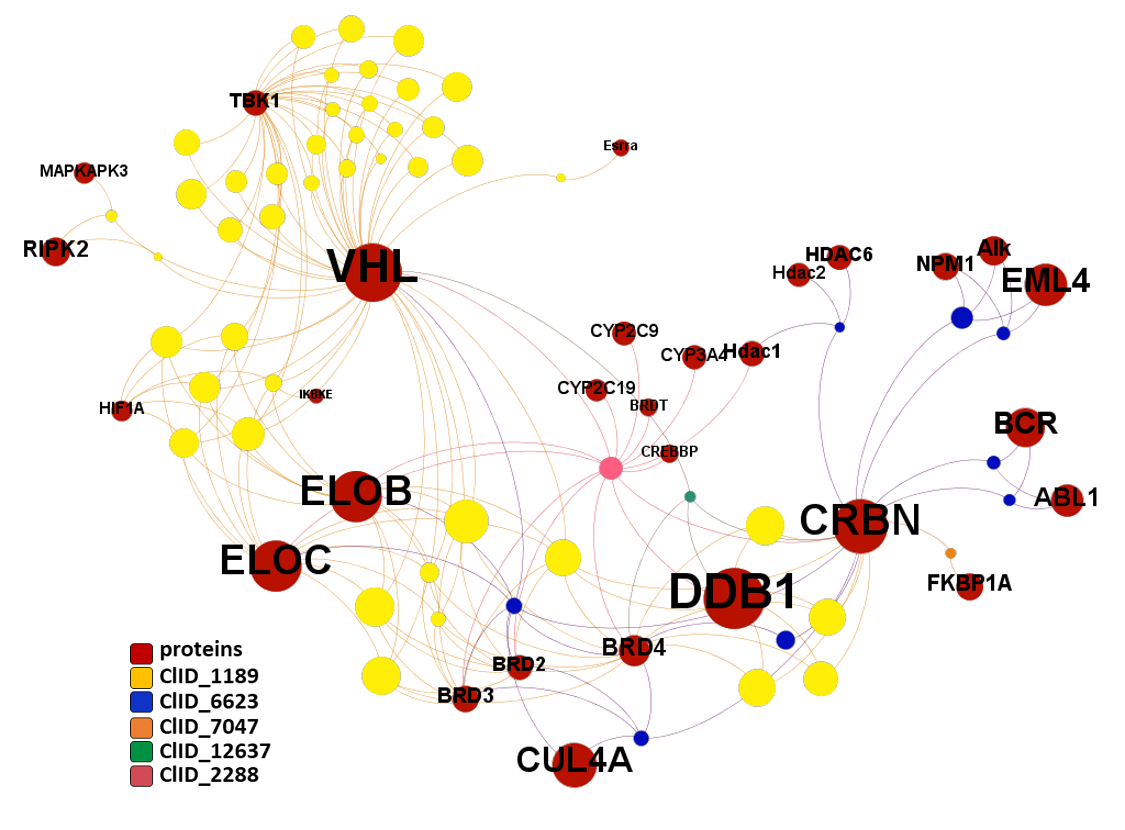
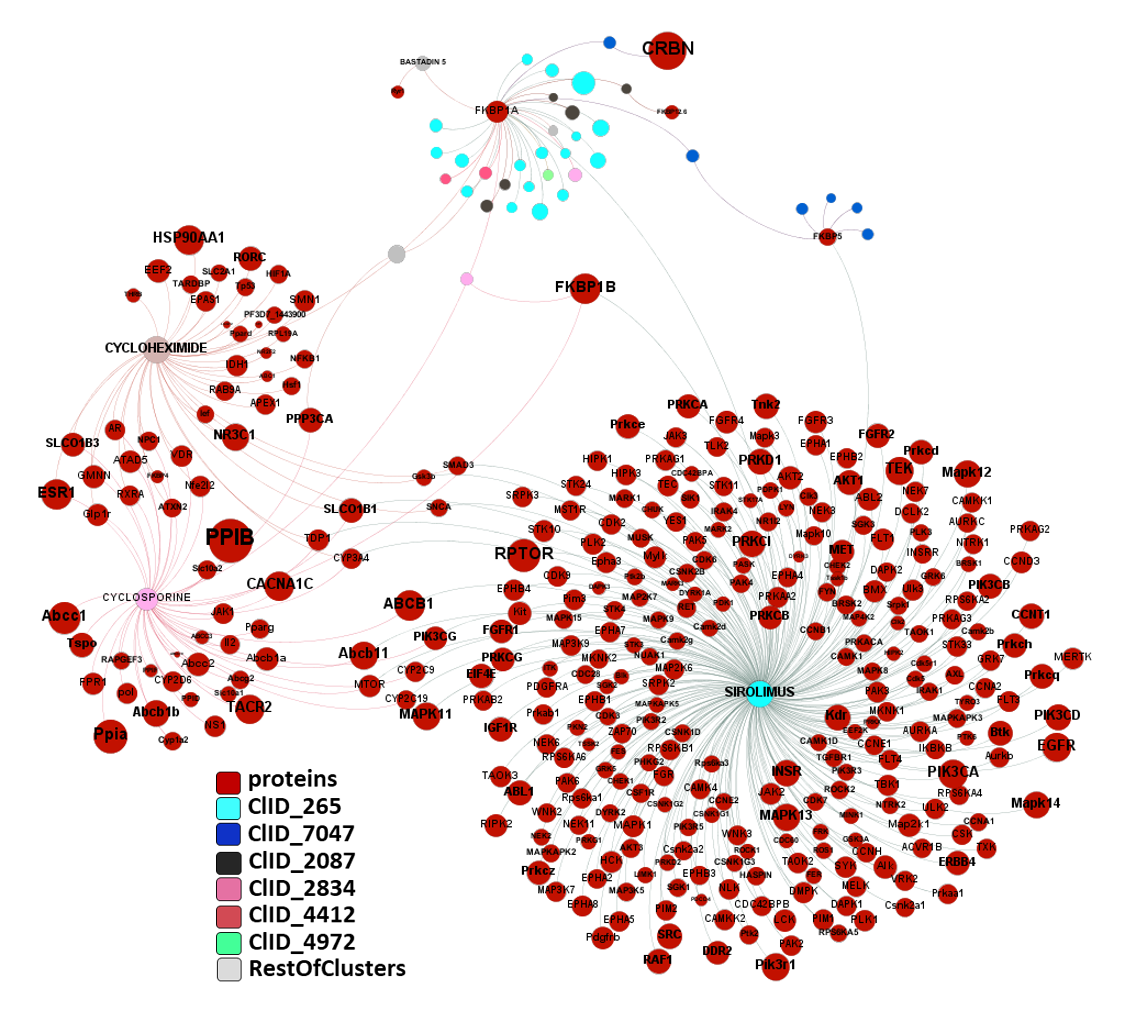
Let’s examine the compounds-proteins-pathways interactions in a network graph. Compounds connecting the Ub-ligase complex elements with BRDs and FKBPs are our PROTACs. It is also worth to observe how many of these proteins are associated to the “Reactome pathways Potential therapeutics for SARS” and “Replication of the SARS-CoV-1 genome”, besides on many other related to HIV replication and, more specifically, elongation.
