The aim of this publication is to identify viral and host pathways involved in coronavirus infection.
For this purpose, we have aggregated at the protein/gene level the predicted activity scores of 1.7M molecules on SARS/MERS viral assays, on the basis of their molecular interactions with target proteins recorded in ChEMBL DB. Click on the links above to access to detailed information about such procedures.
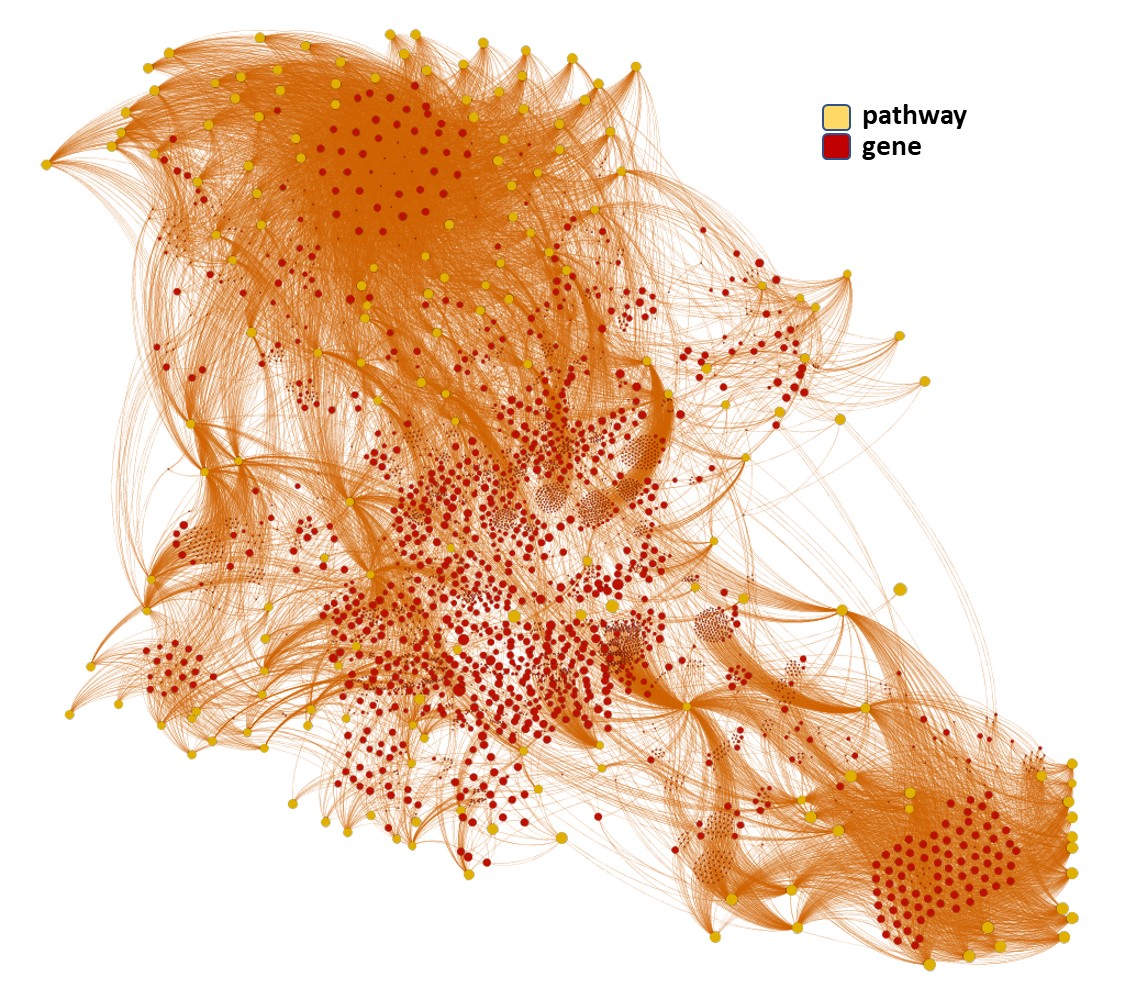
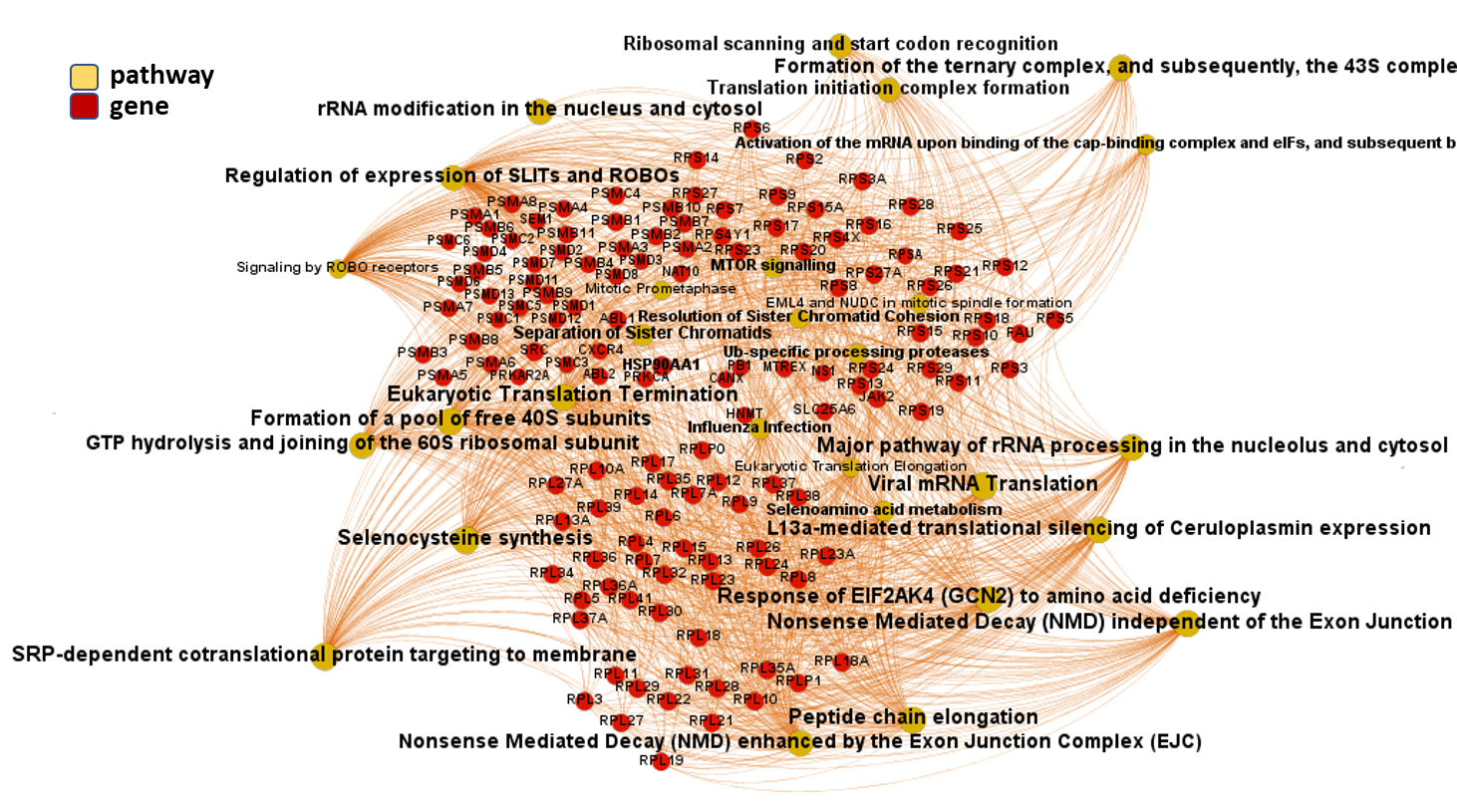
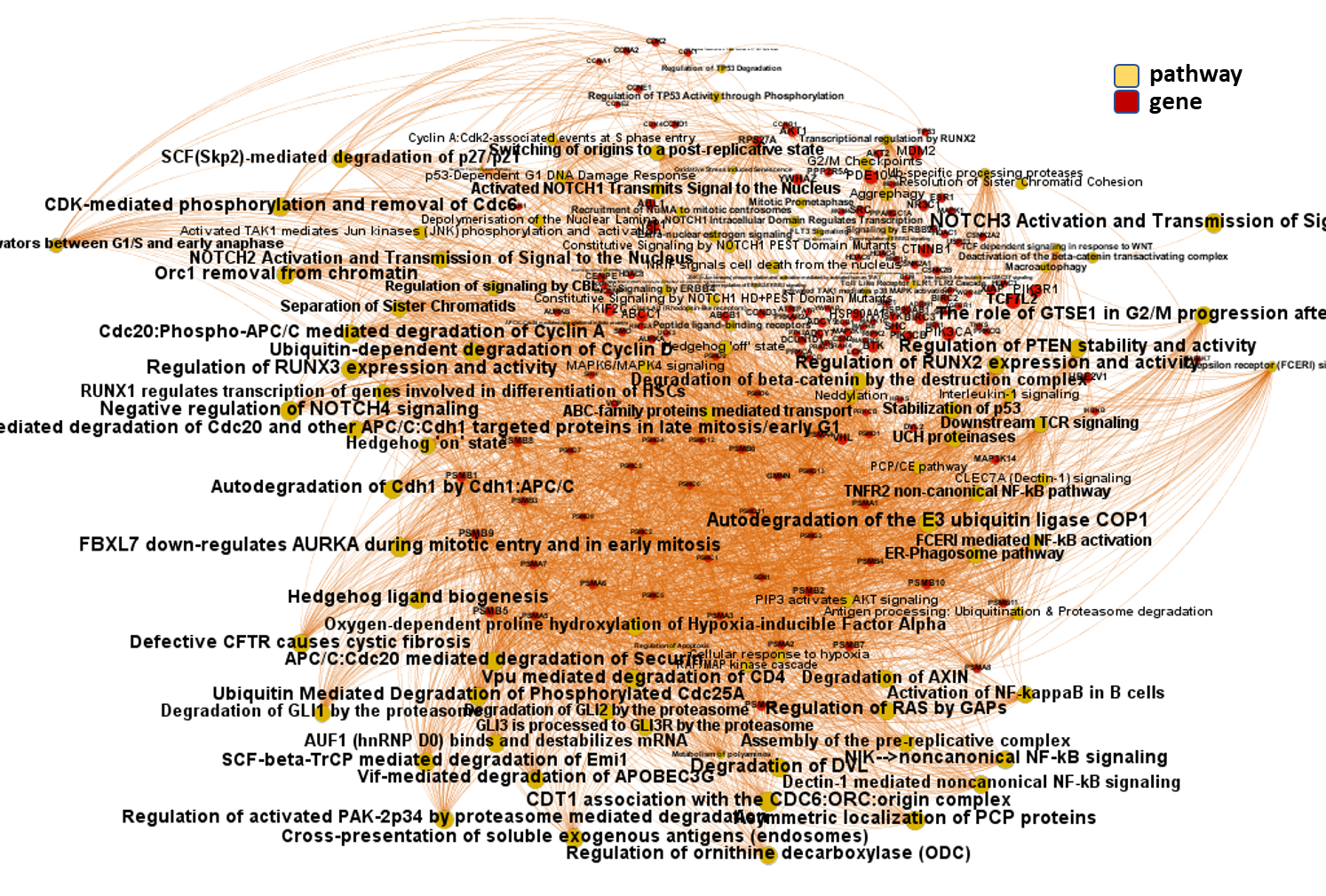
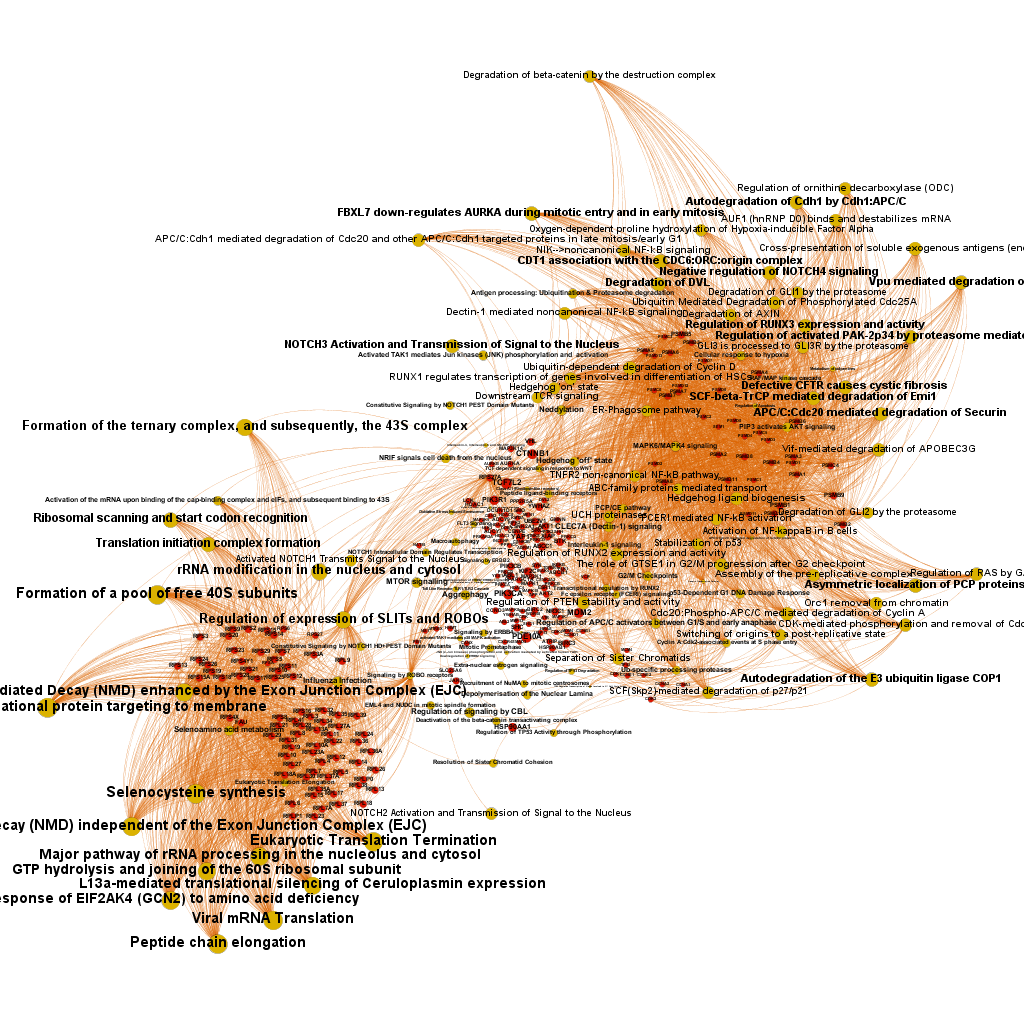
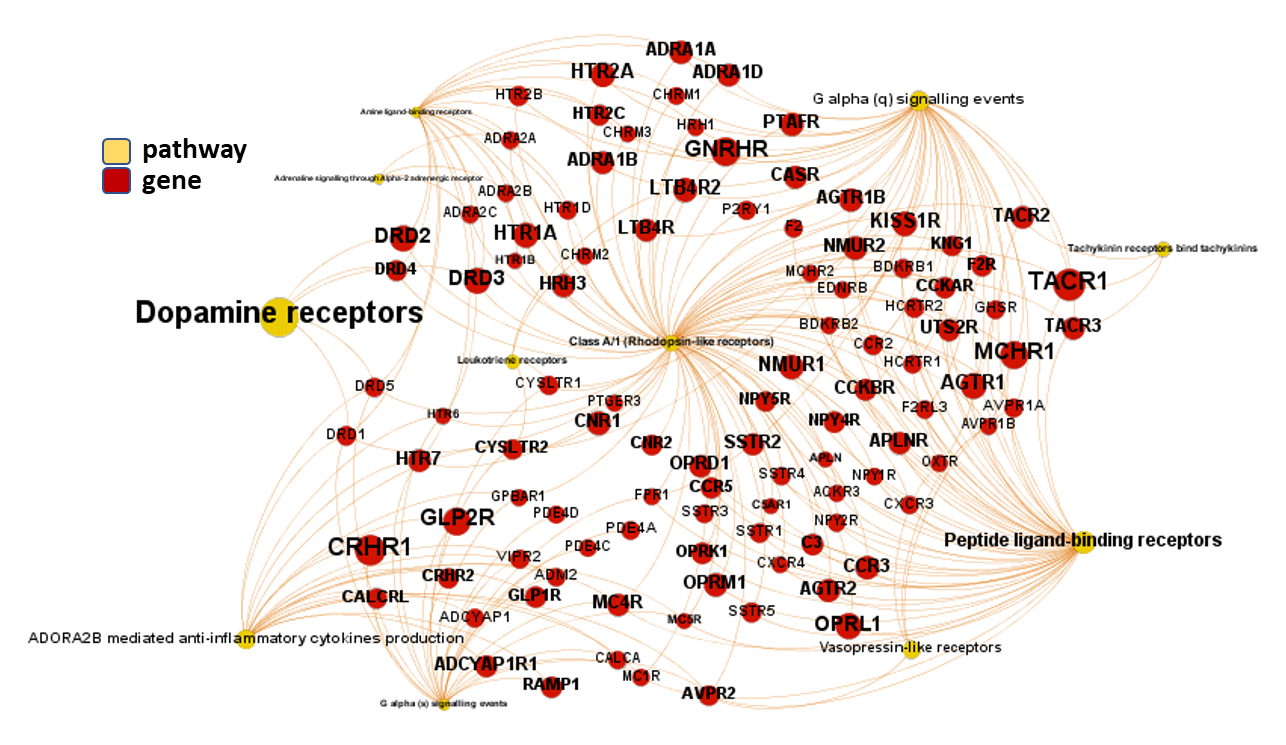
We can use low level pathways from Reactome Pathways DB to trace a gene/pathways association map as below.
We can use this gene/pathways association map to generate, by aggregation of values at the pathway level, several indexes that act as descriptors of the pathway involvement in coronavirus infection.
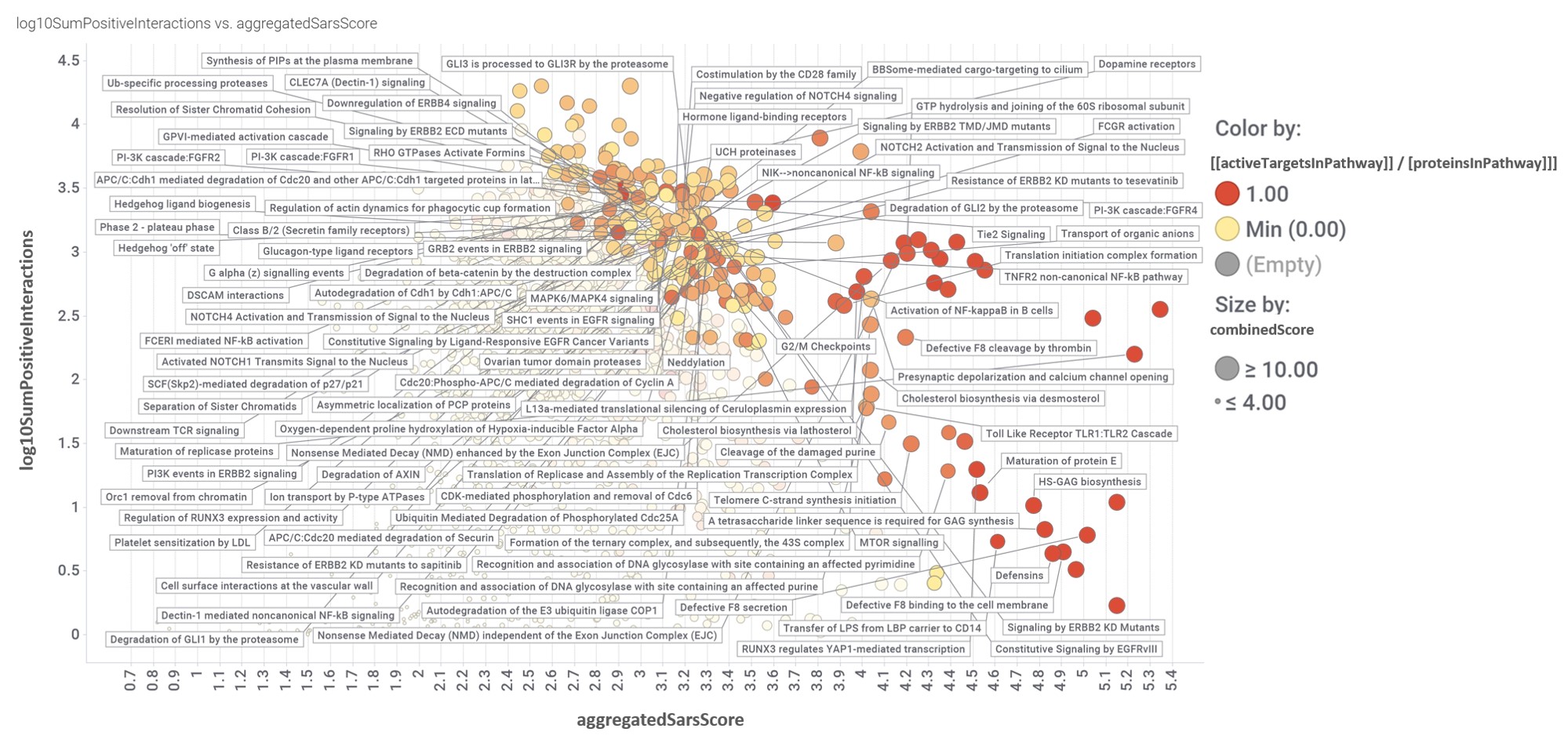
For the scatter plot below we have defined:
- log10SumPositiveInteractions.- As the logged addition of all active interactions of predicted SARS active molecules with all the proteins belonging each pathway.
- aggregatedSarsScore.- As the average value of predicted activity against SARS viruses aggregated at the pathway level from the values assigned to each protein/gene.
- [[activeTargetsInPathway]] / [proteinsInPathway]]].- Number of proteins considered relevant for SARS coronavirus infection in a particular pathway vs the total number of protein of that pathway referred in the ChEMBL DB.
- combinedScore.- Just as linear combination of the number of evidences supporting a score (log10SumPositiveInteractions) and the predicted score from aggregation for a pathway (aggregatedSarsScore).
And we also can use the gene scores and their association to their respective pathways to perform pathways enrichment, thus getting a n statistical indicator of enrichment (enrichmentScore) and a pValue of confidence (FDR: false discovery rate).
In this particular case, we have used Guangchuang Yu methodology published in An R package for Reactome Pathway Analysis. Additional details about the procedure can be obtained accessing to Guangchuang’s published page through the link above.
Now, we can see both, enrichment scores and aggregation scores together in a bar chart.
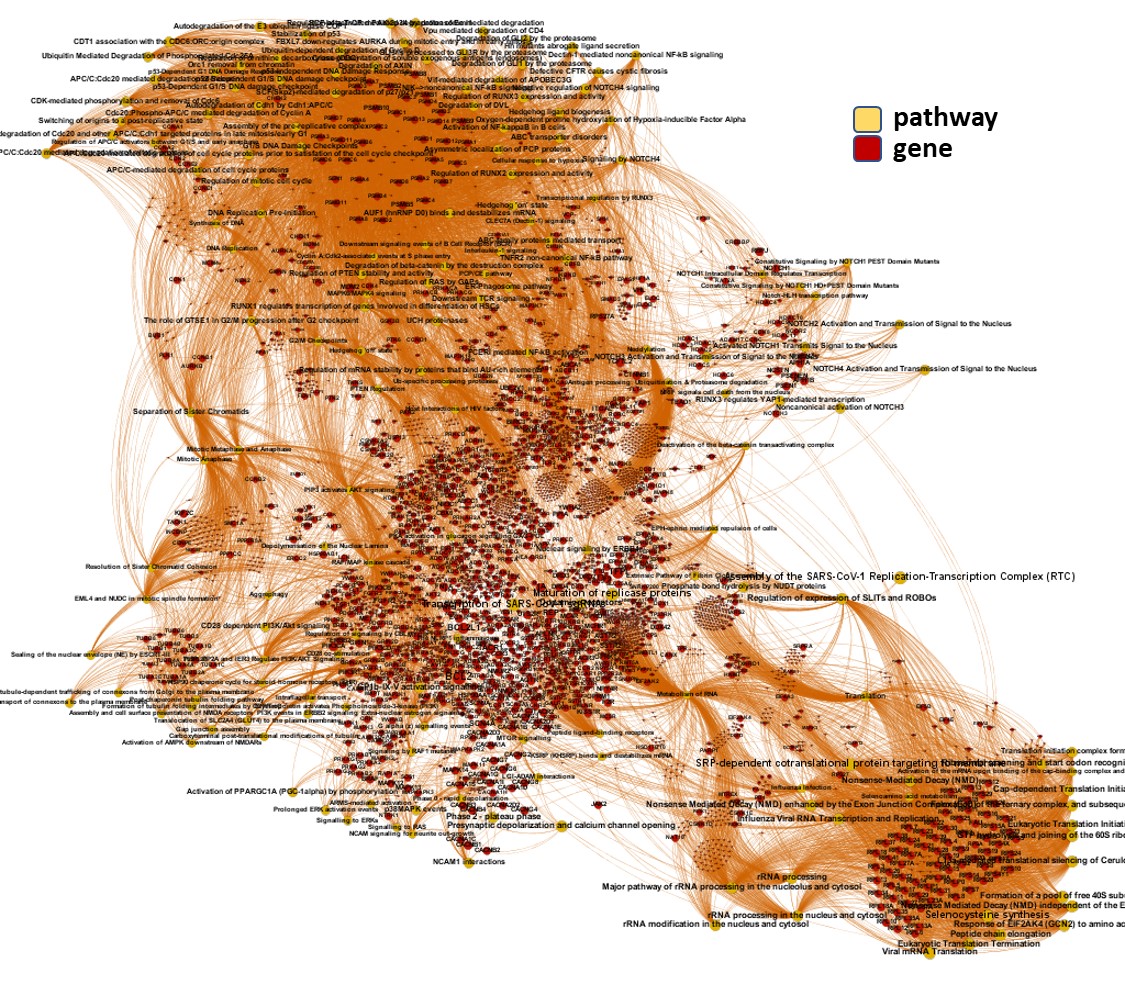
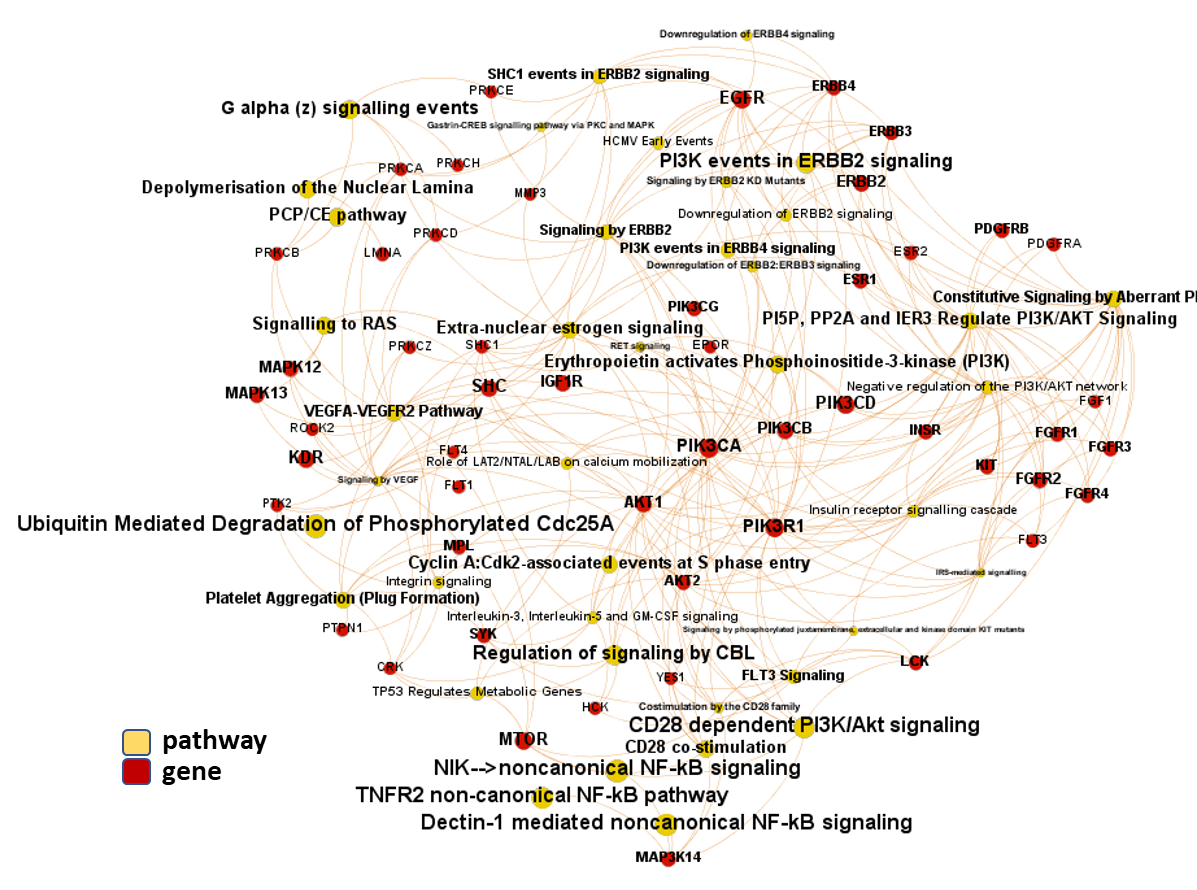
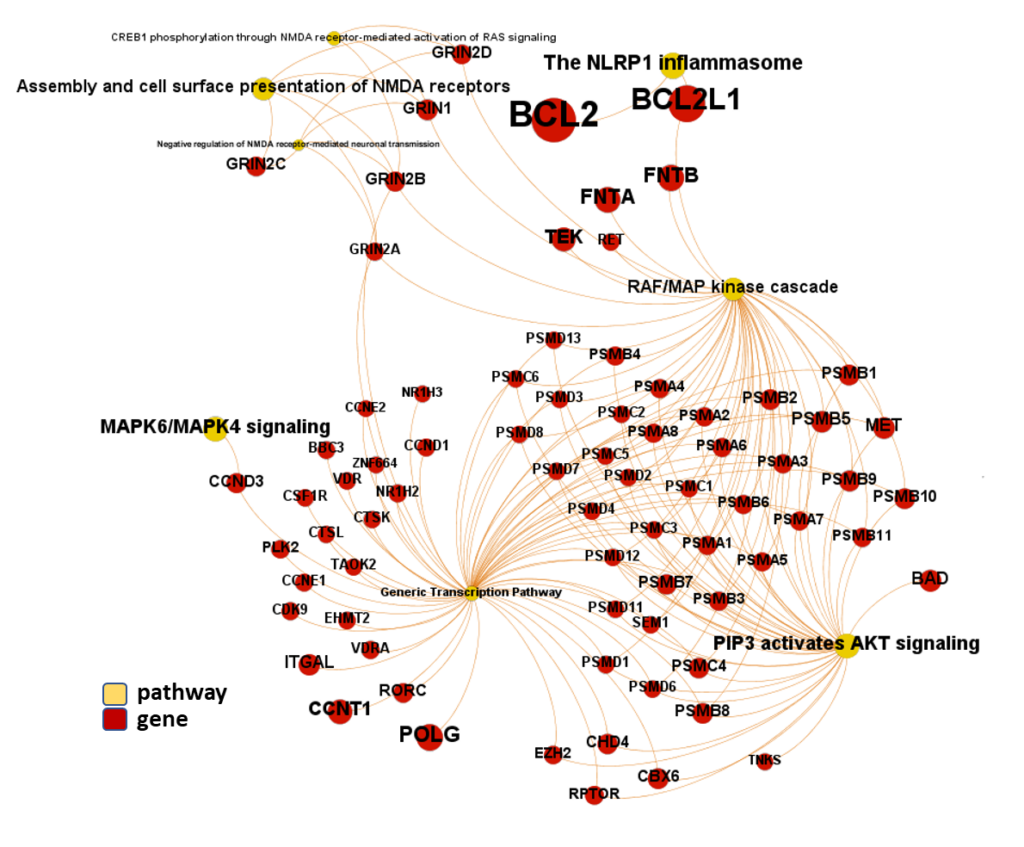
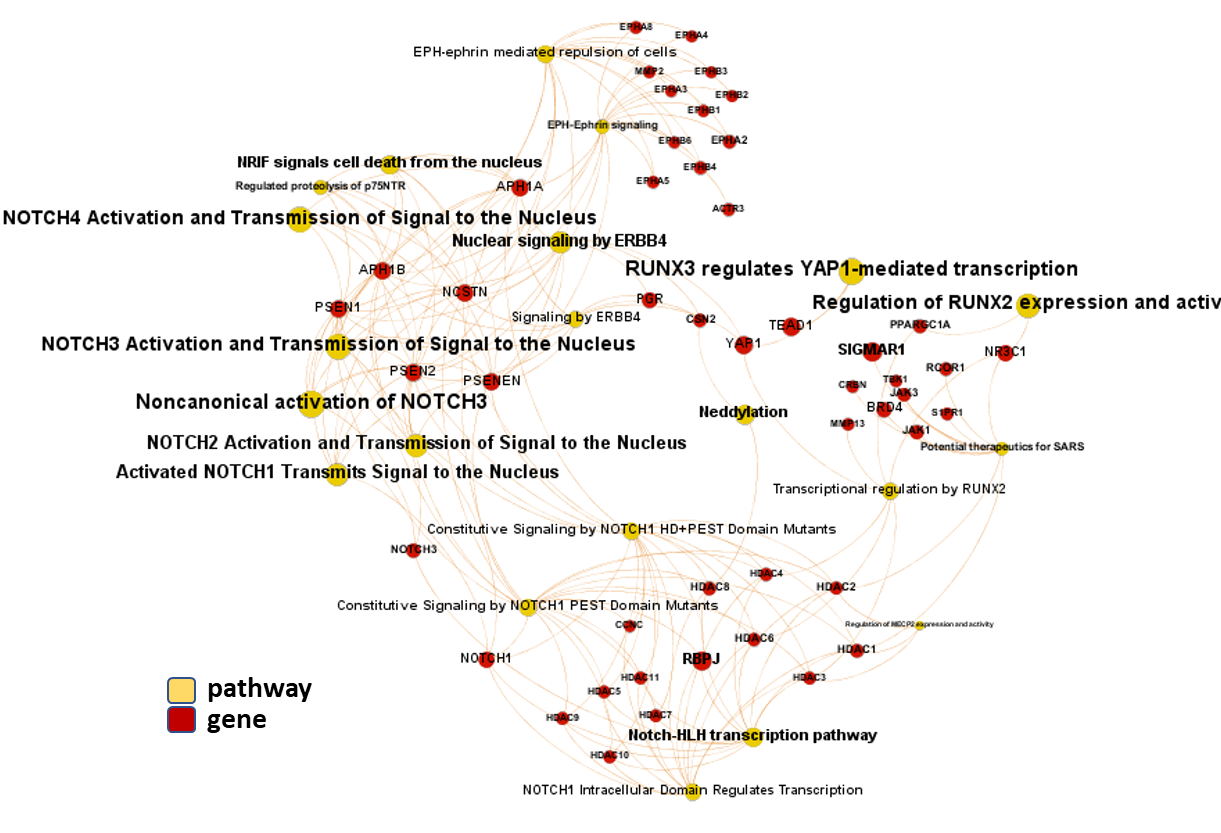
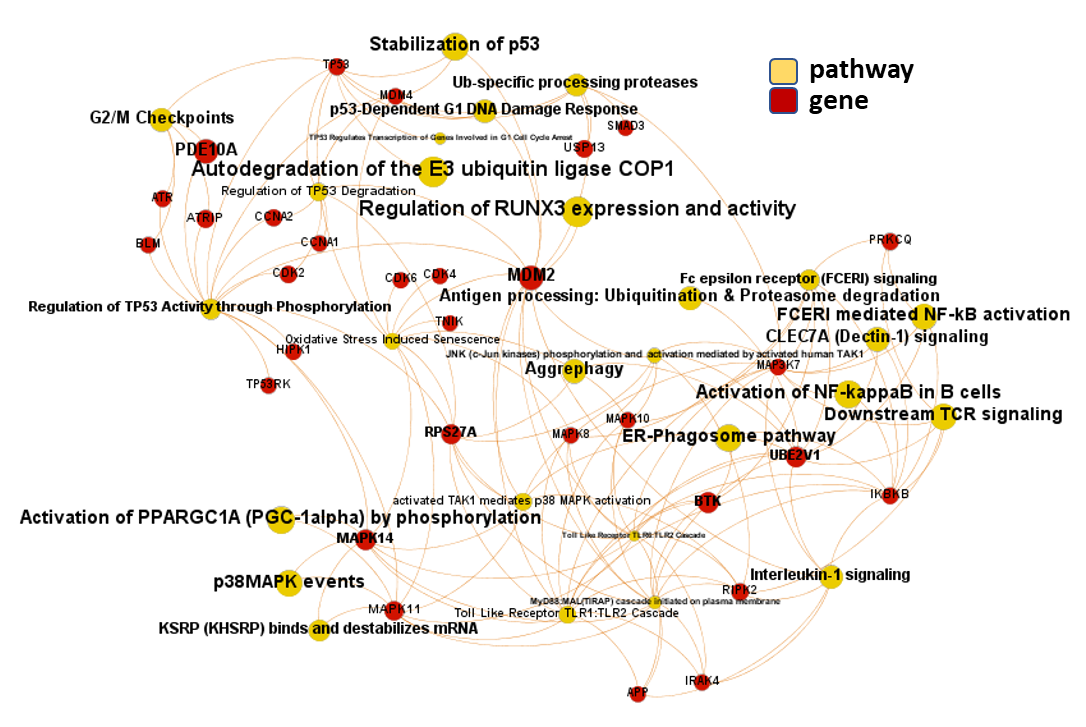
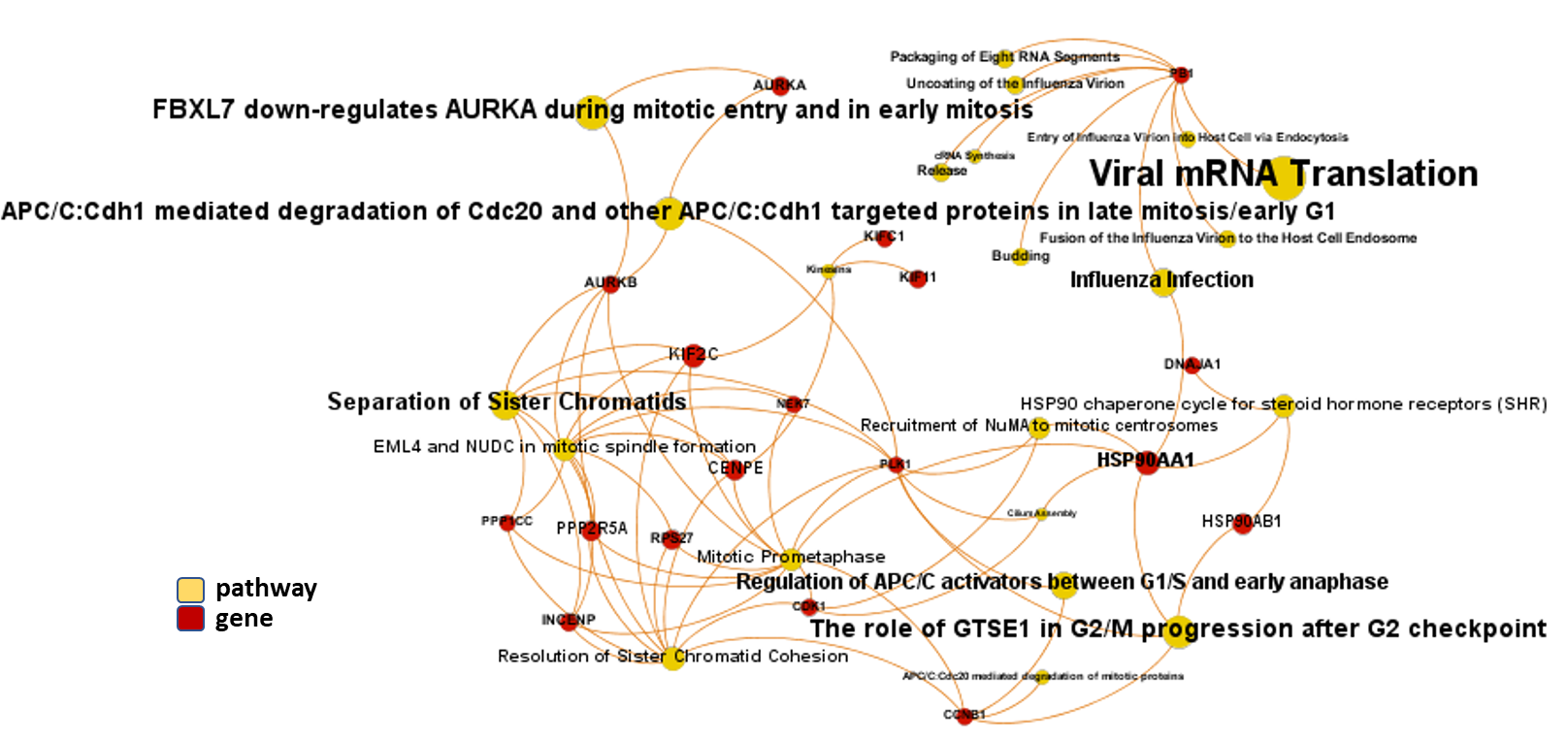
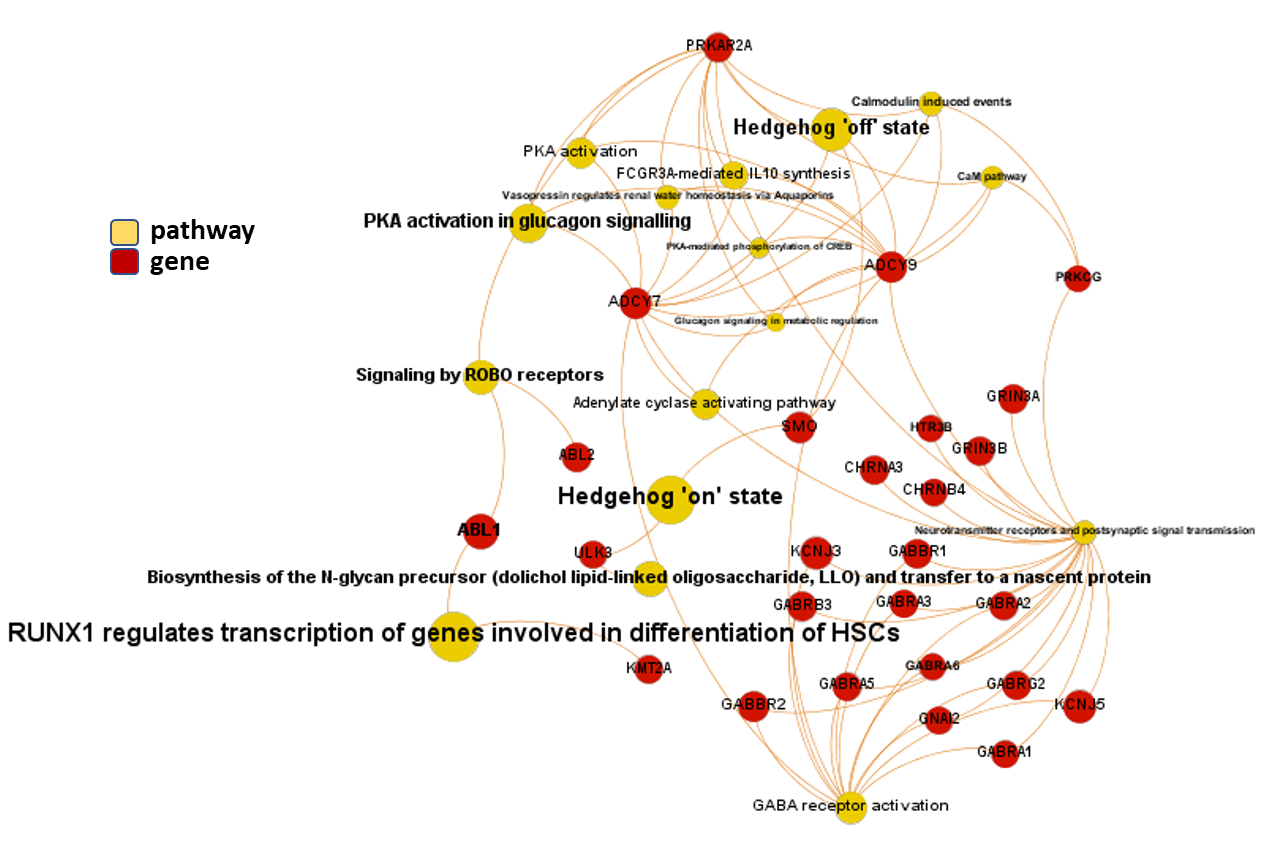
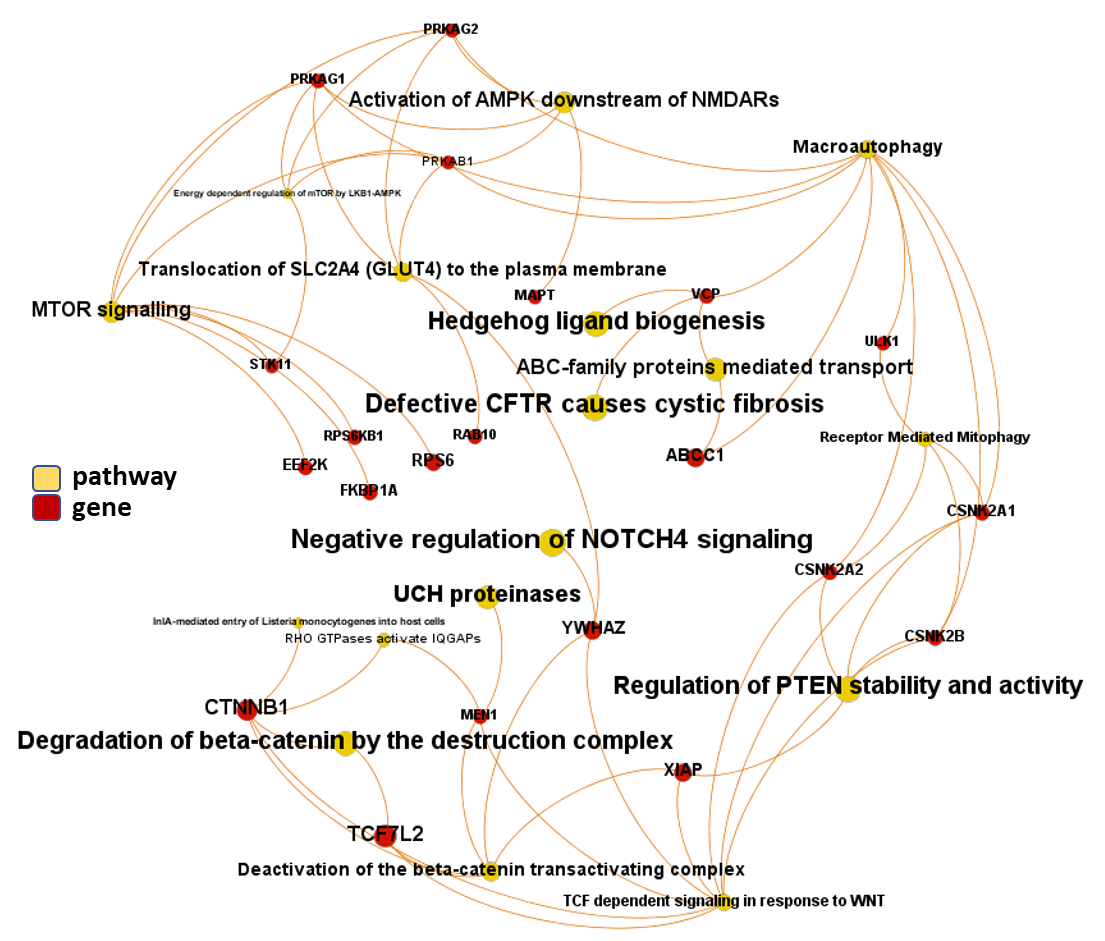