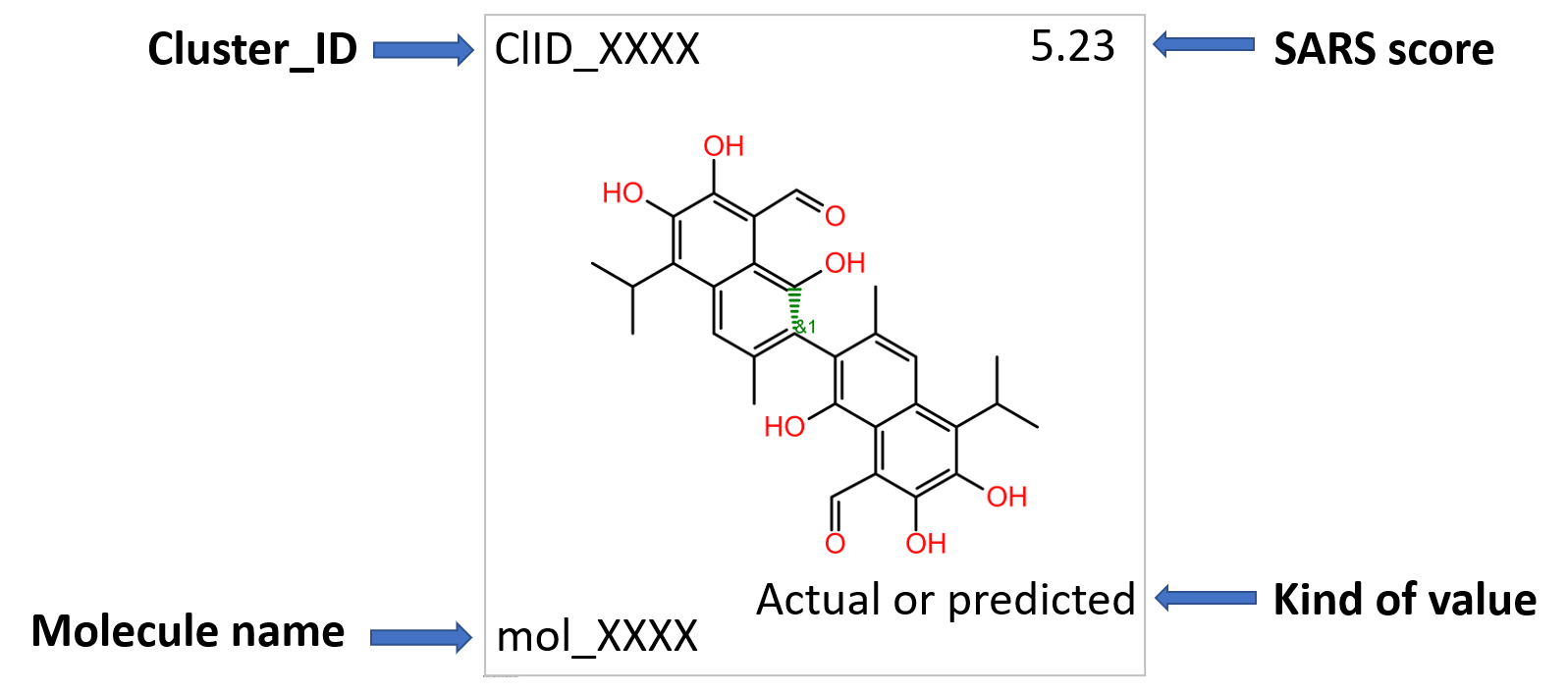
Clustering molecules by structural similarity can allow us to observe the specificity of the interactions within this module…
Extending the interactions network of these molecules to all interactions recorded in ChEMBL we can observe their degree of promiscuity. Mostly restricted to SIROLIMUS, CURCUMIN, TAMOXIFEN, NOCODAZOLE, PAPAVERINE, CYCLOSPORINE, CYCLOHEXIMIDE, DINACICLIB and GS-15943.
Each molecular cluster specificity for this module can be visualized with a simple tree map, where each cluster is sized according to its number of elements and its color graded by the ratio between the number of cluster elements hitting on module 557 proteins and the cluster size. See legend.
And now, combining molecular clustering with modularity of interactions we can analyze separately the specificity of compound-protein interactions for the three groups of proteins: Ub-ligases, BRDs and FKBPs.
Just click on each tab to get the corresponding views for specific interactions and compound structures.