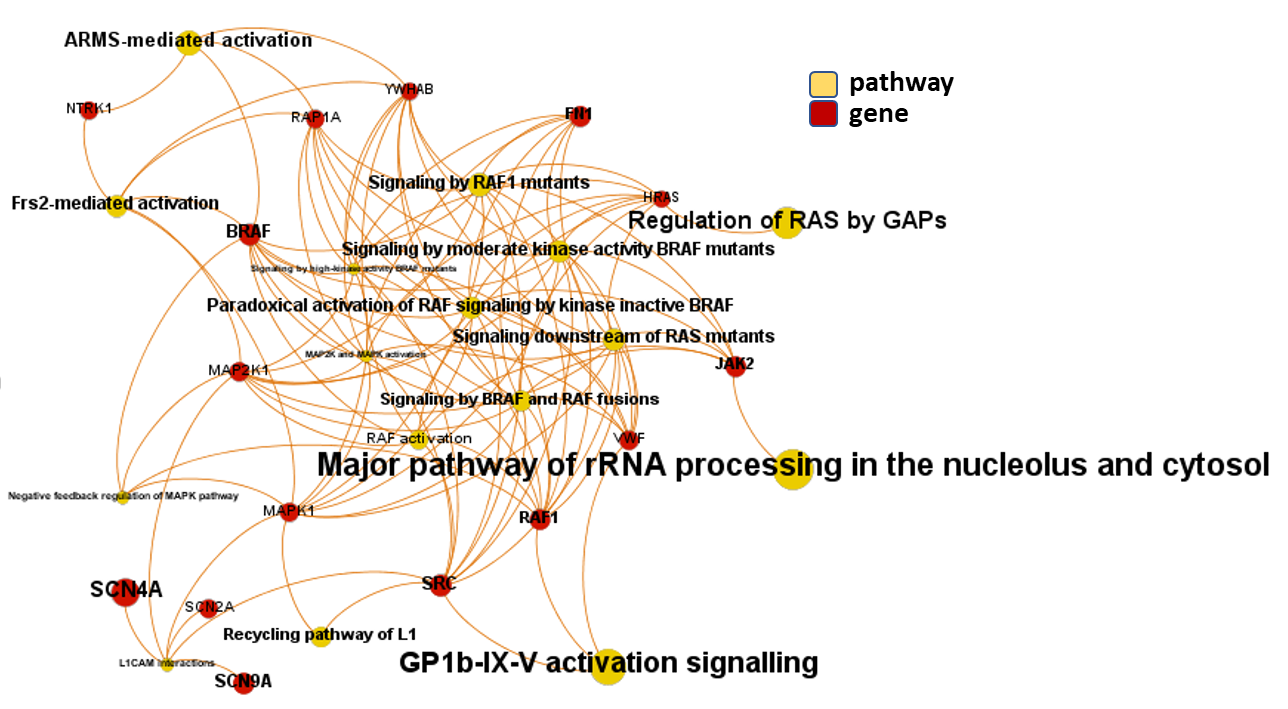
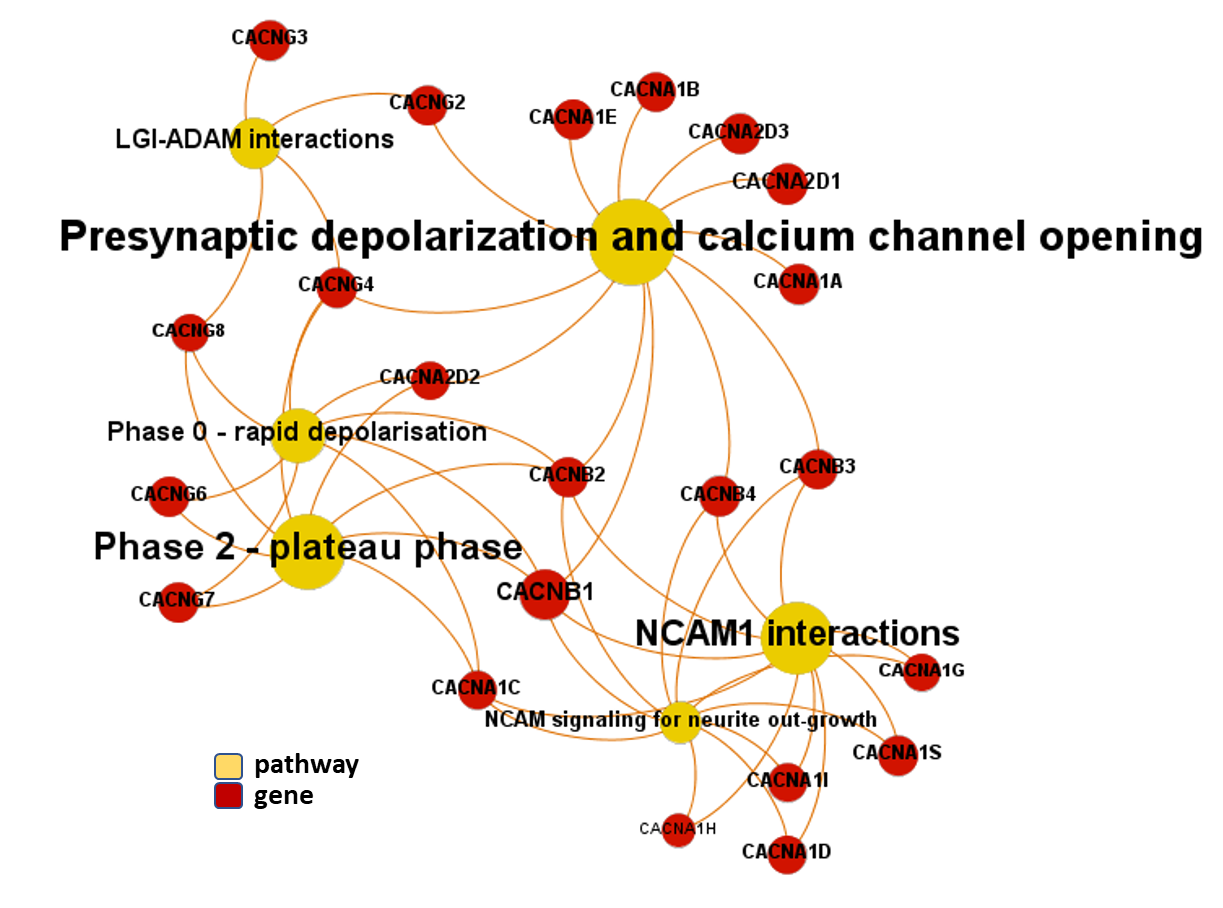
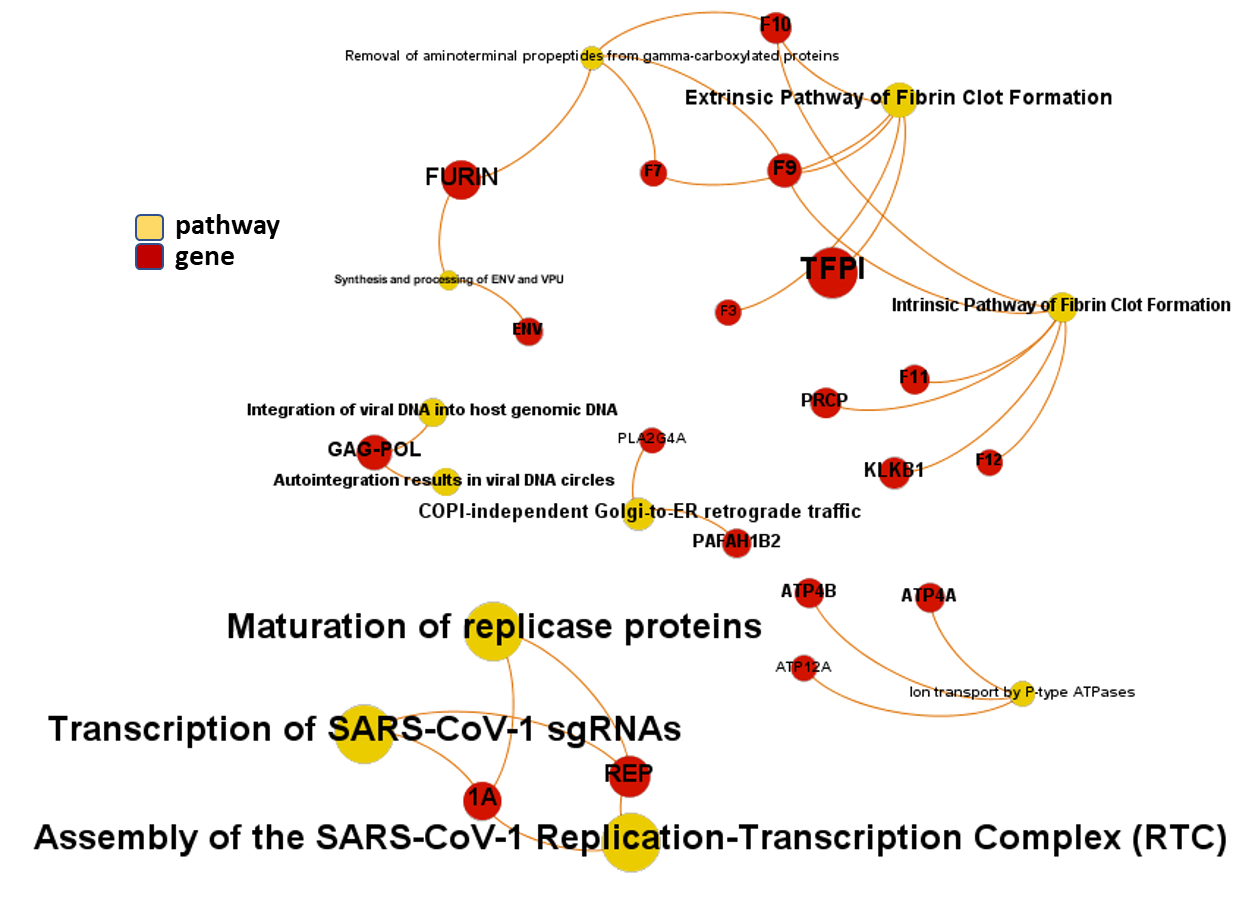
And now, we can incorporate gene and pathway names and scores to the gene/pathway association network graph.
It may be a nice view, although there is not much info we can get from this view nor conclusions to infer.
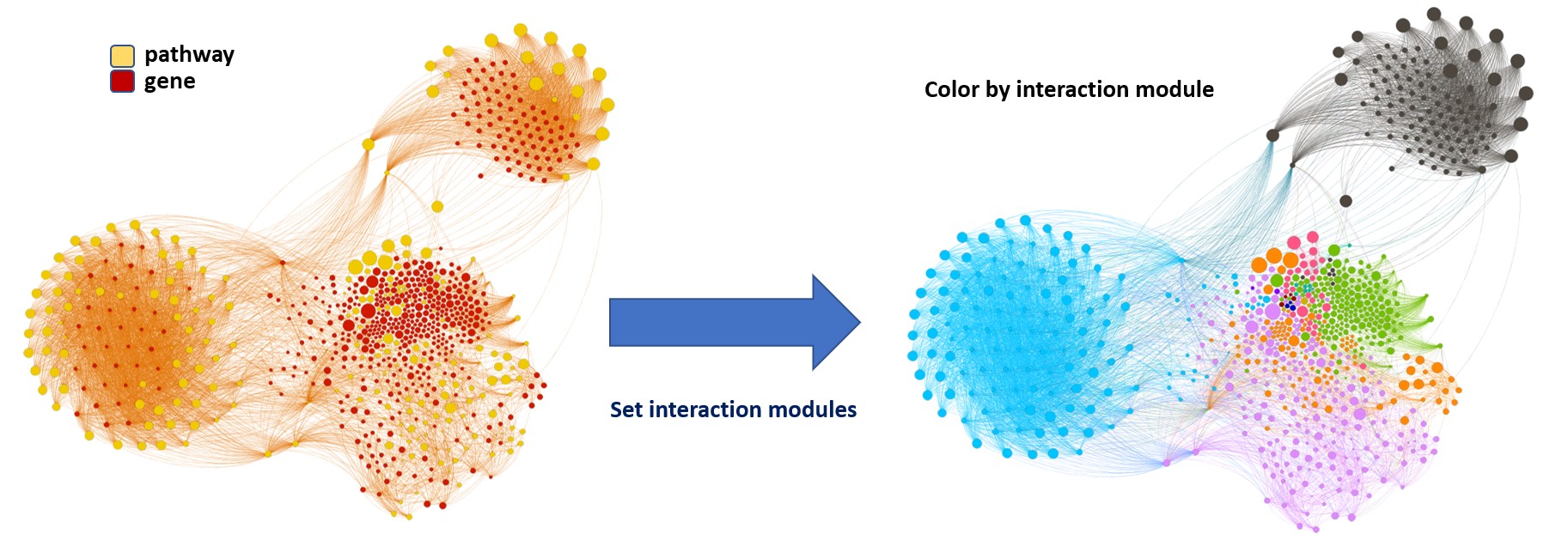
So, let’s do two things: First, select most significant associations selecting best combinations of their aggregation and enrichment scores. Next, subset the network in different areas on the basis of the closeness of their respective interactions in order to define different independent network modules to perform a deeper analysis.
Now, we can look at the different modules closer. Just click on the network graphs below to expand views and on the yellow buttons to see a more complete analysis.
See most relevant gene/pathways associations and scores
| pathwayname | geneName | proteinName | pathwayScoreFromAggregation | enrichmentscore | -log10FDR | pathwayScore | geneScore |
|---|---|---|---|---|---|---|---|
| ABC-family proteins mediated transport | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.515416 |
| ABC-family proteins mediated transport | VCP | 15S Mg(2+)-ATPase p97 subunit, Transitional endoplasmic reticulum ATPase | 9.439972 | 0.656891 | 4 | 0.695944 | 0.526633 |
| ABC-family proteins mediated transport | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.57244 |
| ABC-family proteins mediated transport | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.515416 |
| ABC-family proteins mediated transport | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.515416 |
| ABC-family proteins mediated transport | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.515416 |
| ABC-family proteins mediated transport | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.54735 |
| ABC-family proteins mediated transport | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.565601 |
| ABC-family proteins mediated transport | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.54735 |
| ABC-family proteins mediated transport | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.439972 | 0.656891 | 4 | 0.695944 | 0.515416 |
| ABC-family proteins mediated transport | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.515416 |
| ABC-family proteins mediated transport | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.439972 | 0.656891 | 4 | 0.695944 | 0.515416 |
| ABC-family proteins mediated transport | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.54735 |
| ABC-family proteins mediated transport | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.577847 |
| ABC-family proteins mediated transport | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.54735 |
| ABC-family proteins mediated transport | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.54735 |
| ABC-family proteins mediated transport | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.5197 |
| ABC-family proteins mediated transport | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.547953 |
| ABC-family proteins mediated transport | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.54735 |
| ABC-family proteins mediated transport | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.515416 |
| ABC-family proteins mediated transport | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.515416 |
| ABC-family proteins mediated transport | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.515416 |
| ABC-family proteins mediated transport | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.54735 |
| ABC-family proteins mediated transport | ABCB1 | ABC transporter B family member 1, ATP binding cassette subfamily B member 1, ATP-binding cassette subfamily B member 1, ATP-dependent translocase ABCB1, Multidrug resistance protein 1, Uncharacterized protein | 9.439972 | 0.656891 | 4 | 0.695944 | 0.565653 |
| ABC-family proteins mediated transport | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.439972 | 0.656891 | 4 | 0.695944 | 0.579609 |
| ABC-family proteins mediated transport | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.577481 |
| ABC-family proteins mediated transport | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.515416 |
| ABC-family proteins mediated transport | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.515416 |
| ABC-family proteins mediated transport | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.515416 |
| ABC-family proteins mediated transport | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.58858 |
| ABC-family proteins mediated transport | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.439972 | 0.656891 | 4 | 0.695944 | 0.568645 |
| ABC-family proteins mediated transport | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.54735 |
| ABC-family proteins mediated transport | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.515416 |
| ABC-family proteins mediated transport | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.54735 |
| ABC-family proteins mediated transport | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.572294 |
| ABC-family proteins mediated transport | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.573417 |
| ABC-family proteins mediated transport | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.54735 |
| ABC-family proteins mediated transport | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.546881 |
| ABC-family proteins mediated transport | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.515416 |
| ABC-family proteins mediated transport | ABCC1 | ABC transporter C family member 1, ATP-binding cassette, sub-family C, Multidrug resistance-associated protein 1 | 9.439972 | 0.656891 | 4 | 0.695944 | 0.610269 |
| Activated NOTCH1 Transmits Signal to the Nucleus | NCSTN | Nicastrin | 9.290181 | 0.703247 | 4 | 0.711394 | 0.60749 |
| Activated NOTCH1 Transmits Signal to the Nucleus | NOTCH1 | Neurogenic locus notch homolog protein 1 | 9.290181 | 0.703247 | 4 | 0.711394 | 0.591875 |
| Activated NOTCH1 Transmits Signal to the Nucleus | APH1A | Anterior pharynx defective 1 homolog A, Anterior pharynx defective 1a homolog, Aph-1 homolog A, gamma-secretase subunit, Gamma-secretase subunit APH-1A, Uncharacterized protein | 9.290181 | 0.703247 | 4 | 0.711394 | 0.60749 |
| Activated NOTCH1 Transmits Signal to the Nucleus | PSEN1 | Presenilin, Presenilin-1 | 9.290181 | 0.703247 | 4 | 0.711394 | 0.611222 |
| Activated NOTCH1 Transmits Signal to the Nucleus | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.290181 | 0.703247 | 4 | 0.711394 | 0.579609 |
| Activated NOTCH1 Transmits Signal to the Nucleus | PSENEN | Gamma-secretase subunit PEN-2 | 9.290181 | 0.703247 | 4 | 0.711394 | 0.60725 |
| Activated NOTCH1 Transmits Signal to the Nucleus | APH1B | Aph-1 homolog B, gamma-secretase subunit, APH-1B', Gamma-secretase subunit APH-1B, Uncharacterized protein | 9.290181 | 0.703247 | 4 | 0.711394 | 0.60749 |
| Activated NOTCH1 Transmits Signal to the Nucleus | PSEN2 | Presenilin, Presenilin-2 | 9.290181 | 0.703247 | 4 | 0.711394 | 0.607393 |
| Activated TAK1 mediates Jun kinases (JNK) phosphorylation and activation | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 8.810858 | 0.651931 | 0.579609 | ||
| activated TAK1 mediates p38 MAPK activation | MAPK14 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 14 | 8.877314 | 0.575533 | 3.406086 | 0.533255 | 0.591941 |
| activated TAK1 mediates p38 MAPK activation | MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | 8.877314 | 0.575533 | 3.406086 | 0.533255 | 0.519182 |
| activated TAK1 mediates p38 MAPK activation | MDM2 | E3 ubiquitin-protein ligase Mdm2 | 8.877314 | 0.575533 | 3.406086 | 0.533255 | 0.659771 |
| activated TAK1 mediates p38 MAPK activation | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 8.877314 | 0.575533 | 3.406086 | 0.533255 | 0.579609 |
| activated TAK1 mediates p38 MAPK activation | MAPK11 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 11 | 8.877314 | 0.575533 | 3.406086 | 0.533255 | 0.567539 |
| activated TAK1 mediates p38 MAPK activation | RIPK2 | Receptor-interacting serine-threonine kinase 2, Receptor-interacting serine/threonine-protein kinase 2 | 8.877314 | 0.575533 | 3.406086 | 0.533255 | 0.530344 |
| activated TAK1 mediates p38 MAPK activation | UBE2V1 | Ubiquitin conjugating enzyme E2 V1, Ubiquitin-conjugating enzyme E2 variant 1 | 8.877314 | 0.575533 | 3.406086 | 0.533255 | 0.593364 |
| Activation of AMPK downstream of NMDARs | PRKAG1 | 5'-AMP-activated protein kinase subunit gamma-1, Protein kinase, AMP-activated, gamma 1 non-catalytic subunit | 8.621227 | 0.650693 | 4 | 0.650606 | 0.515113 |
| Activation of AMPK downstream of NMDARs | PRKAG2 | 5'-AMP-activated protein kinase subunit gamma-2, 5'-AMP-activated protein kinase subunit gamma-2 isoform X8, Protein kinase AMP-activated non-catalytic subunit gamma 2, Protein kinase, AMP-activated, gamma 2 non-catalytic subunit, Uncharacterized protein | 8.621227 | 0.650693 | 4 | 0.650606 | 0.510193 |
| Activation of AMPK downstream of NMDARs | PRKAB1 | 5'-AMP-activated protein kinase subunit beta-1, 5'AMP-activated protein kinase beta-1 non-catalytic subunit, Protein kinase AMP-activated non-catalytic subunit beta 1, Protein kinase, AMP-activated, beta 1 non-catalytic subunit | 8.621227 | 0.650693 | 4 | 0.650606 | 0.500481 |
| Activation of AMPK downstream of NMDARs | MAPT | Microtubule-associated protein, Microtubule-associated protein tau | 8.621227 | 0.650693 | 4 | 0.650606 | 0.507322 |
| Activation of NF-kappaB in B cells | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.387858 | 0.721214 | 4 | 0.725416 | 0.579609 |
| Activation of NF-kappaB in B cells | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.387858 | 0.721214 | 4 | 0.725416 | 0.568645 |
| Activation of NF-kappaB in B cells | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.54735 |
| Activation of NF-kappaB in B cells | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.515416 |
| Activation of NF-kappaB in B cells | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.54735 |
| Activation of NF-kappaB in B cells | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.515416 |
| Activation of NF-kappaB in B cells | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.577481 |
| Activation of NF-kappaB in B cells | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.515416 |
| Activation of NF-kappaB in B cells | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.54735 |
| Activation of NF-kappaB in B cells | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.54735 |
| Activation of NF-kappaB in B cells | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.515416 |
| Activation of NF-kappaB in B cells | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.58858 |
| Activation of NF-kappaB in B cells | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.565601 |
| Activation of NF-kappaB in B cells | MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.519182 |
| Activation of NF-kappaB in B cells | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.547953 |
| Activation of NF-kappaB in B cells | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.57244 |
| Activation of NF-kappaB in B cells | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.515416 |
| Activation of NF-kappaB in B cells | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.546881 |
| Activation of NF-kappaB in B cells | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.515416 |
| Activation of NF-kappaB in B cells | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.515416 |
| Activation of NF-kappaB in B cells | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.387858 | 0.721214 | 4 | 0.725416 | 0.515416 |
| Activation of NF-kappaB in B cells | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.515416 |
| Activation of NF-kappaB in B cells | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.387858 | 0.721214 | 4 | 0.725416 | 0.515416 |
| Activation of NF-kappaB in B cells | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.54735 |
| Activation of NF-kappaB in B cells | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.54735 |
| Activation of NF-kappaB in B cells | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.515416 |
| Activation of NF-kappaB in B cells | PRKCB | Protein kinase C, Protein kinase C beta type | 9.387858 | 0.721214 | 4 | 0.725416 | 0.520598 |
| Activation of NF-kappaB in B cells | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.577847 |
| Activation of NF-kappaB in B cells | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.5197 |
| Activation of NF-kappaB in B cells | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.54735 |
| Activation of NF-kappaB in B cells | IKBKB | I-kappa-B kinase, Inhibitor of nuclear factor kappa-B kinase subunit beta | 9.387858 | 0.721214 | 4 | 0.725416 | 0.518113 |
| Activation of NF-kappaB in B cells | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.515416 |
| Activation of NF-kappaB in B cells | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.54735 |
| Activation of NF-kappaB in B cells | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.515416 |
| Activation of NF-kappaB in B cells | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.54735 |
| Activation of NF-kappaB in B cells | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.573417 |
| Activation of NF-kappaB in B cells | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.515416 |
| Activation of NF-kappaB in B cells | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.515416 |
| Activation of NF-kappaB in B cells | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.54735 |
| Activation of NF-kappaB in B cells | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.387858 | 0.721214 | 4 | 0.725416 | 0.572294 |
| Activation of PPARGC1A (PGC-1alpha) by phosphorylation | MAPK14 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 14 | 8.559475 | 0.788303 | 4 | 0.716225 | 0.591941 |
| Activation of PPARGC1A (PGC-1alpha) by phosphorylation | PRKAG1 | 5'-AMP-activated protein kinase subunit gamma-1, Protein kinase, AMP-activated, gamma 1 non-catalytic subunit | 8.559475 | 0.788303 | 4 | 0.716225 | 0.515113 |
| Activation of PPARGC1A (PGC-1alpha) by phosphorylation | PPARGC1A | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | 8.559475 | 0.788303 | 4 | 0.716225 | 0.539093 |
| Activation of PPARGC1A (PGC-1alpha) by phosphorylation | PRKAG2 | 5'-AMP-activated protein kinase subunit gamma-2, 5'-AMP-activated protein kinase subunit gamma-2 isoform X8, Protein kinase AMP-activated non-catalytic subunit gamma 2, Protein kinase, AMP-activated, gamma 2 non-catalytic subunit, Uncharacterized protein | 8.559475 | 0.788303 | 4 | 0.716225 | 0.510193 |
| Activation of PPARGC1A (PGC-1alpha) by phosphorylation | PRKAB1 | 5'-AMP-activated protein kinase subunit beta-1, 5'AMP-activated protein kinase beta-1 non-catalytic subunit, Protein kinase AMP-activated non-catalytic subunit beta 1, Protein kinase, AMP-activated, beta 1 non-catalytic subunit | 8.559475 | 0.788303 | 4 | 0.716225 | 0.500481 |
| Activation of PPARGC1A (PGC-1alpha) by phosphorylation | MAPK12 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 12 | 8.559475 | 0.788303 | 4 | 0.716225 | 0.573393 |
| Activation of PPARGC1A (PGC-1alpha) by phosphorylation | MAPK11 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 11 | 8.559475 | 0.788303 | 4 | 0.716225 | 0.567539 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS28 | 40S ribosomal protein S28 | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS20 | 40S ribosomal protein S20 | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS24 | 40S ribosomal protein S24 | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS5 | 40S ribosomal protein S5, 40S ribosomal protein S5 [Cleaved into: 40S ribosomal protein S5, N-terminally processed], 40S ribosomal protein S5-A, Ribosomal protein S5 | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS27 | 40S ribosomal protein S27 | 7.141576 | 0.777154 | 4 | 0.637503 | 0.549907 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS18 | 40S ribosomal protein S18 | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS4Y1 | 40S ribosomal protein S4, 40S ribosomal protein S4, Y isoform 1 | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS15 | 40S ribosomal protein S15 | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS7 | 40S ribosomal protein S7 | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS15A | 40S ribosomal protein S15a | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS29 | 40S ribosomal protein S29 | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS14 | 40S ribosomal protein S14, Ribosomal protein S14, Uncharacterized protein | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS9 | 40S ribosomal protein S9 | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS25 | 40S ribosomal protein S25 | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS12 | 40S ribosomal protein S12 | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS3 | 40S ribosomal protein S3, DNA-(apurinic or apyrimidinic site) lyase | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS17 | 40S ribosomal protein S17 | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS23 | 40S ribosomal protein S23, 40S ribosomal protein S23-A | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS6 | 40S ribosomal protein S6 | 7.141576 | 0.777154 | 4 | 0.637503 | 0.566528 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS16 | 40S ribosomal protein S16, Ribosomal protein S16, Uncharacterized protein | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS4X | 40S ribosomal protein S4, 40S ribosomal protein S4, X isoform | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS19 | 40S ribosomal protein S19, Uncharacterized protein | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS3A | 40S ribosomal protein S3a | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS2 | 40S ribosomal protein S2 | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS13 | 40S ribosomal protein S13 | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPSA | 40S ribosomal protein SA | 7.141576 | 0.777154 | 4 | 0.637503 | 0.521531 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS8 | 40S ribosomal protein S8 | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS26 | 40S ribosomal protein S26 | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS11 | 40S ribosomal protein S11 | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS10 | 40S ribosomal protein S10, Ribosomal protein S10, S10_plectin domain-containing protein | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | RPS21 | 40S ribosomal protein S21 | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | FAU | 40S ribosomal protein S30 | 7.141576 | 0.777154 | 4 | 0.637503 | 0.579609 |
| Adenylate cyclase activating pathway | ADCY9 | Adenylate cyclase 9, Adenylate cyclase type 9 | 8.964115 | 0.69311 | 3.087337 | 0.532183 | 0.550534 |
| Adenylate cyclase activating pathway | ADCY7 | Adenylate cyclase type 7 | 8.964115 | 0.69311 | 3.087337 | 0.532183 | 0.550655 |
| ADORA2B mediated anti-inflammatory cytokines production | GLP1R | Glucagon-like peptide 1 receptor | 9.229113 | 0.392462 | 4 | 0.552851 | 0.586361 |
| ADORA2B mediated anti-inflammatory cytokines production | HTR6 | 5-hydroxytryptamine, 5-hydroxytryptamine receptor 6, G_PROTEIN_RECEP_F1_2 domain-containing protein | 9.229113 | 0.392462 | 4 | 0.552851 | 0.508125 |
| ADORA2B mediated anti-inflammatory cytokines production | CALCRL | Calcitonin gene-related peptide type 1 receptor | 9.229113 | 0.392462 | 4 | 0.552851 | 0.56686 |
| ADORA2B mediated anti-inflammatory cytokines production | MC1R | Melanocyte-stimulating hormone receptor | 9.229113 | 0.392462 | 4 | 0.552851 | 0.526117 |
| ADORA2B mediated anti-inflammatory cytokines production | RAMP1 | Receptor activity modifying protein 1, Receptor activity-modifying protein 1, Zgc:91818 | 9.229113 | 0.392462 | 4 | 0.552851 | 0.582824 |
| ADORA2B mediated anti-inflammatory cytokines production | ADM2 | Adrenomedullin 2, Protein ADM2 | 9.229113 | 0.392462 | 4 | 0.552851 | 0.537164 |
| ADORA2B mediated anti-inflammatory cytokines production | ADCY7 | Adenylate cyclase type 7 | 9.229113 | 0.392462 | 4 | 0.552851 | 0.550655 |
| ADORA2B mediated anti-inflammatory cytokines production | DRD5 | D(1B) dopamine receptor, Dopamine receptor D5, G_PROTEIN_RECEP_F1_2 domain-containing protein | 9.229113 | 0.392462 | 4 | 0.552851 | 0.551596 |
| ADORA2B mediated anti-inflammatory cytokines production | ADCY9 | Adenylate cyclase 9, Adenylate cyclase type 9 | 9.229113 | 0.392462 | 4 | 0.552851 | 0.550534 |
| ADORA2B mediated anti-inflammatory cytokines production | GPBAR1 | G protein-coupled bile acid receptor 1, G-protein coupled bile acid receptor 1 | 9.229113 | 0.392462 | 4 | 0.552851 | 0.520207 |
| ADORA2B mediated anti-inflammatory cytokines production | GLP2R | Glucagon like peptide 2 receptor, Glucagon-like peptide 2 receptor | 9.229113 | 0.392462 | 4 | 0.552851 | 0.677329 |
| ADORA2B mediated anti-inflammatory cytokines production | CYSLTR2 | CYSLTR2 protein, Cysteinyl leukotriene receptor 2, G_PROTEIN_RECEP_F1_2 domain-containing protein | 9.229113 | 0.392462 | 4 | 0.552851 | 0.566351 |
| ADORA2B mediated anti-inflammatory cytokines production | CALCA | Calcitonin, Calcitonin [Cleaved into: Calcitonin; Katacalcin, Calcitonin gene-related peptide 1, Calcitonin/calcitonin-related polypeptide, alpha | 9.229113 | 0.392462 | 4 | 0.552851 | 0.535129 |
| ADORA2B mediated anti-inflammatory cytokines production | VIPR2 | Vasoactive intestinal peptide receptor 2, Vasoactive intestinal polypeptide receptor 2 | 9.229113 | 0.392462 | 4 | 0.552851 | 0.5519 |
| ADORA2B mediated anti-inflammatory cytokines production | ADCYAP1R1 | ADCYAP receptor type I, Pituitary adenylate cyclase-activating polypeptide type I receptor, Type I adenylate cyclase activating polypeptide receptor, Uncharacterized protein | 9.229113 | 0.392462 | 4 | 0.552851 | 0.594673 |
| ADORA2B mediated anti-inflammatory cytokines production | DRD1 | D(1A) dopamine receptor, Dopamine receptor D1, G_PROTEIN_RECEP_F1_2 domain-containing protein | 9.229113 | 0.392462 | 4 | 0.552851 | 0.557448 |
| ADORA2B mediated anti-inflammatory cytokines production | AVPR2 | AVPR V2, Vasopressin V2 receptor | 9.229113 | 0.392462 | 4 | 0.552851 | 0.569629 |
| ADORA2B mediated anti-inflammatory cytokines production | CRHR1 | Corticotropin-releasing factor receptor 1 | 9.229113 | 0.392462 | 4 | 0.552851 | 0.722459 |
| ADORA2B mediated anti-inflammatory cytokines production | GNAI2 | G protein subunit alpha i2, Guanine nucleotide binding protein, Guanine nucleotide-binding protein G(i) subunit alpha-2 | 9.229113 | 0.392462 | 4 | 0.552851 | 0.512918 |
| ADORA2B mediated anti-inflammatory cytokines production | PRKAR2A | cAMP-dependent protein kinase type II-alpha regulatory subunit, Protein kinase cAMP-dependent type II regulatory subunit alpha, Protein kinase, cAMP-dependent, regulatory subunit type II alpha, Uncharacterized protein | 9.229113 | 0.392462 | 4 | 0.552851 | 0.527065 |
| ADORA2B mediated anti-inflammatory cytokines production | CRHR2 | Corticotropin releasing hormone receptor 2, Corticotropin-releasing factor receptor 2, Corticotropin-releasing hormone receptor 2, Uncharacterized protein | 9.229113 | 0.392462 | 4 | 0.552851 | 0.569524 |
| ADORA2B mediated anti-inflammatory cytokines production | HTR7 | 5-hydroxytryptamine receptor 7, 5-hydroxytryptamine receptor 7 transcript variant 1 | 9.229113 | 0.392462 | 4 | 0.552851 | 0.601446 |
| ADORA2B mediated anti-inflammatory cytokines production | MC5R | Melanocortin receptor 5, mRNA cap guanine-N7 methyltransferase | 9.229113 | 0.392462 | 4 | 0.552851 | 0.501364 |
| ADORA2B mediated anti-inflammatory cytokines production | ADCYAP1 | GLUCAGON domain-containing protein, PACAP-related peptide, Pituitary adenylate cyclase-activating polypeptide | 9.229113 | 0.392462 | 4 | 0.552851 | 0.549868 |
| ADORA2B mediated anti-inflammatory cytokines production | MC4R | Melanocortin receptor 4 | 9.229113 | 0.392462 | 4 | 0.552851 | 0.598533 |
| Adrenaline signalling through Alpha-2 adrenergic receptor | ADRA2B | Adrenoceptor alpha 2B, Alpha-2B adrenergic receptor, G_PROTEIN_RECEP_F1_2 domain-containing protein | 8.193664 | 0.865443 | 2.261877 | 0.416142 | 0.524078 |
| Adrenaline signalling through Alpha-2 adrenergic receptor | ADRA2A | Adrenoceptor alpha 2A, Alpha-2A adrenergic receptor | 8.193664 | 0.865443 | 2.261877 | 0.416142 | 0.530746 |
| Adrenaline signalling through Alpha-2 adrenergic receptor | ADRA2C | Adrenoceptor alpha 2C, Alpha-2C adrenergic receptor | 8.193664 | 0.865443 | 2.261877 | 0.416142 | 0.528655 |
| Aggrephagy | ABCC1 | ABC transporter C family member 1, ATP-binding cassette, sub-family C, Multidrug resistance-associated protein 1 | 9.419851 | 0.579865 | 4 | 0.656392 | 0.610269 |
| Aggrephagy | HSP90AA1 | Heat shock protein 90 alpha family class A member 1, Heat shock protein HSP 90-alpha | 9.419851 | 0.579865 | 4 | 0.656392 | 0.624325 |
| Aggrephagy | VCP | 15S Mg(2+)-ATPase p97 subunit, Transitional endoplasmic reticulum ATPase | 9.419851 | 0.579865 | 4 | 0.656392 | 0.526633 |
| Aggrephagy | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.419851 | 0.579865 | 4 | 0.656392 | 0.579609 |
| Aggrephagy | UBE2V1 | Ubiquitin conjugating enzyme E2 V1, Ubiquitin-conjugating enzyme E2 variant 1 | 9.419851 | 0.579865 | 4 | 0.656392 | 0.593364 |
| Aggrephagy | HDAC6 | Histone deacetylase 6, Histone deacetylase 6, isoform G | 9.419851 | 0.579865 | 4 | 0.656392 | 0.531859 |
| Amine ligand-binding receptors | HTR2B | 5-hydroxytryptamine receptor 2B | 5.09067 | 0.562419 | 4 | 0.42433 | 0.563255 |
| Amine ligand-binding receptors | HTR2A | 5-hydroxytryptamine receptor 2A | 5.09067 | 0.562419 | 4 | 0.42433 | 0.628347 |
| Amine ligand-binding receptors | HTR1A | 5-hydroxytryptamine, 5-hydroxytryptamine receptor 1A | 5.09067 | 0.562419 | 4 | 0.42433 | 0.638284 |
| Amine ligand-binding receptors | HTR2C | 5-hydroxytryptamine receptor 2C | 5.09067 | 0.562419 | 4 | 0.42433 | 0.576739 |
| Amine ligand-binding receptors | ADRA2A | Adrenoceptor alpha 2A, Alpha-2A adrenergic receptor | 5.09067 | 0.562419 | 4 | 0.42433 | 0.530746 |
| Amine ligand-binding receptors | DRD4 | D(4) dopamine receptor, Dopamine receptor D4, DRD4 | 5.09067 | 0.562419 | 4 | 0.42433 | 0.571927 |
| Amine ligand-binding receptors | ADRA1B | Alpha-1B adrenergic receptor | 5.09067 | 0.562419 | 4 | 0.42433 | 0.601659 |
| Amine ligand-binding receptors | HTR7 | 5-hydroxytryptamine receptor 7, 5-hydroxytryptamine receptor 7 transcript variant 1 | 5.09067 | 0.562419 | 4 | 0.42433 | 0.601446 |
| Amine ligand-binding receptors | DRD5 | D(1B) dopamine receptor, Dopamine receptor D5, G_PROTEIN_RECEP_F1_2 domain-containing protein | 5.09067 | 0.562419 | 4 | 0.42433 | 0.551596 |
| Amine ligand-binding receptors | DRD2 | D(2) dopamine receptor, Dopamine receptor 2 protein transcript variant 1 | 5.09067 | 0.562419 | 4 | 0.42433 | 0.651493 |
| Amine ligand-binding receptors | HRH1 | Histamine H1 receptor | 5.09067 | 0.562419 | 4 | 0.42433 | 0.518086 |
| Amine ligand-binding receptors | ADRA2B | Adrenoceptor alpha 2B, Alpha-2B adrenergic receptor, G_PROTEIN_RECEP_F1_2 domain-containing protein | 5.09067 | 0.562419 | 4 | 0.42433 | 0.524078 |
| Amine ligand-binding receptors | CHRM1 | Muscarinic acetylcholine receptor, Muscarinic acetylcholine receptor M1 | 5.09067 | 0.562419 | 4 | 0.42433 | 0.532293 |
| Amine ligand-binding receptors | DRD3 | D(3) dopamine receptor, Dopamine receptor D3 | 5.09067 | 0.562419 | 4 | 0.42433 | 0.649772 |
| Amine ligand-binding receptors | CHRM2 | Muscarinic acetylcholine receptor, Muscarinic acetylcholine receptor M2 | 5.09067 | 0.562419 | 4 | 0.42433 | 0.519456 |
| Amine ligand-binding receptors | ADRA1A | Alpha-1A adrenergic receptor | 5.09067 | 0.562419 | 4 | 0.42433 | 0.61112 |
| Amine ligand-binding receptors | HTR1B | 5-hydroxytryptamine receptor 1B | 5.09067 | 0.562419 | 4 | 0.42433 | 0.500006 |
| Amine ligand-binding receptors | ADRA2C | Adrenoceptor alpha 2C, Alpha-2C adrenergic receptor | 5.09067 | 0.562419 | 4 | 0.42433 | 0.528655 |
| Amine ligand-binding receptors | DRD1 | D(1A) dopamine receptor, Dopamine receptor D1, G_PROTEIN_RECEP_F1_2 domain-containing protein | 5.09067 | 0.562419 | 4 | 0.42433 | 0.557448 |
| Amine ligand-binding receptors | CHRM3 | Muscarinic acetylcholine receptor, Muscarinic acetylcholine receptor M3 | 5.09067 | 0.562419 | 4 | 0.42433 | 0.510046 |
| Amine ligand-binding receptors | HRH3 | G_PROTEIN_RECEP_F1_2 domain-containing protein, Histamine H3 receptor, Histamine receptor H3 | 5.09067 | 0.562419 | 4 | 0.42433 | 0.608088 |
| Amine ligand-binding receptors | HTR6 | 5-hydroxytryptamine, 5-hydroxytryptamine receptor 6, G_PROTEIN_RECEP_F1_2 domain-containing protein | 5.09067 | 0.562419 | 4 | 0.42433 | 0.508125 |
| Amine ligand-binding receptors | ADRA1D | Alpha-1D adrenergic receptor, Procollagen C-endopeptidase enhancer | 5.09067 | 0.562419 | 4 | 0.42433 | 0.602889 |