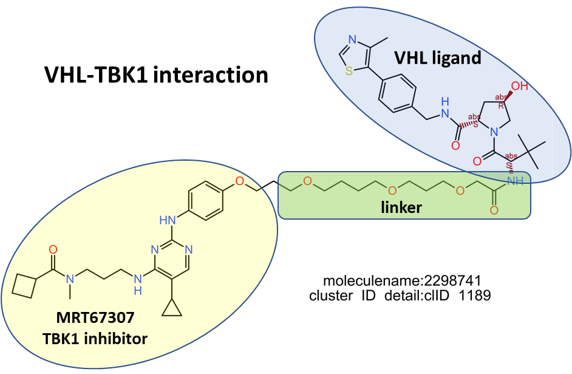
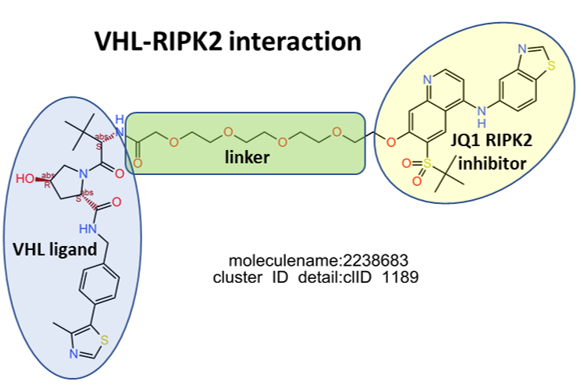
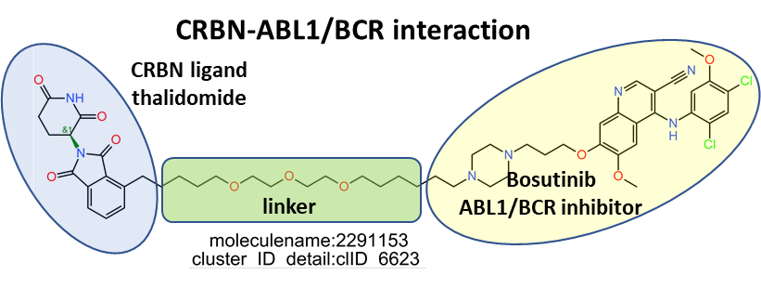
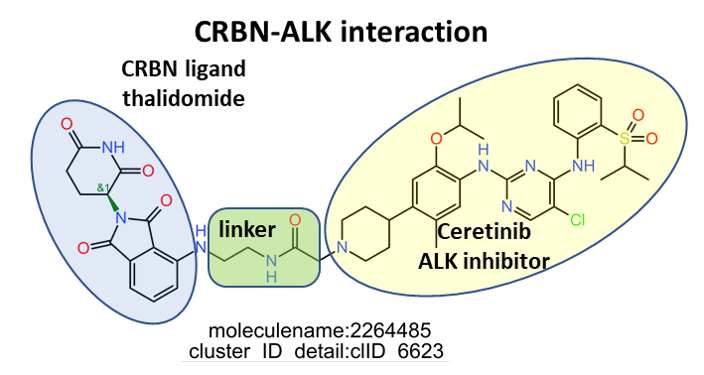
E3 Ub-ligases targeted by SARS predicted active compounds represented in ChEMBL correspond to the VHL receptor – ELOB/C adaptor – CUL2 scaffold and to the CRBN receptor – DDB1 adaptor – CUL4A scaffold complexes. Both superstructures interact with bifunctional molecules (PROTACs) that also bind to BRDs, FKBPs and some other tested in ChEMBL (ALK, ABL1, TBK1, RIPK2 and HDACs).
VHL-ELOB/C-CUL2-RBX2-E2 Ub-ligase complex targets to Hypoxia-induced factors (HIFs).