| Amine ligand-binding receptors | HTR1D | 5-hydroxytryptamine receptor 1D | 5.09067 | 0.562419 | 4 | 0.42433 | 0.542406 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.54735 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.266447 | 0.581555 | 4 | 0.649324 | 0.568645 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.54735 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.54735 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.515416 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.573417 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.515416 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.266447 | 0.581555 | 4 | 0.649324 | 0.515416 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.58858 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.54735 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.54735 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.54735 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.515416 |
| Antigen processing: Ubiquitination & Proteasome degradation | UBE2V1 | Ubiquitin conjugating enzyme E2 V1, Ubiquitin-conjugating enzyme E2 variant 1 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.593364 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.5197 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.577847 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.515416 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.54735 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.515416 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.515416 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.577481 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.515416 |
| Antigen processing: Ubiquitination & Proteasome degradation | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.515416 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.54735 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.515416 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.54735 |
| Antigen processing: Ubiquitination & Proteasome degradation | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.266447 | 0.581555 | 4 | 0.649324 | 0.579609 |
| Antigen processing: Ubiquitination & Proteasome degradation | VHL | Protein Vhl, Uncharacterized protein, von Hippel-Lindau disease tumor suppressor, von Hippel-Lindau disease tumor suppressor isoform 1, von Hippel-Lindau tumor suppressor | 9.266447 | 0.581555 | 4 | 0.649324 | 0.588856 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.515416 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.54735 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.515416 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.515416 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.547953 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.572294 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.546881 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.515416 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.266447 | 0.581555 | 4 | 0.649324 | 0.515416 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.57244 |
| Antigen processing: Ubiquitination & Proteasome degradation | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.266447 | 0.581555 | 4 | 0.649324 | 0.565601 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.515416 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.515416 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.515416 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.54735 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.546881 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | CCNA2 | Cyclin-A2 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.527977 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.5197 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.515416 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | CDK1 | Cell division control protein 2, Cyclin dependent kinase 1, Cyclin-dependent kinase 1 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.514008 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.573417 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.54735 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.515416 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.54735 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.54735 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.515416 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.515416 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.547953 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.515416 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.572294 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.54735 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 4.083668 | 0.766671 | 4 | 0.474506 | 0.515416 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.577847 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.515416 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.54735 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 4.083668 | 0.766671 | 4 | 0.474506 | 0.568645 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.54735 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | CCNB1 | Cyclin B1, G2/mitotic-specific cyclin-B1, Uncharacterized protein | 4.083668 | 0.766671 | 4 | 0.474506 | 0.513205 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.515416 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.565601 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | CCNA1 | Cyclin A1, Cyclin-A1, Cyclin-A2 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.506777 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.54735 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.57244 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.54735 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.54735 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 4.083668 | 0.766671 | 4 | 0.474506 | 0.515416 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.515416 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.577481 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.515416 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.515416 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 4.083668 | 0.766671 | 4 | 0.474506 | 0.58858 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 4.083668 | 0.766671 | 4 | 0.474506 | 0.579609 |
| APC/C:Cdc20 mediated degradation of Securin | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.54735 |
| APC/C:Cdc20 mediated degradation of Securin | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.54735 |
| APC/C:Cdc20 mediated degradation of Securin | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.54735 |
| APC/C:Cdc20 mediated degradation of Securin | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.534484 | 0.799277 | 4 | 0.772012 | 0.568645 |
| APC/C:Cdc20 mediated degradation of Securin | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.54735 |
| APC/C:Cdc20 mediated degradation of Securin | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.573417 |
| APC/C:Cdc20 mediated degradation of Securin | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.58858 |
| APC/C:Cdc20 mediated degradation of Securin | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.515416 |
| APC/C:Cdc20 mediated degradation of Securin | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.515416 |
| APC/C:Cdc20 mediated degradation of Securin | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.515416 |
| APC/C:Cdc20 mediated degradation of Securin | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.515416 |
| APC/C:Cdc20 mediated degradation of Securin | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.515416 |
| APC/C:Cdc20 mediated degradation of Securin | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.54735 |
| APC/C:Cdc20 mediated degradation of Securin | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.515416 |
| APC/C:Cdc20 mediated degradation of Securin | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.5197 |
| APC/C:Cdc20 mediated degradation of Securin | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.515416 |
| APC/C:Cdc20 mediated degradation of Securin | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.515416 |
| APC/C:Cdc20 mediated degradation of Securin | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.515416 |
| APC/C:Cdc20 mediated degradation of Securin | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.57244 |
| APC/C:Cdc20 mediated degradation of Securin | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.54735 |
| APC/C:Cdc20 mediated degradation of Securin | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.515416 |
| APC/C:Cdc20 mediated degradation of Securin | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.515416 |
| APC/C:Cdc20 mediated degradation of Securin | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.534484 | 0.799277 | 4 | 0.772012 | 0.515416 |
| APC/C:Cdc20 mediated degradation of Securin | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.546881 |
| APC/C:Cdc20 mediated degradation of Securin | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.577847 |
| APC/C:Cdc20 mediated degradation of Securin | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.54735 |
| APC/C:Cdc20 mediated degradation of Securin | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.547953 |
| APC/C:Cdc20 mediated degradation of Securin | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.515416 |
| APC/C:Cdc20 mediated degradation of Securin | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.54735 |
| APC/C:Cdc20 mediated degradation of Securin | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.565601 |
| APC/C:Cdc20 mediated degradation of Securin | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.515416 |
| APC/C:Cdc20 mediated degradation of Securin | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.534484 | 0.799277 | 4 | 0.772012 | 0.515416 |
| APC/C:Cdc20 mediated degradation of Securin | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.572294 |
| APC/C:Cdc20 mediated degradation of Securin | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.534484 | 0.799277 | 4 | 0.772012 | 0.579609 |
| APC/C:Cdc20 mediated degradation of Securin | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.54735 |
| APC/C:Cdc20 mediated degradation of Securin | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.577481 |
| APC/C:Cdc20 mediated degradation of Securin | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.534484 | 0.799277 | 4 | 0.772012 | 0.54735 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.5197 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.54735 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.515416 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.515416 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.58858 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.573417 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.54735 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.515416 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.515416 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.515416 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | AURKA | Aurora kinase, Aurora kinase A | 9.647483 | 0.755439 | 4 | 0.755923 | 0.54457 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.547953 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.54735 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.54735 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.647483 | 0.755439 | 4 | 0.755923 | 0.515416 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.515416 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.515416 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.577847 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.54735 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.565601 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.515416 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.54735 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | AURKB | Aurora kinase, Aurora kinase B | 9.647483 | 0.755439 | 4 | 0.755923 | 0.524412 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.54735 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PLK1 | Serine/threonine-protein kinase PLK, Serine/threonine-protein kinase PLK1 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.504187 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.57244 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.515416 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.515416 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.546881 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.54735 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.515416 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.54735 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.515416 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.572294 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.577481 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.647483 | 0.755439 | 4 | 0.755923 | 0.568645 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.647483 | 0.755439 | 4 | 0.755923 | 0.579609 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.515416 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.647483 | 0.755439 | 4 | 0.755923 | 0.54735 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.647483 | 0.755439 | 4 | 0.755923 | 0.515416 |
| ARMS-mediated activation | BRAF | Non-specific serine/threonine protein kinase, Serine/threonine-protein kinase B-raf | 9.358388 | 0.802273 | 3.390943 | 0.648031 | 0.590074 |
| ARMS-mediated activation | YWHAB | 14-3-3 protein beta/alpha, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta | 9.358388 | 0.802273 | 3.390943 | 0.648031 | 0.513843 |
| ARMS-mediated activation | NTRK1 | High affinity nerve growth factor receptor, Tyrosine-protein kinase receptor | 9.358388 | 0.802273 | 3.390943 | 0.648031 | 0.527885 |
| ARMS-mediated activation | CRK | 14_3_3 domain-containing protein, Adapter molecule crk, Adapter molecule Crk, CRK proto-oncogene, adaptor protein, Uncharacterized protein, V-crk avian sarcoma virus CT10 oncogene homolog | 9.358388 | 0.802273 | 3.390943 | 0.648031 | 0.513369 |
| ARMS-mediated activation | RAP1A | RAP1A, member of RAS oncogene family, Ras-related protein Rap-1A, Uncharacterized protein | 9.358388 | 0.802273 | 3.390943 | 0.648031 | 0.530823 |
| Assembly and cell surface presentation of NMDA receptors | GRIN3A | Glutamate receptor, Glutamate receptor ionotropic, NMDA 3A | 8.728157 | 0.547829 | 4 | 0.60469 | 0.524922 |
| Assembly and cell surface presentation of NMDA receptors | GRIN1 | Glutamate receptor, Glutamate receptor ionotropic, NMDA 1 | 8.728157 | 0.547829 | 4 | 0.60469 | 0.573676 |
| Assembly and cell surface presentation of NMDA receptors | GRIN2C | Glutamate receptor, Glutamate receptor ionotropic, NMDA 2C | 8.728157 | 0.547829 | 4 | 0.60469 | 0.557006 |
| Assembly and cell surface presentation of NMDA receptors | GRIN2A | Glutamate receptor, Glutamate receptor ionotropic, NMDA 2A | 8.728157 | 0.547829 | 4 | 0.60469 | 0.527756 |
| Assembly and cell surface presentation of NMDA receptors | GRIN3B | Glutamate receptor ionotropic, NMDA 3B | 8.728157 | 0.547829 | 4 | 0.60469 | 0.524714 |
| Assembly and cell surface presentation of NMDA receptors | GRIN2B | Glutamate receptor, Glutamate receptor ionotropic, NMDA 2B | 8.728157 | 0.547829 | 4 | 0.60469 | 0.56443 |
| Assembly and cell surface presentation of NMDA receptors | GRIN2D | Glutamate receptor, Glutamate receptor ionotropic, NMDA 2D | 8.728157 | 0.547829 | 4 | 0.60469 | 0.556318 |
| Assembly of the pre-replicative complex | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.565601 |
| Assembly of the pre-replicative complex | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.515416 |
| Assembly of the pre-replicative complex | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.515416 |
| Assembly of the pre-replicative complex | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.57244 |
| Assembly of the pre-replicative complex | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.5197 |
| Assembly of the pre-replicative complex | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.515416 |
| Assembly of the pre-replicative complex | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.54735 |
| Assembly of the pre-replicative complex | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.54735 |
| Assembly of the pre-replicative complex | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.515416 |
| Assembly of the pre-replicative complex | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.577481 |
| Assembly of the pre-replicative complex | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.54735 |
| Assembly of the pre-replicative complex | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.54735 |
| Assembly of the pre-replicative complex | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.515416 |
| Assembly of the pre-replicative complex | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.515416 |
| Assembly of the pre-replicative complex | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 8.757997 | 0.799396 | 4 | 0.732014 | 0.568645 |
| Assembly of the pre-replicative complex | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.54735 |
| Assembly of the pre-replicative complex | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.58858 |
| Assembly of the pre-replicative complex | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.572294 |
| Assembly of the pre-replicative complex | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 8.757997 | 0.799396 | 4 | 0.732014 | 0.515416 |
| Assembly of the pre-replicative complex | GMNN | Geminin, Geminin DNA replication inhibitor, Gmnn protein, GMNN protein, Uncharacterized protein | 8.757997 | 0.799396 | 4 | 0.732014 | 0.542818 |
| Assembly of the pre-replicative complex | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.54735 |
| Assembly of the pre-replicative complex | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.515416 |
| Assembly of the pre-replicative complex | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.573417 |
| Assembly of the pre-replicative complex | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.546881 |
| Assembly of the pre-replicative complex | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.515416 |
| Assembly of the pre-replicative complex | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.577847 |
| Assembly of the pre-replicative complex | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 8.757997 | 0.799396 | 4 | 0.732014 | 0.515416 |
| Assembly of the pre-replicative complex | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.515416 |
| Assembly of the pre-replicative complex | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.515416 |
| Assembly of the pre-replicative complex | MCM4 | DNA helicase, DNA replication licensing factor mcm4, DNA replication licensing factor MCM4 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.515269 |
| Assembly of the pre-replicative complex | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.54735 |
| Assembly of the pre-replicative complex | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.547953 |
| Assembly of the pre-replicative complex | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 8.757997 | 0.799396 | 4 | 0.732014 | 0.579609 |
| Assembly of the pre-replicative complex | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.54735 |
| Assembly of the pre-replicative complex | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.54735 |
| Assembly of the pre-replicative complex | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.515416 |
| Assembly of the pre-replicative complex | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.54735 |
| Assembly of the pre-replicative complex | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.515416 |
| Assembly of the pre-replicative complex | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 8.757997 | 0.799396 | 4 | 0.732014 | 0.515416 |
| Assembly of the SARS-CoV-1 Replication-Transcription Complex (RTC) | 1A | Replicase polyprotein 1a | 12.767545 | 1 | 0.701304 | ||
| Assembly of the SARS-CoV-1 Replication-Transcription Complex (RTC) | REP | Rab proteins geranylgeranyltransferase component A, Replicase polyprotein 1ab | 12.767545 | 1 | 0.746505 | ||
| Asymmetric localization of PCP proteins | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.515416 |
| Asymmetric localization of PCP proteins | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.515416 |
| Asymmetric localization of PCP proteins | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.515416 |
| Asymmetric localization of PCP proteins | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.54735 |
| Asymmetric localization of PCP proteins | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.675609 | 0.800355 | 4 | 0.779832 | 0.515416 |
| Asymmetric localization of PCP proteins | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.547953 |
| Asymmetric localization of PCP proteins | DVL2 | Dishevelled segment polarity protein 2, Segment polarity protein dishevelled homolog DVL-2, Uncharacterized protein | 9.675609 | 0.800355 | 4 | 0.779832 | 0.529242 |
| Asymmetric localization of PCP proteins | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.515416 |
| Asymmetric localization of PCP proteins | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.573417 |
| Asymmetric localization of PCP proteins | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.572294 |
| Asymmetric localization of PCP proteins | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.577847 |
| Asymmetric localization of PCP proteins | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.54735 |
| Asymmetric localization of PCP proteins | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.54735 |
| Asymmetric localization of PCP proteins | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.515416 |
| Asymmetric localization of PCP proteins | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.515416 |
| Asymmetric localization of PCP proteins | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.57244 |
| Asymmetric localization of PCP proteins | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.565601 |
| Asymmetric localization of PCP proteins | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.54735 |
| Asymmetric localization of PCP proteins | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.675609 | 0.800355 | 4 | 0.779832 | 0.515416 |
| Asymmetric localization of PCP proteins | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.515416 |
| Asymmetric localization of PCP proteins | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.515416 |
| Asymmetric localization of PCP proteins | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.54735 |
| Asymmetric localization of PCP proteins | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.515416 |
| Asymmetric localization of PCP proteins | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.577481 |
| Asymmetric localization of PCP proteins | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.54735 |
| Asymmetric localization of PCP proteins | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.515416 |
| Asymmetric localization of PCP proteins | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.675609 | 0.800355 | 4 | 0.779832 | 0.568645 |
| Asymmetric localization of PCP proteins | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.54735 |
| Asymmetric localization of PCP proteins | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.54735 |
| Asymmetric localization of PCP proteins | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.515416 |
| Asymmetric localization of PCP proteins | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.54735 |
| Asymmetric localization of PCP proteins | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.5197 |
| Asymmetric localization of PCP proteins | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.58858 |
| Asymmetric localization of PCP proteins | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.515416 |
| Asymmetric localization of PCP proteins | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.675609 | 0.800355 | 4 | 0.779832 | 0.579609 |
| Asymmetric localization of PCP proteins | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.546881 |
| Asymmetric localization of PCP proteins | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.515416 |
| Asymmetric localization of PCP proteins | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.675609 | 0.800355 | 4 | 0.779832 | 0.54735 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.515416 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.577847 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.54735 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.57244 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.515416 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.515416 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.515416 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.573417 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.54735 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.577481 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.400058 | 0.759684 | 4 | 0.745281 | 0.579609 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.54735 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.58858 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.515416 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.572294 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.5197 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.515416 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.54735 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.515416 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.400058 | 0.759684 | 4 | 0.745281 | 0.515416 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.547953 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.400058 | 0.759684 | 4 | 0.745281 | 0.515416 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.54735 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.565601 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.54735 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.515416 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.546881 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.515416 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.515416 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.515416 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.515416 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.54735 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.54735 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.54735 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.54735 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.400058 | 0.759684 | 4 | 0.745281 | 0.515416 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.400058 | 0.759684 | 4 | 0.745281 | 0.568645 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.54735 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.515416 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.520935 | 0.795888 | 4 | 0.769619 | 0.515416 |
| Autodegradation of Cdh1 by Cdh1:APC/C | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.515416 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.572294 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.515416 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.54735 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.54735 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.54735 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.546881 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.515416 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.515416 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.515416 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.547953 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.515416 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.577847 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.515416 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.573417 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.520935 | 0.795888 | 4 | 0.769619 | 0.515416 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.54735 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.54735 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.515416 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.54735 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.577481 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.515416 |
| Autodegradation of Cdh1 by Cdh1:APC/C | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.520935 | 0.795888 | 4 | 0.769619 | 0.579609 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.520935 | 0.795888 | 4 | 0.769619 | 0.568645 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.515416 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.5197 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.565601 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.515416 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.515416 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.54735 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.58858 |
| Autodegradation of Cdh1 by Cdh1:APC/C | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.520935 | 0.795888 | 4 | 0.769619 | 0.57244 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.515416 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.515416 |
| Autodegradation of the E3 ubiquitin ligase COP1 | TP53 | Cellular tumor antigen p53 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.525937 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.577481 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.515416 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.54735 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.686656 | 0.804353 | 4 | 0.782401 | 0.515416 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.54735 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.686656 | 0.804353 | 4 | 0.782401 | 0.515416 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.58858 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.686656 | 0.804353 | 4 | 0.782401 | 0.568645 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.54735 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.573417 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.515416 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.5197 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.54735 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.515416 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.515416 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.515416 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.515416 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.515416 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.54735 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.54735 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.54735 |
| Autodegradation of the E3 ubiquitin ligase COP1 | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.515416 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.565601 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.54735 |
| Autodegradation of the E3 ubiquitin ligase COP1 | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.686656 | 0.804353 | 4 | 0.782401 | 0.579609 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.515416 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.515416 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.572294 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.547953 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.54735 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.577847 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.515416 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.57244 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.54735 |
| Autodegradation of the E3 ubiquitin ligase COP1 | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.686656 | 0.804353 | 4 | 0.782401 | 0.546881 |
| Autointegration results in viral DNA circles | GAG-POL | Gag-Pol polyprotein | 7.548022 | 0.54084 | 0.64835 | ||
| Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein | ULK3 | Serine/threonine-protein kinase ULK3, Unc-51-like kinase 3 | -0.587555 | 4 | 0.587555 | 0.512263 | |
| Budding | PB1 | Protein PB1-F2, RNA-directed RNA polymerase catalytic subunit | 7.405237 | 0.528279 | 0.508942 | ||
| Calmodulin induced events | ADCY9 | Adenylate cyclase 9, Adenylate cyclase type 9 | 8.158705 | 0.484032 | 3.478698 | 0.472594 | 0.550534 |
| Calmodulin induced events | PRKCD | Protein kinase C, Protein kinase C delta type | 8.158705 | 0.484032 | 3.478698 | 0.472594 | 0.520528 |
| Calmodulin induced events | PRKCG | Protein kinase C, Protein kinase C gamma type, Sperm-associated antigen 9 | 8.158705 | 0.484032 | 3.478698 | 0.472594 | 0.500344 |
| Calmodulin induced events | PRKAR2A | cAMP-dependent protein kinase type II-alpha regulatory subunit, Protein kinase cAMP-dependent type II regulatory subunit alpha, Protein kinase, cAMP-dependent, regulatory subunit type II alpha, Uncharacterized protein | 8.158705 | 0.484032 | 3.478698 | 0.472594 | 0.527065 |
| Calmodulin induced events | ADCY7 | Adenylate cyclase type 7 | 8.158705 | 0.484032 | 3.478698 | 0.472594 | 0.550655 |
| Calmodulin induced events | PRKCA | Protein kinase C, Protein kinase C alpha type, Protein kinase C, alpha | 8.158705 | 0.484032 | 3.478698 | 0.472594 | 0.52202 |
| CaM pathway | ADCY7 | Adenylate cyclase type 7 | 7.87986 | 0.484032 | 3.478698 | 0.460083 | 0.550655 |
| CaM pathway | PRKAR2A | cAMP-dependent protein kinase type II-alpha regulatory subunit, Protein kinase cAMP-dependent type II regulatory subunit alpha, Protein kinase, cAMP-dependent, regulatory subunit type II alpha, Uncharacterized protein | 7.87986 | 0.484032 | 3.478698 | 0.460083 | 0.527065 |
| CaM pathway | PRKCG | Protein kinase C, Protein kinase C gamma type, Sperm-associated antigen 9 | 7.87986 | 0.484032 | 3.478698 | 0.460083 | 0.500344 |
| CaM pathway | ADCY9 | Adenylate cyclase 9, Adenylate cyclase type 9 | 7.87986 | 0.484032 | 3.478698 | 0.460083 | 0.550534 |
| CaM pathway | PRKCD | Protein kinase C, Protein kinase C delta type | 7.87986 | 0.484032 | 3.478698 | 0.460083 | 0.520528 |
| CaM pathway | PRKCA | Protein kinase C, Protein kinase C alpha type, Protein kinase C, alpha | 7.87986 | 0.484032 | 3.478698 | 0.460083 | 0.52202 |
| CD28 co-stimulation | MTOR | Nucleoprotein TPR, Serine/threonine-protein kinase mTOR, Serine/threonine-protein kinase TOR | 8.820281 | 0.545473 | 4 | 0.608265 | 0.619026 |
| CD28 co-stimulation | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 8.820281 | 0.545473 | 4 | 0.608265 | 0.589663 |
| CD28 co-stimulation | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 8.820281 | 0.545473 | 4 | 0.608265 | 0.594807 |
| CD28 co-stimulation | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 8.820281 | 0.545473 | 4 | 0.608265 | 0.646924 |
| CD28 co-stimulation | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 8.820281 | 0.545473 | 4 | 0.608265 | 0.552041 |
| CD28 co-stimulation | LCK | Proto-oncogene tyrosine-protein kinase LCK, Tyrosine-protein kinase, Tyrosine-protein kinase Lck | 8.820281 | 0.545473 | 4 | 0.608265 | 0.543278 |
| CD28 co-stimulation | YES1 | Tyrosine-protein kinase, Tyrosine-protein kinase yes, Tyrosine-protein kinase Yes | 8.820281 | 0.545473 | 4 | 0.608265 | 0.506641 |
| CD28 co-stimulation | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 8.820281 | 0.545473 | 4 | 0.608265 | 0.660919 |
| CD28 co-stimulation | MAP3K14 | Mitogen-activated protein kinase kinase kinase 14 | 8.820281 | 0.545473 | 4 | 0.608265 | 0.556123 |
| CD28 dependent PI3K/Akt signaling | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.236302 | 0.686942 | 4 | 0.700462 | 0.660919 |
| CD28 dependent PI3K/Akt signaling | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 9.236302 | 0.686942 | 4 | 0.700462 | 0.646924 |
| CD28 dependent PI3K/Akt signaling | MTOR | Nucleoprotein TPR, Serine/threonine-protein kinase mTOR, Serine/threonine-protein kinase TOR | 9.236302 | 0.686942 | 4 | 0.700462 | 0.619026 |
| CD28 dependent PI3K/Akt signaling | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 9.236302 | 0.686942 | 4 | 0.700462 | 0.552041 |
| CD28 dependent PI3K/Akt signaling | LCK | Proto-oncogene tyrosine-protein kinase LCK, Tyrosine-protein kinase, Tyrosine-protein kinase Lck | 9.236302 | 0.686942 | 4 | 0.700462 | 0.543278 |
| CD28 dependent PI3K/Akt signaling | MAP3K14 | Mitogen-activated protein kinase kinase kinase 14 | 9.236302 | 0.686942 | 4 | 0.700462 | 0.556123 |
| CD28 dependent PI3K/Akt signaling | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 9.236302 | 0.686942 | 4 | 0.700462 | 0.589663 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.577481 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.547953 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.515416 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.573417 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | CDK1 | Cell division control protein 2, Cyclin dependent kinase 1, Cyclin-dependent kinase 1 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.514008 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.515416 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.515416 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.543784 | 0.783947 | 4 | 0.764827 | 0.515416 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.54735 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.515416 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | CCNA2 | Cyclin-A2 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.527977 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.515416 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.515416 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.543784 | 0.783947 | 4 | 0.764827 | 0.579609 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.57244 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.515416 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.572294 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.515416 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.5197 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.515416 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.515416 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.54735 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.565601 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.577847 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.54735 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.58858 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.54735 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.543784 | 0.783947 | 4 | 0.764827 | 0.515416 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | CCNA1 | Cyclin A1, Cyclin-A1, Cyclin-A2 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.506777 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.54735 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.54735 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.54735 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.54735 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.54735 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.54735 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.546881 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.515416 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.543784 | 0.783947 | 4 | 0.764827 | 0.568645 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.515416 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.543784 | 0.783947 | 4 | 0.764827 | 0.515416 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.54735 |
| CDK-mediated phosphorylation and removal of Cdc6 | CDK2 | Cyclin dependent kinase 2, Cyclin-dependent kinase 2, Protein kinase domain-containing protein, Uncharacterized protein | 9.481822 | 0.774146 | 4 | 0.75673 | 0.532011 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.481822 | 0.774146 | 4 | 0.75673 | 0.515416 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.577481 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.515416 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.515416 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.515416 |
| CDK-mediated phosphorylation and removal of Cdc6 | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.481822 | 0.774146 | 4 | 0.75673 | 0.579609 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.577847 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.515416 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.546881 |
| CDK-mediated phosphorylation and removal of Cdc6 | CCNA2 | Cyclin-A2 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.527977 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.5197 |
| CDK-mediated phosphorylation and removal of Cdc6 | CCNE2 | Cyclin E2, Cyclin N-terminal domain-containing protein, G1/S-specific cyclin-E2 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.503983 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.481822 | 0.774146 | 4 | 0.75673 | 0.568645 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.515416 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.573417 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.54735 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.572294 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.58858 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.54735 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.57244 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.515416 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.54735 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.515416 |
| CDK-mediated phosphorylation and removal of Cdc6 | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.515416 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.515416 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.54735 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.54735 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.515416 |
| CDK-mediated phosphorylation and removal of Cdc6 | CDK1 | Cell division control protein 2, Cyclin dependent kinase 1, Cyclin-dependent kinase 1 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.514008 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.515416 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.565601 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.515416 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.515416 |
| CDK-mediated phosphorylation and removal of Cdc6 | CCNE1 | Cyclin E1, Cyclin N-terminal domain-containing protein, G1/S-specific cyclin-E1 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.526741 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.54735 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.54735 |
| CDK-mediated phosphorylation and removal of Cdc6 | CCNA1 | Cyclin A1, Cyclin-A1, Cyclin-A2 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.506777 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.54735 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.547953 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.481822 | 0.774146 | 4 | 0.75673 | 0.54735 |
| CDK-mediated phosphorylation and removal of Cdc6 | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.481822 | 0.774146 | 4 | 0.75673 | 0.515416 |
| CDT1 association with the CDC6:ORC:origin complex | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.515416 |
| CDT1 association with the CDC6:ORC:origin complex | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.515416 |
| CDT1 association with the CDC6:ORC:origin complex | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.54735 |
| CDT1 association with the CDC6:ORC:origin complex | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.573417 |
| CDT1 association with the CDC6:ORC:origin complex | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.577847 |
| CDT1 association with the CDC6:ORC:origin complex | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.546881 |
| CDT1 association with the CDC6:ORC:origin complex | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.54735 |
| CDT1 association with the CDC6:ORC:origin complex | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.58858 |
| CDT1 association with the CDC6:ORC:origin complex | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.515416 |
| CDT1 association with the CDC6:ORC:origin complex | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.5197 |
| CDT1 association with the CDC6:ORC:origin complex | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.54735 |
| CDT1 association with the CDC6:ORC:origin complex | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.515416 |
| CDT1 association with the CDC6:ORC:origin complex | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.54735 |
| CDT1 association with the CDC6:ORC:origin complex | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.515416 |
| CDT1 association with the CDC6:ORC:origin complex | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.577481 |
| CDT1 association with the CDC6:ORC:origin complex | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.57244 |
| CDT1 association with the CDC6:ORC:origin complex | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.82128 | 0.81513 | 4 | 0.794734 | 0.515416 |
| CDT1 association with the CDC6:ORC:origin complex | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.54735 |
| CDT1 association with the CDC6:ORC:origin complex | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.515416 |
| CDT1 association with the CDC6:ORC:origin complex | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.515416 |
| CDT1 association with the CDC6:ORC:origin complex | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.54735 |
| CDT1 association with the CDC6:ORC:origin complex | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.515416 |
| CDT1 association with the CDC6:ORC:origin complex | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.82128 | 0.81513 | 4 | 0.794734 | 0.515416 |
| CDT1 association with the CDC6:ORC:origin complex | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.515416 |
| CDT1 association with the CDC6:ORC:origin complex | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.54735 |
| CDT1 association with the CDC6:ORC:origin complex | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.82128 | 0.81513 | 4 | 0.794734 | 0.579609 |
| CDT1 association with the CDC6:ORC:origin complex | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.515416 |
| CDT1 association with the CDC6:ORC:origin complex | GMNN | Geminin, Geminin DNA replication inhibitor, Gmnn protein, GMNN protein, Uncharacterized protein | 9.82128 | 0.81513 | 4 | 0.794734 | 0.542818 |
| CDT1 association with the CDC6:ORC:origin complex | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.54735 |
| CDT1 association with the CDC6:ORC:origin complex | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.54735 |
| CDT1 association with the CDC6:ORC:origin complex | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.565601 |
| CDT1 association with the CDC6:ORC:origin complex | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.572294 |
| CDT1 association with the CDC6:ORC:origin complex | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.515416 |
| CDT1 association with the CDC6:ORC:origin complex | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.547953 |
| CDT1 association with the CDC6:ORC:origin complex | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.82128 | 0.81513 | 4 | 0.794734 | 0.568645 |
| CDT1 association with the CDC6:ORC:origin complex | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.515416 |
| CDT1 association with the CDC6:ORC:origin complex | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.54735 |
| CDT1 association with the CDC6:ORC:origin complex | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.82128 | 0.81513 | 4 | 0.794734 | 0.515416 |
| Cellular response to hypoxia | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.54735 |
| Cellular response to hypoxia | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.57244 |
| Cellular response to hypoxia | VHL | Protein Vhl, Uncharacterized protein, von Hippel-Lindau disease tumor suppressor, von Hippel-Lindau disease tumor suppressor isoform 1, von Hippel-Lindau tumor suppressor | 8.458167 | 0.6372 | 4 | 0.635448 | 0.588856 |
| Cellular response to hypoxia | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.515416 |
| Cellular response to hypoxia | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.54735 |
| Cellular response to hypoxia | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 8.458167 | 0.6372 | 4 | 0.635448 | 0.515416 |
| Cellular response to hypoxia | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.577481 |
| Cellular response to hypoxia | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.515416 |
| Cellular response to hypoxia | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.515416 |
| Cellular response to hypoxia | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.54735 |
| Cellular response to hypoxia | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.515416 |
| Cellular response to hypoxia | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.547953 |
| Cellular response to hypoxia | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.577847 |
| Cellular response to hypoxia | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.54735 |
| Cellular response to hypoxia | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.58858 |
| Cellular response to hypoxia | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.54735 |
| Cellular response to hypoxia | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.515416 |
| Cellular response to hypoxia | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.54735 |
| Cellular response to hypoxia | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.54735 |
| Cellular response to hypoxia | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.54735 |
| Cellular response to hypoxia | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.515416 |
| Cellular response to hypoxia | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.54735 |
| Cellular response to hypoxia | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 8.458167 | 0.6372 | 4 | 0.635448 | 0.515416 |
| Cellular response to hypoxia | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.572294 |
| Cellular response to hypoxia | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.515416 |
| Cellular response to hypoxia | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 8.458167 | 0.6372 | 4 | 0.635448 | 0.579609 |
| Cellular response to hypoxia | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.515416 |
| Cellular response to hypoxia | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.515416 |
| Cellular response to hypoxia | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.565601 |
| Cellular response to hypoxia | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.54735 |
| Cellular response to hypoxia | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.515416 |
| Cellular response to hypoxia | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.515416 |
| Cellular response to hypoxia | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 8.458167 | 0.6372 | 4 | 0.635448 | 0.568645 |
| Cellular response to hypoxia | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.515416 |
| Cellular response to hypoxia | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.5197 |
| Cellular response to hypoxia | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.515416 |
| Cellular response to hypoxia | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.546881 |
| Cellular response to hypoxia | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 8.458167 | 0.6372 | 4 | 0.635448 | 0.573417 |
| Cilium Assembly | PLK1 | Serine/threonine-protein kinase PLK, Serine/threonine-protein kinase PLK1 | 8.732952 | 0.433193 | 3.200728 | 0.438196 | 0.504187 |
| Cilium Assembly | HDAC6 | Histone deacetylase 6, Histone deacetylase 6, isoform G | 8.732952 | 0.433193 | 3.200728 | 0.438196 | 0.531859 |
| Cilium Assembly | SSTR3 | Somatostatin receptor 3, Somatostatin receptor type 3 | 8.732952 | 0.433193 | 3.200728 | 0.438196 | 0.543809 |
| Cilium Assembly | MCHR1 | G-protein coupled receptor 24, Melanin-concentrating hormone receptor 1 | 8.732952 | 0.433193 | 3.200728 | 0.438196 | 0.678762 |
| Cilium Assembly | CDK1 | Cell division control protein 2, Cyclin dependent kinase 1, Cyclin-dependent kinase 1 | 8.732952 | 0.433193 | 3.200728 | 0.438196 | 0.514008 |
| Cilium Assembly | HSP90AA1 | Heat shock protein 90 alpha family class A member 1, Heat shock protein HSP 90-alpha | 8.732952 | 0.433193 | 3.200728 | 0.438196 | 0.624325 |
| Cilium Assembly | CRK | 14_3_3 domain-containing protein, Adapter molecule crk, Adapter molecule Crk, CRK proto-oncogene, adaptor protein, Uncharacterized protein, V-crk avian sarcoma virus CT10 oncogene homolog | 8.732952 | 0.433193 | 3.200728 | 0.438196 | 0.513369 |
| Cilium Assembly | SMO | Protein smoothened, Smoothened homolog, Smoothened, frizzled class receptor | 8.732952 | 0.433193 | 3.200728 | 0.438196 | 0.547562 |
| Class A/1 (Rhodopsin-like receptors) | HTR2B | 5-hydroxytryptamine receptor 2B | 8.546785 | 0.366878 | 4 | 0.504858 | 0.563255 |
| Class A/1 (Rhodopsin-like receptors) | HTR1A | 5-hydroxytryptamine, 5-hydroxytryptamine receptor 1A | 8.546785 | 0.366878 | 4 | 0.504858 | 0.638284 |
| Class A/1 (Rhodopsin-like receptors) | SSTR2 | Somatostatin receptor type 2 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.595919 |
| Class A/1 (Rhodopsin-like receptors) | P2RY1 | P2Y purinoceptor 1 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.550301 |
| Class A/1 (Rhodopsin-like receptors) | F2 | Activation peptide fragment 1, Prothrombin | 8.546785 | 0.366878 | 4 | 0.504858 | 0.5402 |
| Class A/1 (Rhodopsin-like receptors) | CHRM3 | Muscarinic acetylcholine receptor, Muscarinic acetylcholine receptor M3 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.510046 |
| Class A/1 (Rhodopsin-like receptors) | CXCR4 | C-X-C chemokine receptor type 4, G_PROTEIN_RECEP_F1_2 domain-containing protein | 8.546785 | 0.366878 | 4 | 0.504858 | 0.512583 |
| Class A/1 (Rhodopsin-like receptors) | HTR1D | 5-hydroxytryptamine receptor 1D | 8.546785 | 0.366878 | 4 | 0.504858 | 0.542406 |
| Class A/1 (Rhodopsin-like receptors) | SSTR5 | G_PROTEIN_RECEP_F1_2 domain-containing protein, Somatostatin receptor 5, Somatostatin receptor type 5 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.563799 |
| Class A/1 (Rhodopsin-like receptors) | DRD2 | D(2) dopamine receptor, Dopamine receptor 2 protein transcript variant 1 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.651493 |
| Class A/1 (Rhodopsin-like receptors) | C3 | C3-beta-c, Complement C3, Complement C3 [Cleaved into: Complement C3 beta chain; C3-beta-c, Uncharacterized protein | 8.546785 | 0.366878 | 4 | 0.504858 | 0.586567 |
| Class A/1 (Rhodopsin-like receptors) | ADRA1B | Alpha-1B adrenergic receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.601659 |
| Class A/1 (Rhodopsin-like receptors) | HCRTR2 | Hypocretin receptor type 2, Orexin receptor type 2 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.558726 |
| Class A/1 (Rhodopsin-like receptors) | CCR5 | C-C chemokine receptor type 5, Chemokine receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.581451 |
| Class A/1 (Rhodopsin-like receptors) | DRD3 | D(3) dopamine receptor, Dopamine receptor D3 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.649772 |
| Class A/1 (Rhodopsin-like receptors) | APP | ABPP, Amyloid-beta A4 protein, Amyloid-beta precursor protein | 8.546785 | 0.366878 | 4 | 0.504858 | 0.506343 |
| Class A/1 (Rhodopsin-like receptors) | CCKAR | Cholecystokinin receptor type A, Cholecystokinin-2 receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.582813 |
| Class A/1 (Rhodopsin-like receptors) | AGTR2 | Angiotensin II receptor, type 2, Angiotensin II type-2 receptor, Type-2 angiotensin II receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.611168 |
| Class A/1 (Rhodopsin-like receptors) | HTR6 | 5-hydroxytryptamine, 5-hydroxytryptamine receptor 6, G_PROTEIN_RECEP_F1_2 domain-containing protein | 8.546785 | 0.366878 | 4 | 0.504858 | 0.508125 |
| Class A/1 (Rhodopsin-like receptors) | F2RL3 | F2R like thrombin or trypsin receptor 3, F2R-like thrombin or trypsin receptor 3, G_PROTEIN_RECEP_F1_2 domain-containing protein, Proteinase-activated receptor 4 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.537847 |
| Class A/1 (Rhodopsin-like receptors) | EDNRB | Endothelin receptor non-selective type, Endothelin receptor type B | 8.546785 | 0.366878 | 4 | 0.504858 | 0.518386 |
| Class A/1 (Rhodopsin-like receptors) | HTR1B | 5-hydroxytryptamine receptor 1B | 8.546785 | 0.366878 | 4 | 0.504858 | 0.500006 |
| Class A/1 (Rhodopsin-like receptors) | HTR2C | 5-hydroxytryptamine receptor 2C | 8.546785 | 0.366878 | 4 | 0.504858 | 0.576739 |
| Class A/1 (Rhodopsin-like receptors) | CHRM2 | Muscarinic acetylcholine receptor, Muscarinic acetylcholine receptor M2 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.519456 |