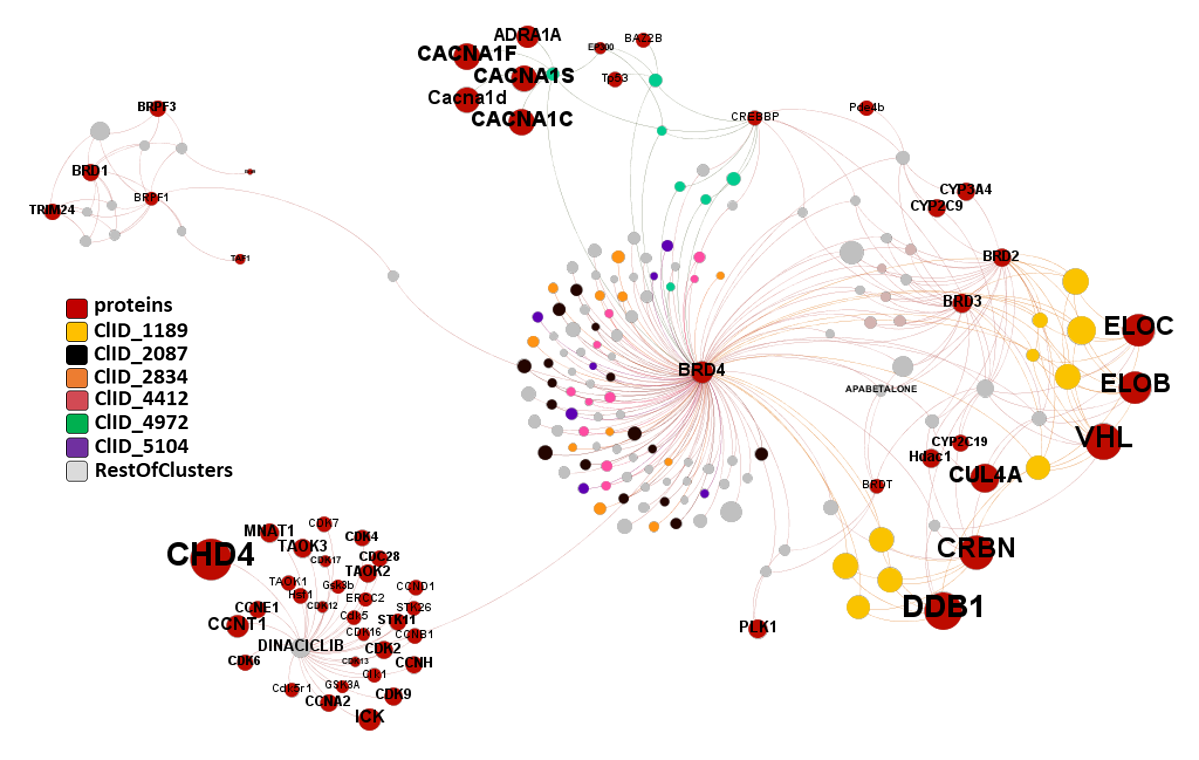
And here is the network graph of SARS active molecules hitting Ub-Ligases with all their interactions with any other protein recorded in ChEMBL. The graph includes genuine Ub-Ligase inhibitors plus bifunctional PROTACs targeting BRDs, FKBPs and some other proteins.
Let’s see here most representative examples of the proteolysis targeted chimeras (PROTACs).
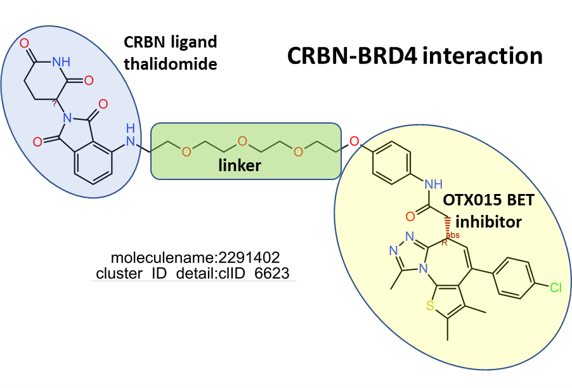
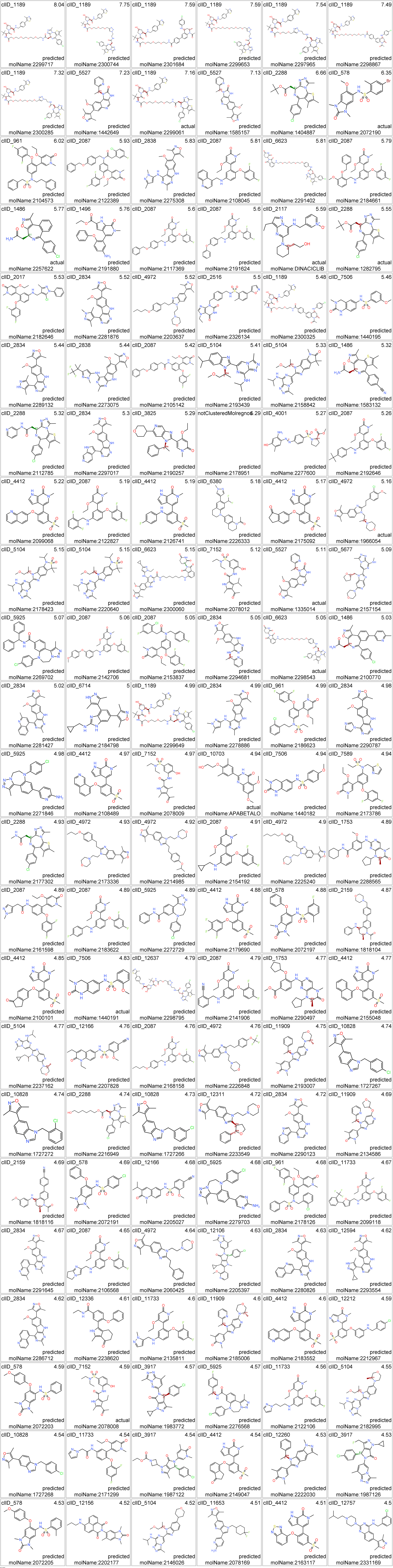
The network graph represents SARS active molecules hitting bromodomains proteins with all their interactions with any other protein recorded in ChEMBL. While ClIDs 2087, 2834, 4412 and 5104 seem to be BRD4-specific, ClID4972 shows multiple interactions with CREBBP and calcium channels, and ClID_1189 manifests a strong connection with Ub-ligase components due to its composition as BRD4-VHL/CRBN bifunctional ligands. Less represented clusters show some crossed relation with additional BRDs.
Below, two representative examples of the proteolysis targeted chimeras (PROTACs) linking VHL/CRBN to BRDs.