| Class A/1 (Rhodopsin-like receptors) | HTR2A | 5-hydroxytryptamine receptor 2A | 8.546785 | 0.366878 | 4 | 0.504858 | 0.628347 |
| Class A/1 (Rhodopsin-like receptors) | MCHR1 | G-protein coupled receptor 24, Melanin-concentrating hormone receptor 1 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.678762 |
| Class A/1 (Rhodopsin-like receptors) | C5AR1 | C5a anaphylatoxin chemotactic receptor, C5a anaphylatoxin chemotactic receptor 1 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.500177 |
| Class A/1 (Rhodopsin-like receptors) | CCKBR | Cholecystokinin-2 receptor, Gastrin/cholecystokinin type B receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.593267 |
| Class A/1 (Rhodopsin-like receptors) | OPRK1 | Kappa-type opioid receptor, Opioid receptor kappa 1, Opioid receptor, kappa 1 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.590275 |
| Class A/1 (Rhodopsin-like receptors) | GHSR | GH-releasing peptide receptor, Growth hormone secretagogue receptor type 1 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.542668 |
| Class A/1 (Rhodopsin-like receptors) | TACR3 | Neuromedin-K receptor, NK-3 receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.602621 |
| Class A/1 (Rhodopsin-like receptors) | LTB4R2 | Leukotriene B4 receptor 2 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.627297 |
| Class A/1 (Rhodopsin-like receptors) | NMUR2 | Neuromedin U receptor 2, Neuromedin U receptor type 2, Neuromedin-U receptor 2 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.612605 |
| Class A/1 (Rhodopsin-like receptors) | MCHR2 | Melanin concentrating hormone receptor 2, Melanin concentrating hormone receptor 2a, Melanin-concentrating hormone receptor 2 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.520996 |
| Class A/1 (Rhodopsin-like receptors) | CCR2 | C-C chemokine receptor type 2, C-C chemokine receptor type 2 isoform B, C-C motif chemokine receptor 2, Chemokine, G_PROTEIN_RECEP_F1_2 domain-containing protein | 8.546785 | 0.366878 | 4 | 0.504858 | 0.562183 |
| Class A/1 (Rhodopsin-like receptors) | KNG1 | Bradykinin, Kininogen 1, Kininogen-1, Kininogen-1 [Cleaved into: Kininogen-1 heavy chain; Bradykinin; Kininogen-1 light chain], Uncharacterized protein | 8.546785 | 0.366878 | 4 | 0.504858 | 0.566192 |
| Class A/1 (Rhodopsin-like receptors) | BDKRB2 | B1 bradykinin receptor, B2 bradykinin receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.518514 |
| Class A/1 (Rhodopsin-like receptors) | OPRL1 | Kappa-type 3 opioid receptor, Nociceptin receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.654503 |
| Class A/1 (Rhodopsin-like receptors) | PTAFR | Platelet-activating factor receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.612734 |
| Class A/1 (Rhodopsin-like receptors) | MC1R | Melanocyte-stimulating hormone receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.526117 |
| Class A/1 (Rhodopsin-like receptors) | UTS2R | G_PROTEIN_RECEP_F1_2 domain-containing protein, Urotensin II receptor, Urotensin-2 receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.594282 |
| Class A/1 (Rhodopsin-like receptors) | SSTR3 | Somatostatin receptor 3, Somatostatin receptor type 3 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.543809 |
| Class A/1 (Rhodopsin-like receptors) | HTR7 | 5-hydroxytryptamine receptor 7, 5-hydroxytryptamine receptor 7 transcript variant 1 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.601446 |
| Class A/1 (Rhodopsin-like receptors) | NPY5R | Neuropeptide Y receptor type 5, NPY-Y5 receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.567119 |
| Class A/1 (Rhodopsin-like receptors) | NPY4R | Neuropeptide Y receptor 4, Neuropeptide Y receptor type 1, Neuropeptide Y receptor type 4 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.586341 |
| Class A/1 (Rhodopsin-like receptors) | AVPR1B | G_PROTEIN_RECEP_F1_2 domain-containing protein, Vasopressin V1b receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.514485 |
| Class A/1 (Rhodopsin-like receptors) | AVPR2 | AVPR V2, Vasopressin V2 receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.569629 |
| Class A/1 (Rhodopsin-like receptors) | NPY2R | Neuropeptide Y receptor type 2, NPY-Y2 receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.534429 |
| Class A/1 (Rhodopsin-like receptors) | ADRA2C | Adrenoceptor alpha 2C, Alpha-2C adrenergic receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.528655 |
| Class A/1 (Rhodopsin-like receptors) | FPR1 | FK506-binding protein 1, fMet-Leu-Phe receptor, Formyl peptide receptor 1 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.511829 |
| Class A/1 (Rhodopsin-like receptors) | HRH1 | Histamine H1 receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.518086 |
| Class A/1 (Rhodopsin-like receptors) | CHRM1 | Muscarinic acetylcholine receptor, Muscarinic acetylcholine receptor M1 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.532293 |
| Class A/1 (Rhodopsin-like receptors) | GPBAR1 | G protein-coupled bile acid receptor 1, G-protein coupled bile acid receptor 1 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.520207 |
| Class A/1 (Rhodopsin-like receptors) | S1PR1 | Sphingosine 1-phosphate receptor 1 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.519942 |
| Class A/1 (Rhodopsin-like receptors) | NPY1R | Neuropeptide Y receptor type 1 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.527768 |
| Class A/1 (Rhodopsin-like receptors) | ADRA2B | Adrenoceptor alpha 2B, Alpha-2B adrenergic receptor, G_PROTEIN_RECEP_F1_2 domain-containing protein | 8.546785 | 0.366878 | 4 | 0.504858 | 0.524078 |
| Class A/1 (Rhodopsin-like receptors) | F2R | CWC22 homolog, spliceosome-associated protein, Proteinase-activated receptor 1 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.58059 |
| Class A/1 (Rhodopsin-like receptors) | ADRA2A | Adrenoceptor alpha 2A, Alpha-2A adrenergic receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.530746 |
| Class A/1 (Rhodopsin-like receptors) | GNRHR | Gonadotropin-releasing hormone receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.689059 |
| Class A/1 (Rhodopsin-like receptors) | DRD5 | D(1B) dopamine receptor, Dopamine receptor D5, G_PROTEIN_RECEP_F1_2 domain-containing protein | 8.546785 | 0.366878 | 4 | 0.504858 | 0.551596 |
| Class A/1 (Rhodopsin-like receptors) | HCRTR1 | Hypocretin receptor 1, Orexin receptor type 1 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.541349 |
| Class A/1 (Rhodopsin-like receptors) | PTGER3 | Prostaglandin E2 receptor EP3 subtype | 8.546785 | 0.366878 | 4 | 0.504858 | 0.513867 |
| Class A/1 (Rhodopsin-like receptors) | AGTR1B | Angiotensin II receptor, type 1b | 8.546785 | 0.366878 | 4 | 0.504858 | 0.614033 |
| Class A/1 (Rhodopsin-like receptors) | APLN | Apelin, APJ endogenous ligand | 8.546785 | 0.366878 | 4 | 0.504858 | 0.502238 |
| Class A/1 (Rhodopsin-like receptors) | CCR3 | C-C chemokine receptor type 3, C-C motif chemokine receptor 3, Probable C-C chemokine receptor type 3 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.617373 |
| Class A/1 (Rhodopsin-like receptors) | APLNR | Apelin receptor, Apelin receptor 1, APLNR protein | 8.546785 | 0.366878 | 4 | 0.504858 | 0.600344 |
| Class A/1 (Rhodopsin-like receptors) | AGTR1 | Agtr1.2 protein, Angiotensin II receptor type 1, Type-1 angiotensin II receptor, Type-1A angiotensin II receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.648107 |
| Class A/1 (Rhodopsin-like receptors) | MC5R | Melanocortin receptor 5, mRNA cap guanine-N7 methyltransferase | 8.546785 | 0.366878 | 4 | 0.504858 | 0.501364 |
| Class A/1 (Rhodopsin-like receptors) | CNR1 | Cannabinoid receptor, Cannabinoid receptor 1 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.618554 |
| Class A/1 (Rhodopsin-like receptors) | AVPR1A | Arginine vasopressin receptor 1A, G_PROTEIN_RECEP_F1_2 domain-containing protein, Vasopressin V1a receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.54747 |
| Class A/1 (Rhodopsin-like receptors) | MC4R | Melanocortin receptor 4 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.598533 |
| Class A/1 (Rhodopsin-like receptors) | ACKR3 | Atypical chemokine receptor 3 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.521317 |
| Class A/1 (Rhodopsin-like receptors) | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 8.546785 | 0.366878 | 4 | 0.504858 | 0.515416 |
| Class A/1 (Rhodopsin-like receptors) | HRH3 | G_PROTEIN_RECEP_F1_2 domain-containing protein, Histamine H3 receptor, Histamine receptor H3 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.608088 |
| Class A/1 (Rhodopsin-like receptors) | LTB4R | Leukotriene B4 receptor, Leukotriene B4 receptor 1 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.604494 |
| Class A/1 (Rhodopsin-like receptors) | CNR2 | Cannabinod receptor, Cannabinoid receptor 2, G_PROTEIN_RECEP_F1_2 domain-containing protein | 8.546785 | 0.366878 | 4 | 0.504858 | 0.575385 |
| Class A/1 (Rhodopsin-like receptors) | OPRM1 | Mu-type opioid receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.614279 |
| Class A/1 (Rhodopsin-like receptors) | DRD4 | D(4) dopamine receptor, Dopamine receptor D4, DRD4 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.571927 |
| Class A/1 (Rhodopsin-like receptors) | BDKRB1 | B1 bradykinin receptor, Bradykinin receptor B1 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.547577 |
| Class A/1 (Rhodopsin-like receptors) | SSTR1 | Somatostatin receptor type 1 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.557663 |
| Class A/1 (Rhodopsin-like receptors) | KISS1R | G protein-coupled receptor 54, KiSS-1 receptor, KISS1 receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.63445 |
| Class A/1 (Rhodopsin-like receptors) | ADRA1D | Alpha-1D adrenergic receptor, Procollagen C-endopeptidase enhancer | 8.546785 | 0.366878 | 4 | 0.504858 | 0.602889 |
| Class A/1 (Rhodopsin-like receptors) | OPRD1 | Delta-type opioid receptor, G_PROTEIN_RECEP_F1_2 domain-containing protein, Opioid receptor delta 1 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.598587 |
| Class A/1 (Rhodopsin-like receptors) | NMUR1 | Neuromedin U receptor 1, Neuromedin U receptor type 1, Neuromedin-U receptor 1 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.62859 |
| Class A/1 (Rhodopsin-like receptors) | DRD1 | D(1A) dopamine receptor, Dopamine receptor D1, G_PROTEIN_RECEP_F1_2 domain-containing protein | 8.546785 | 0.366878 | 4 | 0.504858 | 0.557448 |
| Class A/1 (Rhodopsin-like receptors) | CYSLTR2 | CYSLTR2 protein, Cysteinyl leukotriene receptor 2, G_PROTEIN_RECEP_F1_2 domain-containing protein | 8.546785 | 0.366878 | 4 | 0.504858 | 0.566351 |
| Class A/1 (Rhodopsin-like receptors) | CXCR3 | C-X-C chemokine receptor type 3 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.537539 |
| Class A/1 (Rhodopsin-like receptors) | ADRA1A | Alpha-1A adrenergic receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.61112 |
| Class A/1 (Rhodopsin-like receptors) | TACR1 | NK-1 receptor, Substance-P receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.738091 |
| Class A/1 (Rhodopsin-like receptors) | TACR2 | NK-2 receptor, Substance-K receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.601846 |
| Class A/1 (Rhodopsin-like receptors) | SSTR4 | G_PROTEIN_RECEP_F1_2 domain-containing protein, Somatostatin receptor type 4 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.53719 |
| Class A/1 (Rhodopsin-like receptors) | CYSLTR1 | Cysteinyl leukotriene receptor 1 | 8.546785 | 0.366878 | 4 | 0.504858 | 0.552802 |
| Class A/1 (Rhodopsin-like receptors) | OXTR | Isotocin receptor-like 2, Oxytocin receptor, Oxytocin-like receptor | 8.546785 | 0.366878 | 4 | 0.504858 | 0.526619 |
| CLEC7A (Dectin-1) signaling | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 9.463749 | 0.575113 | 4 | 0.656281 | 0.594807 |
| CLEC7A (Dectin-1) signaling | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.577481 |
| CLEC7A (Dectin-1) signaling | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.54735 |
| CLEC7A (Dectin-1) signaling | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.515416 |
| CLEC7A (Dectin-1) signaling | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.572294 |
| CLEC7A (Dectin-1) signaling | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.463749 | 0.575113 | 4 | 0.656281 | 0.579609 |
| CLEC7A (Dectin-1) signaling | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.547953 |
| CLEC7A (Dectin-1) signaling | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.515416 |
| CLEC7A (Dectin-1) signaling | PRKCD | Protein kinase C, Protein kinase C delta type | 9.463749 | 0.575113 | 4 | 0.656281 | 0.520528 |
| CLEC7A (Dectin-1) signaling | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.54735 |
| CLEC7A (Dectin-1) signaling | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.565601 |
| CLEC7A (Dectin-1) signaling | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.54735 |
| CLEC7A (Dectin-1) signaling | SYK | Tyrosine-protein kinase, Tyrosine-protein kinase SYK | 9.463749 | 0.575113 | 4 | 0.656281 | 0.557587 |
| CLEC7A (Dectin-1) signaling | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.463749 | 0.575113 | 4 | 0.656281 | 0.515416 |
| CLEC7A (Dectin-1) signaling | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.54735 |
| CLEC7A (Dectin-1) signaling | IKBKB | I-kappa-B kinase, Inhibitor of nuclear factor kappa-B kinase subunit beta | 9.463749 | 0.575113 | 4 | 0.656281 | 0.518113 |
| CLEC7A (Dectin-1) signaling | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.515416 |
| CLEC7A (Dectin-1) signaling | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.463749 | 0.575113 | 4 | 0.656281 | 0.515416 |
| CLEC7A (Dectin-1) signaling | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.515416 |
| CLEC7A (Dectin-1) signaling | UBE2V1 | Ubiquitin conjugating enzyme E2 V1, Ubiquitin-conjugating enzyme E2 variant 1 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.593364 |
| CLEC7A (Dectin-1) signaling | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.515416 |
| CLEC7A (Dectin-1) signaling | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.58858 |
| CLEC7A (Dectin-1) signaling | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.54735 |
| CLEC7A (Dectin-1) signaling | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.515416 |
| CLEC7A (Dectin-1) signaling | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.54735 |
| CLEC7A (Dectin-1) signaling | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.515416 |
| CLEC7A (Dectin-1) signaling | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.463749 | 0.575113 | 4 | 0.656281 | 0.568645 |
| CLEC7A (Dectin-1) signaling | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.515416 |
| CLEC7A (Dectin-1) signaling | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.515416 |
| CLEC7A (Dectin-1) signaling | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.573417 |
| CLEC7A (Dectin-1) signaling | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.5197 |
| CLEC7A (Dectin-1) signaling | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.54735 |
| CLEC7A (Dectin-1) signaling | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.515416 |
| CLEC7A (Dectin-1) signaling | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.54735 |
| CLEC7A (Dectin-1) signaling | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.546881 |
| CLEC7A (Dectin-1) signaling | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.577847 |
| CLEC7A (Dectin-1) signaling | MAP3K14 | Mitogen-activated protein kinase kinase kinase 14 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.556123 |
| CLEC7A (Dectin-1) signaling | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.54735 |
| CLEC7A (Dectin-1) signaling | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.515416 |
| CLEC7A (Dectin-1) signaling | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.515416 |
| CLEC7A (Dectin-1) signaling | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.54735 |
| CLEC7A (Dectin-1) signaling | MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.519182 |
| CLEC7A (Dectin-1) signaling | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.57244 |
| CLEC7A (Dectin-1) signaling | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.463749 | 0.575113 | 4 | 0.656281 | 0.515416 |
| Constitutive Signaling by Aberrant PI3K in Cancer | ERBB4 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-4 | 9.045328 | 0.504623 | 4 | 0.59945 | 0.550399 |
| Constitutive Signaling by Aberrant PI3K in Cancer | FLT3 | Receptor protein-tyrosine kinase, Receptor-type tyrosine-protein kinase FLT3 | 9.045328 | 0.504623 | 4 | 0.59945 | 0.529563 |
| Constitutive Signaling by Aberrant PI3K in Cancer | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 9.045328 | 0.504623 | 4 | 0.59945 | 0.646924 |
| Constitutive Signaling by Aberrant PI3K in Cancer | FGFR1 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 1 | 9.045328 | 0.504623 | 4 | 0.59945 | 0.55821 |
| Constitutive Signaling by Aberrant PI3K in Cancer | PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.045328 | 0.504623 | 4 | 0.59945 | 0.589385 |
| Constitutive Signaling by Aberrant PI3K in Cancer | ERBB2 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-2 | 9.045328 | 0.504623 | 4 | 0.59945 | 0.589497 |
| Constitutive Signaling by Aberrant PI3K in Cancer | LCK | Proto-oncogene tyrosine-protein kinase LCK, Tyrosine-protein kinase, Tyrosine-protein kinase Lck | 9.045328 | 0.504623 | 4 | 0.59945 | 0.543278 |
| Constitutive Signaling by Aberrant PI3K in Cancer | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 9.045328 | 0.504623 | 4 | 0.59945 | 0.594807 |
| Constitutive Signaling by Aberrant PI3K in Cancer | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.045328 | 0.504623 | 4 | 0.59945 | 0.660919 |
| Constitutive Signaling by Aberrant PI3K in Cancer | PIK3CD | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.045328 | 0.504623 | 4 | 0.59945 | 0.632077 |
| Constitutive Signaling by Aberrant PI3K in Cancer | PDGFRB | Platelet-derived growth factor receptor beta, Receptor protein-tyrosine kinase | 9.045328 | 0.504623 | 4 | 0.59945 | 0.550695 |
| Constitutive Signaling by Aberrant PI3K in Cancer | FGF1 | Fibroblast growth factor 1, Multifunctional fusion protein [Includes: Fibroblast growth factor 1, Putative fibroblast growth factor 1 | 9.045328 | 0.504623 | 4 | 0.59945 | 0.509844 |
| Constitutive Signaling by Aberrant PI3K in Cancer | FGFR2 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 2 | 9.045328 | 0.504623 | 4 | 0.59945 | 0.577012 |
| Constitutive Signaling by Aberrant PI3K in Cancer | EGFR | Epidermal growth factor receptor, Receptor protein-tyrosine kinase | 9.045328 | 0.504623 | 4 | 0.59945 | 0.625817 |
| Constitutive Signaling by Aberrant PI3K in Cancer | MET | Hepatocyte growth factor receptor, HGF/SF receptor | 9.045328 | 0.504623 | 4 | 0.59945 | 0.5914 |
| Constitutive Signaling by Aberrant PI3K in Cancer | FGFR4 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 4 | 9.045328 | 0.504623 | 4 | 0.59945 | 0.555253 |
| Constitutive Signaling by Aberrant PI3K in Cancer | ESR1 | Estrogen receptor | 9.045328 | 0.504623 | 4 | 0.59945 | 0.561602 |
| Constitutive Signaling by Aberrant PI3K in Cancer | KIT | Mast/stem cell growth factor receptor, Mast/stem cell growth factor receptor Kit | 9.045328 | 0.504623 | 4 | 0.59945 | 0.562353 |
| Constitutive Signaling by Aberrant PI3K in Cancer | ESR2 | Estrogen receptor 2, Estrogen receptor beta | 9.045328 | 0.504623 | 4 | 0.59945 | 0.53484 |
| Constitutive Signaling by Aberrant PI3K in Cancer | PDGFRA | Platelet-derived growth factor receptor alpha | 9.045328 | 0.504623 | 4 | 0.59945 | 0.532138 |
| Constitutive Signaling by Aberrant PI3K in Cancer | FGFR3 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 3 | 9.045328 | 0.504623 | 4 | 0.59945 | 0.560785 |
| Constitutive Signaling by Aberrant PI3K in Cancer | ERBB3 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-3 | 9.045328 | 0.504623 | 4 | 0.59945 | 0.551662 |
| Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants | HDAC9 | Histone deacetylase, Histone deacetylase 9 | 8.797722 | 0.554377 | 4 | 0.611554 | 0.525623 |
| Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants | NOTCH1 | Neurogenic locus notch homolog protein 1 | 8.797722 | 0.554377 | 4 | 0.611554 | 0.591875 |
| Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants | HDAC2 | Histone deacetylase 2 | 8.797722 | 0.554377 | 4 | 0.611554 | 0.52963 |
| Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants | APH1A | Anterior pharynx defective 1 homolog A, Anterior pharynx defective 1a homolog, Aph-1 homolog A, gamma-secretase subunit, Gamma-secretase subunit APH-1A, Uncharacterized protein | 8.797722 | 0.554377 | 4 | 0.611554 | 0.60749 |
| Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants | HDAC6 | Histone deacetylase 6, Histone deacetylase 6, isoform G | 8.797722 | 0.554377 | 4 | 0.611554 | 0.531859 |
| Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants | NCSTN | Nicastrin | 8.797722 | 0.554377 | 4 | 0.611554 | 0.60749 |
| Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants | HDAC8 | Histone deacetylase, Histone deacetylase 8, WD repeat and SOCS box containing 2 | 8.797722 | 0.554377 | 4 | 0.611554 | 0.535733 |
| Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants | HDAC1 | Histone deacetylase 1, Histone deacetylase HDAC1 | 8.797722 | 0.554377 | 4 | 0.611554 | 0.540105 |
| Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 8.797722 | 0.554377 | 4 | 0.611554 | 0.579609 |
| Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants | HDAC4 | Histone deacetylase, Histone deacetylase 4 | 8.797722 | 0.554377 | 4 | 0.611554 | 0.528897 |
| Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants | PSENEN | Gamma-secretase subunit PEN-2 | 8.797722 | 0.554377 | 4 | 0.611554 | 0.60725 |
| Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants | CCNC | Cyclin-C | 8.797722 | 0.554377 | 4 | 0.611554 | 0.503295 |
| Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants | PSEN1 | Presenilin, Presenilin-1 | 8.797722 | 0.554377 | 4 | 0.611554 | 0.611222 |
| Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants | APH1B | Aph-1 homolog B, gamma-secretase subunit, APH-1B', Gamma-secretase subunit APH-1B, Uncharacterized protein | 8.797722 | 0.554377 | 4 | 0.611554 | 0.60749 |
| Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants | HDAC10 | Histone deacetylase 10, Polyamine deacetylase HDAC10 | 8.797722 | 0.554377 | 4 | 0.611554 | 0.525881 |
| Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants | HDAC3 | Histone deacetylase, Histone deacetylase 3 | 8.797722 | 0.554377 | 4 | 0.611554 | 0.528637 |
| Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants | HDAC7 | Hist_deacetyl domain-containing protein, Histone deacetylase, Histone deacetylase 7 | 8.797722 | 0.554377 | 4 | 0.611554 | 0.522826 |
| Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants | RBPJ | Recombination signal binding protein for immunoglobulin kappa J region, Recombination signal-binding protein for immunoglobulin kappa J region, Recombining binding protein suppressor of hairless, Uncharacterized protein | 8.797722 | 0.554377 | 4 | 0.611554 | 0.647854 |
| Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants | PSEN2 | Presenilin, Presenilin-2 | 8.797722 | 0.554377 | 4 | 0.611554 | 0.607393 |
| Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants | HDAC11 | Hdac11 protein, Hist_deacetyl domain-containing protein, Histone deacetylase 11, Histone deacetylase 11, isoform A | 8.797722 | 0.554377 | 4 | 0.611554 | 0.524904 |
| Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants | HDAC5 | Histone deacetylase, Histone deacetylase 5 | 8.797722 | 0.554377 | 4 | 0.611554 | 0.523224 |
| Constitutive Signaling by NOTCH1 PEST Domain Mutants | HDAC4 | Histone deacetylase, Histone deacetylase 4 | 8.797722 | 0.554377 | 4 | 0.611554 | 0.528897 |
| Constitutive Signaling by NOTCH1 PEST Domain Mutants | HDAC1 | Histone deacetylase 1, Histone deacetylase HDAC1 | 8.797722 | 0.554377 | 4 | 0.611554 | 0.540105 |
| Constitutive Signaling by NOTCH1 PEST Domain Mutants | HDAC3 | Histone deacetylase, Histone deacetylase 3 | 8.797722 | 0.554377 | 4 | 0.611554 | 0.528637 |
| Constitutive Signaling by NOTCH1 PEST Domain Mutants | HDAC11 | Hdac11 protein, Hist_deacetyl domain-containing protein, Histone deacetylase 11, Histone deacetylase 11, isoform A | 8.797722 | 0.554377 | 4 | 0.611554 | 0.524904 |
| Constitutive Signaling by NOTCH1 PEST Domain Mutants | PSENEN | Gamma-secretase subunit PEN-2 | 8.797722 | 0.554377 | 4 | 0.611554 | 0.60725 |
| Constitutive Signaling by NOTCH1 PEST Domain Mutants | NCSTN | Nicastrin | 8.797722 | 0.554377 | 4 | 0.611554 | 0.60749 |
| Constitutive Signaling by NOTCH1 PEST Domain Mutants | HDAC6 | Histone deacetylase 6, Histone deacetylase 6, isoform G | 8.797722 | 0.554377 | 4 | 0.611554 | 0.531859 |
| Constitutive Signaling by NOTCH1 PEST Domain Mutants | HDAC5 | Histone deacetylase, Histone deacetylase 5 | 8.797722 | 0.554377 | 4 | 0.611554 | 0.523224 |
| Constitutive Signaling by NOTCH1 PEST Domain Mutants | CCNC | Cyclin-C | 8.797722 | 0.554377 | 4 | 0.611554 | 0.503295 |
| Constitutive Signaling by NOTCH1 PEST Domain Mutants | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 8.797722 | 0.554377 | 4 | 0.611554 | 0.579609 |
| Constitutive Signaling by NOTCH1 PEST Domain Mutants | HDAC2 | Histone deacetylase 2 | 8.797722 | 0.554377 | 4 | 0.611554 | 0.52963 |
| Constitutive Signaling by NOTCH1 PEST Domain Mutants | PSEN2 | Presenilin, Presenilin-2 | 8.797722 | 0.554377 | 4 | 0.611554 | 0.607393 |
| Constitutive Signaling by NOTCH1 PEST Domain Mutants | APH1B | Aph-1 homolog B, gamma-secretase subunit, APH-1B', Gamma-secretase subunit APH-1B, Uncharacterized protein | 8.797722 | 0.554377 | 4 | 0.611554 | 0.60749 |
| Constitutive Signaling by NOTCH1 PEST Domain Mutants | RBPJ | Recombination signal binding protein for immunoglobulin kappa J region, Recombination signal-binding protein for immunoglobulin kappa J region, Recombining binding protein suppressor of hairless, Uncharacterized protein | 8.797722 | 0.554377 | 4 | 0.611554 | 0.647854 |
| Constitutive Signaling by NOTCH1 PEST Domain Mutants | HDAC8 | Histone deacetylase, Histone deacetylase 8, WD repeat and SOCS box containing 2 | 8.797722 | 0.554377 | 4 | 0.611554 | 0.535733 |
| Constitutive Signaling by NOTCH1 PEST Domain Mutants | HDAC10 | Histone deacetylase 10, Polyamine deacetylase HDAC10 | 8.797722 | 0.554377 | 4 | 0.611554 | 0.525881 |
| Constitutive Signaling by NOTCH1 PEST Domain Mutants | HDAC9 | Histone deacetylase, Histone deacetylase 9 | 8.797722 | 0.554377 | 4 | 0.611554 | 0.525623 |
| Constitutive Signaling by NOTCH1 PEST Domain Mutants | PSEN1 | Presenilin, Presenilin-1 | 8.797722 | 0.554377 | 4 | 0.611554 | 0.611222 |
| Constitutive Signaling by NOTCH1 PEST Domain Mutants | NOTCH1 | Neurogenic locus notch homolog protein 1 | 8.797722 | 0.554377 | 4 | 0.611554 | 0.591875 |
| Constitutive Signaling by NOTCH1 PEST Domain Mutants | APH1A | Anterior pharynx defective 1 homolog A, Anterior pharynx defective 1a homolog, Aph-1 homolog A, gamma-secretase subunit, Gamma-secretase subunit APH-1A, Uncharacterized protein | 8.797722 | 0.554377 | 4 | 0.611554 | 0.60749 |
| Constitutive Signaling by NOTCH1 PEST Domain Mutants | HDAC7 | Hist_deacetyl domain-containing protein, Histone deacetylase, Histone deacetylase 7 | 8.797722 | 0.554377 | 4 | 0.611554 | 0.522826 |
| COPI-independent Golgi-to-ER retrograde traffic | PLA2G4A | Cytosolic phospholipase A2, Phospholipase A2 | 8.525271 | 0.556862 | 4 | 0.59874 | 0.503807 |
| COPI-independent Golgi-to-ER retrograde traffic | PAFAH1B2 | Platelet activating factor acetylhydrolase 1b catalytic subunit 2, Platelet-activating factor acetylhydrolase 1b, catalytic subunit 2, Platelet-activating factor acetylhydrolase IB subunit alpha2 | 8.525271 | 0.556862 | 4 | 0.59874 | 0.555559 |
| Costimulation by the CD28 family | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 9.397792 | 0.446686 | 3.030316 | 0.44596 | 0.594807 |
| Costimulation by the CD28 family | MTOR | Nucleoprotein TPR, Serine/threonine-protein kinase mTOR, Serine/threonine-protein kinase TOR | 9.397792 | 0.446686 | 3.030316 | 0.44596 | 0.619026 |
| Costimulation by the CD28 family | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 9.397792 | 0.446686 | 3.030316 | 0.44596 | 0.552041 |
| Costimulation by the CD28 family | MAP3K14 | Mitogen-activated protein kinase kinase kinase 14 | 9.397792 | 0.446686 | 3.030316 | 0.44596 | 0.556123 |