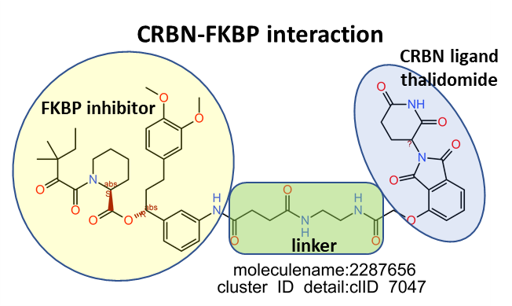
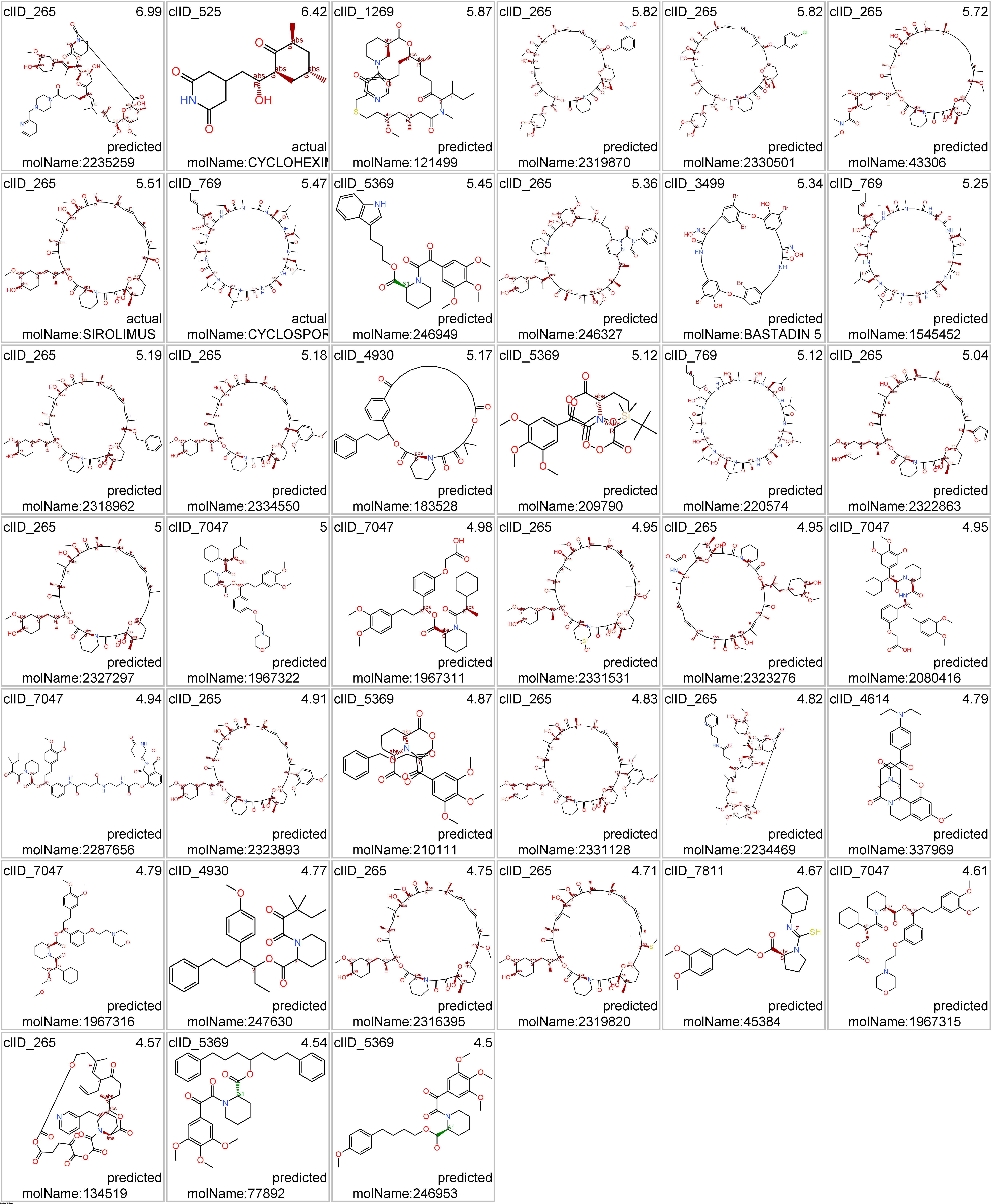
The network graph represents SARS active molecules hitting FK506 binding proteins with all their interactions with any other protein recorded in ChEMBL. ClIDs 265 2087 and 2834 seem to perform as FKBP1 specific, with the notable exception of sirolimus and cyclosporin, which shows high promiscuity interacting with many protein types. ClID_7047 molecules interact mainly with FKBP5, with occasional FKBP1A hits and CRBN, in this last case it’s a double FKBP1A/CRBN interaction by the corresponding PROTAC.
Below, arepresentative examples of the proteolysis targeted chimeras (PROTACs) linking CRBN to FKBPs.
PROTAC examples referenced in this review below.
Comput Struct Biotechnol J. 2019; 17: 160–176.
Bivalent Ligands for Protein Degradation in Drug Discovery
Abstract
Targeting the “undruggable” proteome remains one of the big challenges in drug discovery. Recent innovations in the field of targeted protein degradation and manipulation of the ubiquitin-proteasome system open up new therapeutic approaches for disorders that cannot be targeted with conventional inhibitor paradigms. Proteolysis targeting chimeras (PROTACs) are bivalent ligands in which a compound that binds to the protein target of interest is connected to a second molecule that binds an E3 ligase via a linker. The E3 protein is usually either Cereblon or Von Hippel-Lindau. Several examples of selective PROTAC molecules with potent effect in cells and in vivo models have been reported. The degradation of specific proteins via these bivalent molecules is already allowing for the study of biochemical pathways and cell biology with more specificity than was possible with inhibitor compounds. In this review, we provide a comprehensive overview of recent developments in the field of small molecule mediated protein degradation, including transcription factors, kinases and nuclear receptors. We discuss the potential benefits of protein degradation over inhibition as well as the challenges that need to be overcome.