| Costimulation by the CD28 family | YES1 | Tyrosine-protein kinase, Tyrosine-protein kinase yes, Tyrosine-protein kinase Yes | 9.397792 | 0.446686 | 3.030316 | 0.44596 | 0.506641 |
| Costimulation by the CD28 family | LCK | Proto-oncogene tyrosine-protein kinase LCK, Tyrosine-protein kinase, Tyrosine-protein kinase Lck | 9.397792 | 0.446686 | 3.030316 | 0.44596 | 0.543278 |
| Costimulation by the CD28 family | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 9.397792 | 0.446686 | 3.030316 | 0.44596 | 0.589663 |
| Costimulation by the CD28 family | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 9.397792 | 0.446686 | 3.030316 | 0.44596 | 0.646924 |
| Costimulation by the CD28 family | PPP2R5A | Serine/threonine protein phosphatase 2A regulatory subunit, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform | 9.397792 | 0.446686 | 3.030316 | 0.44596 | 0.559184 |
| Costimulation by the CD28 family | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.397792 | 0.446686 | 3.030316 | 0.44596 | 0.660919 |
| CREB1 phosphorylation through NMDA receptor-mediated activation of RAS signaling | GRIN1 | Glutamate receptor, Glutamate receptor ionotropic, NMDA 1 | 7.430249 | 0.551035 | 3.363099 | 0.45346 | 0.573676 |
| CREB1 phosphorylation through NMDA receptor-mediated activation of RAS signaling | GRIN2B | Glutamate receptor, Glutamate receptor ionotropic, NMDA 2B | 7.430249 | 0.551035 | 3.363099 | 0.45346 | 0.56443 |
| CREB1 phosphorylation through NMDA receptor-mediated activation of RAS signaling | GRIN2D | Glutamate receptor, Glutamate receptor ionotropic, NMDA 2D | 7.430249 | 0.551035 | 3.363099 | 0.45346 | 0.556318 |
| CREB1 phosphorylation through NMDA receptor-mediated activation of RAS signaling | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 7.430249 | 0.551035 | 3.363099 | 0.45346 | 0.541254 |
| CREB1 phosphorylation through NMDA receptor-mediated activation of RAS signaling | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 7.430249 | 0.551035 | 3.363099 | 0.45346 | 0.505988 |
| cRNA Synthesis | PB1 | Protein PB1-F2, RNA-directed RNA polymerase catalytic subunit | 6.606128 | 0.457982 | 0.508942 | ||
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.577847 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.515416 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.547953 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.54735 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.546881 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.565601 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.54735 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.515416 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.515416 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.54735 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.515416 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.572294 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.54735 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.54735 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.54735 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.515416 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.515416 |
| Cross-presentation of soluble exogenous antigens (endosomes) | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.515416 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.515416 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.515416 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.498314 | 0.794739 | 4 | 0.767877 | 0.568645 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.498314 | 0.794739 | 4 | 0.767877 | 0.515416 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.515416 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.54735 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.573417 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.58858 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.54735 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.5197 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.57244 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.54735 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.498314 | 0.794739 | 4 | 0.767877 | 0.515416 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.515416 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.515416 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.577481 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.54735 |
| Cross-presentation of soluble exogenous antigens (endosomes) | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.498314 | 0.794739 | 4 | 0.767877 | 0.515416 |
| Cyclin A:Cdk2-associated events at S phase entry | CCNA2 | Cyclin-A2 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.527977 |
| Cyclin A:Cdk2-associated events at S phase entry | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.589663 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.515416 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 8.174292 | 0.611324 | 4 | 0.607865 | 0.568645 |
| Cyclin A:Cdk2-associated events at S phase entry | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.515416 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.515416 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.515416 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.54735 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.57244 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.577847 |
| Cyclin A:Cdk2-associated events at S phase entry | CCNE2 | Cyclin E2, Cyclin N-terminal domain-containing protein, G1/S-specific cyclin-E2 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.503983 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.54735 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.547953 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.515416 |
| Cyclin A:Cdk2-associated events at S phase entry | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 8.174292 | 0.611324 | 4 | 0.607865 | 0.579609 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 8.174292 | 0.611324 | 4 | 0.607865 | 0.515416 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.5197 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.546881 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.54735 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.565601 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.515416 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.58858 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.572294 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.515416 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.515416 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.54735 |
| Cyclin A:Cdk2-associated events at S phase entry | CCND1 | Cyclin D1, G1/S-specific cyclin-D1 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.525654 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.515416 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.573417 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.515416 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.54735 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.515416 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.577481 |
| Cyclin A:Cdk2-associated events at S phase entry | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 8.174292 | 0.611324 | 4 | 0.607865 | 0.552041 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.54735 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.515416 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.54735 |
| Cyclin A:Cdk2-associated events at S phase entry | CCNE1 | Cyclin E1, Cyclin N-terminal domain-containing protein, G1/S-specific cyclin-E1 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.526741 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.54735 |
| Cyclin A:Cdk2-associated events at S phase entry | CDK2 | Cyclin dependent kinase 2, Cyclin-dependent kinase 2, Protein kinase domain-containing protein, Uncharacterized protein | 8.174292 | 0.611324 | 4 | 0.607865 | 0.532011 |
| Cyclin A:Cdk2-associated events at S phase entry | CCNA1 | Cyclin A1, Cyclin-A1, Cyclin-A2 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.506777 |
| Cyclin A:Cdk2-associated events at S phase entry | CDK1 | Cell division control protein 2, Cyclin dependent kinase 1, Cyclin-dependent kinase 1 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.514008 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 8.174292 | 0.611324 | 4 | 0.607865 | 0.515416 |
| Cyclin A:Cdk2-associated events at S phase entry | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 8.174292 | 0.611324 | 4 | 0.607865 | 0.515416 |
| Cyclin A:Cdk2-associated events at S phase entry | CDK4 | Cyclin dependent kinase 4, Cyclin-dependent kinase 4, Cyclin-dependent kinase 4, isoform A | 8.174292 | 0.611324 | 4 | 0.607865 | 0.511296 |
| Deactivation of the beta-catenin transactivating complex | YWHAZ | 14_3_3 domain-containing protein, 14-3-3 protein zeta, 14-3-3 protein zeta/delta, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta | 8.887697 | 0.600224 | 3.786034 | 0.604931 | 0.602005 |
| Deactivation of the beta-catenin transactivating complex | HDAC1 | Histone deacetylase 1, Histone deacetylase HDAC1 | 8.887697 | 0.600224 | 3.786034 | 0.604931 | 0.540105 |
| Deactivation of the beta-catenin transactivating complex | MEN1 | Actin like 6A, Menin | 8.887697 | 0.600224 | 3.786034 | 0.604931 | 0.517021 |
| Deactivation of the beta-catenin transactivating complex | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 8.887697 | 0.600224 | 3.786034 | 0.604931 | 0.589663 |
| Deactivation of the beta-catenin transactivating complex | CTNNB1 | Catenin, Catenin beta-1, Uncharacterized protein | 8.887697 | 0.600224 | 3.786034 | 0.604931 | 0.657916 |
| Deactivation of the beta-catenin transactivating complex | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 8.887697 | 0.600224 | 3.786034 | 0.604931 | 0.579609 |
| Deactivation of the beta-catenin transactivating complex | TCF7L2 | HMG box domain-containing protein, Transcription factor 7 like 2, Transcription factor 7-like 2 | 8.887697 | 0.600224 | 3.786034 | 0.604931 | 0.688556 |
| Deactivation of the beta-catenin transactivating complex | XIAP | E3 ubiquitin-protein ligase XIAP, RING-type domain-containing protein, X-linked inhibitor of apoptosis | 8.887697 | 0.600224 | 3.786034 | 0.604931 | 0.589894 |
| Deactivation of the beta-catenin transactivating complex | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 8.887697 | 0.600224 | 3.786034 | 0.604931 | 0.552041 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.515416 |
| Dectin-1 mediated noncanonical NF-kB signaling | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.515416 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.54735 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.54735 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.54735 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.515416 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.515416 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.57244 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.457649 | 0.740666 | 4 | 0.738743 | 0.515416 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.515416 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.54735 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.565601 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.5197 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.515416 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.54735 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.572294 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.515416 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.54735 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.577481 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.54735 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.58858 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.457649 | 0.740666 | 4 | 0.738743 | 0.568645 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.515416 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.54735 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.573417 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.515416 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.54735 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.547953 |
| Dectin-1 mediated noncanonical NF-kB signaling | MAP3K14 | Mitogen-activated protein kinase kinase kinase 14 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.556123 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.577847 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.515416 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.515416 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.54735 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.515416 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.457649 | 0.740666 | 4 | 0.738743 | 0.515416 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.546881 |
| Dectin-1 mediated noncanonical NF-kB signaling | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.457649 | 0.740666 | 4 | 0.738743 | 0.515416 |
| Dectin-1 mediated noncanonical NF-kB signaling | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.457649 | 0.740666 | 4 | 0.738743 | 0.579609 |
| Defective CFTR causes cystic fibrosis | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.515416 |
| Defective CFTR causes cystic fibrosis | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.515416 |
| Defective CFTR causes cystic fibrosis | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.57244 |
| Defective CFTR causes cystic fibrosis | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.515416 |
| Defective CFTR causes cystic fibrosis | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.515416 |
| Defective CFTR causes cystic fibrosis | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.503911 | 0.802874 | 4 | 0.772233 | 0.579609 |
| Defective CFTR causes cystic fibrosis | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.515416 |
| Defective CFTR causes cystic fibrosis | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.54735 |
| Defective CFTR causes cystic fibrosis | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.515416 |
| Defective CFTR causes cystic fibrosis | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.577481 |
| Defective CFTR causes cystic fibrosis | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.547953 |
| Defective CFTR causes cystic fibrosis | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.58858 |
| Defective CFTR causes cystic fibrosis | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.515416 |
| Defective CFTR causes cystic fibrosis | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.54735 |
| Defective CFTR causes cystic fibrosis | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.54735 |
| Defective CFTR causes cystic fibrosis | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.54735 |
| Defective CFTR causes cystic fibrosis | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.573417 |
| Defective CFTR causes cystic fibrosis | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.515416 |
| Defective CFTR causes cystic fibrosis | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.54735 |
| Defective CFTR causes cystic fibrosis | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.54735 |
| Defective CFTR causes cystic fibrosis | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.503911 | 0.802874 | 4 | 0.772233 | 0.515416 |
| Defective CFTR causes cystic fibrosis | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.5197 |
| Defective CFTR causes cystic fibrosis | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.577847 |
| Defective CFTR causes cystic fibrosis | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.54735 |
| Defective CFTR causes cystic fibrosis | VCP | 15S Mg(2+)-ATPase p97 subunit, Transitional endoplasmic reticulum ATPase | 9.503911 | 0.802874 | 4 | 0.772233 | 0.526633 |
| Defective CFTR causes cystic fibrosis | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.515416 |
| Defective CFTR causes cystic fibrosis | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.54735 |
| Defective CFTR causes cystic fibrosis | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.565601 |
| Defective CFTR causes cystic fibrosis | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.546881 |
| Defective CFTR causes cystic fibrosis | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.515416 |
| Defective CFTR causes cystic fibrosis | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.515416 |
| Defective CFTR causes cystic fibrosis | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.503911 | 0.802874 | 4 | 0.772233 | 0.515416 |
| Defective CFTR causes cystic fibrosis | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.572294 |
| Defective CFTR causes cystic fibrosis | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.503911 | 0.802874 | 4 | 0.772233 | 0.568645 |
| Defective CFTR causes cystic fibrosis | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.515416 |
| Defective CFTR causes cystic fibrosis | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.515416 |
| Defective CFTR causes cystic fibrosis | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.54735 |
| Defective CFTR causes cystic fibrosis | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.503911 | 0.802874 | 4 | 0.772233 | 0.54735 |
| Degradation of AXIN | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.515416 |
| Degradation of AXIN | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.57244 |
| Degradation of AXIN | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.515416 |
| Degradation of AXIN | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.515416 |
| Degradation of AXIN | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.547953 |
| Degradation of AXIN | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.572294 |
| Degradation of AXIN | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.515416 |
| Degradation of AXIN | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.54735 |
| Degradation of AXIN | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.54735 |
| Degradation of AXIN | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.515416 |
| Degradation of AXIN | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.515416 |
| Degradation of AXIN | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.546881 |
| Degradation of AXIN | TNKS | Poly [ADP-ribose] polymerase, Poly [ADP-ribose] polymerase tankyrase, Poly [ADP-ribose] polymerase tankyrase-1 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.506213 |
| Degradation of AXIN | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.462267 | 0.756078 | 4 | 0.746687 | 0.515416 |
| Degradation of AXIN | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.54735 |
| Degradation of AXIN | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.577481 |
| Degradation of AXIN | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.54735 |
| Degradation of AXIN | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.54735 |
| Degradation of AXIN | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.515416 |
| Degradation of AXIN | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.515416 |
| Degradation of AXIN | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.515416 |
| Degradation of AXIN | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.515416 |
| Degradation of AXIN | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.54735 |
| Degradation of AXIN | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.54735 |
| Degradation of AXIN | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.573417 |
| Degradation of AXIN | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.54735 |
| Degradation of AXIN | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.5197 |
| Degradation of AXIN | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.462267 | 0.756078 | 4 | 0.746687 | 0.579609 |
| Degradation of AXIN | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.462267 | 0.756078 | 4 | 0.746687 | 0.568645 |
| Degradation of AXIN | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.54735 |
| Degradation of AXIN | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.515416 |
| Degradation of AXIN | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.515416 |
| Degradation of AXIN | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.54735 |
| Degradation of AXIN | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.462267 | 0.756078 | 4 | 0.746687 | 0.515416 |
| Degradation of AXIN | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.577847 |
| Degradation of AXIN | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.565601 |
| Degradation of AXIN | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.58858 |
| Degradation of AXIN | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.462267 | 0.756078 | 4 | 0.746687 | 0.515416 |
| Degradation of beta-catenin by the destruction complex | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.54735 |
| Degradation of beta-catenin by the destruction complex | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.54735 |
| Degradation of beta-catenin by the destruction complex | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.54735 |
| Degradation of beta-catenin by the destruction complex | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.54735 |
| Degradation of beta-catenin by the destruction complex | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.515416 |
| Degradation of beta-catenin by the destruction complex | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.58858 |
| Degradation of beta-catenin by the destruction complex | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.54735 |
| Degradation of beta-catenin by the destruction complex | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.515416 |
| Degradation of beta-catenin by the destruction complex | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.490748 | 0.716563 | 4 | 0.728399 | 0.515416 |
| Degradation of beta-catenin by the destruction complex | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.515416 |
| Degradation of beta-catenin by the destruction complex | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.490748 | 0.716563 | 4 | 0.728399 | 0.568645 |
| Degradation of beta-catenin by the destruction complex | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.515416 |
| Degradation of beta-catenin by the destruction complex | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.5197 |
| Degradation of beta-catenin by the destruction complex | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.547953 |
| Degradation of beta-catenin by the destruction complex | HDAC1 | Histone deacetylase 1, Histone deacetylase HDAC1 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.540105 |
| Degradation of beta-catenin by the destruction complex | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.515416 |
| Degradation of beta-catenin by the destruction complex | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.515416 |
| Degradation of beta-catenin by the destruction complex | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.54735 |
| Degradation of beta-catenin by the destruction complex | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.572294 |
| Degradation of beta-catenin by the destruction complex | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.515416 |
| Degradation of beta-catenin by the destruction complex | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.515416 |
| Degradation of beta-catenin by the destruction complex | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.577847 |
| Degradation of beta-catenin by the destruction complex | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.54735 |
| Degradation of beta-catenin by the destruction complex | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.565601 |
| Degradation of beta-catenin by the destruction complex | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.515416 |
| Degradation of beta-catenin by the destruction complex | TCF7L2 | HMG box domain-containing protein, Transcription factor 7 like 2, Transcription factor 7-like 2 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.688556 |
| Degradation of beta-catenin by the destruction complex | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.54735 |
| Degradation of beta-catenin by the destruction complex | PPP2R5A | Serine/threonine protein phosphatase 2A regulatory subunit, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform | 9.490748 | 0.716563 | 4 | 0.728399 | 0.559184 |
| Degradation of beta-catenin by the destruction complex | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.577481 |
| Degradation of beta-catenin by the destruction complex | CTNNB1 | Catenin, Catenin beta-1, Uncharacterized protein | 9.490748 | 0.716563 | 4 | 0.728399 | 0.657916 |
| Degradation of beta-catenin by the destruction complex | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.490748 | 0.716563 | 4 | 0.728399 | 0.579609 |
| Degradation of beta-catenin by the destruction complex | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.515416 |
| Degradation of beta-catenin by the destruction complex | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.515416 |
| Degradation of beta-catenin by the destruction complex | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.54735 |
| Degradation of beta-catenin by the destruction complex | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.573417 |
| Degradation of beta-catenin by the destruction complex | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.57244 |
| Degradation of beta-catenin by the destruction complex | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.54735 |
| Degradation of beta-catenin by the destruction complex | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.546881 |
| Degradation of beta-catenin by the destruction complex | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.515416 |
| Degradation of beta-catenin by the destruction complex | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.490748 | 0.716563 | 4 | 0.728399 | 0.515416 |
| Degradation of beta-catenin by the destruction complex | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.490748 | 0.716563 | 4 | 0.728399 | 0.515416 |
| Degradation of DVL | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.577847 |
| Degradation of DVL | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.718012 | 0.789374 | 4 | 0.776529 | 0.515416 |
| Degradation of DVL | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.565601 |
| Degradation of DVL | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.54735 |
| Degradation of DVL | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.54735 |
| Degradation of DVL | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.515416 |
| Degradation of DVL | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.54735 |
| Degradation of DVL | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.718012 | 0.789374 | 4 | 0.776529 | 0.579609 |
| Degradation of DVL | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.515416 |
| Degradation of DVL | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.58858 |
| Degradation of DVL | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.515416 |
| Degradation of DVL | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.718012 | 0.789374 | 4 | 0.776529 | 0.515416 |
| Degradation of DVL | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.515416 |
| Degradation of DVL | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.515416 |
| Degradation of DVL | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.547953 |
| Degradation of DVL | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.718012 | 0.789374 | 4 | 0.776529 | 0.568645 |
| Degradation of DVL | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.54735 |
| Degradation of DVL | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.515416 |
| Degradation of DVL | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.573417 |
| Degradation of DVL | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.546881 |
| Degradation of DVL | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.577481 |
| Degradation of DVL | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.515416 |
| Degradation of DVL | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.515416 |
| Degradation of DVL | DVL2 | Dishevelled segment polarity protein 2, Segment polarity protein dishevelled homolog DVL-2, Uncharacterized protein | 9.718012 | 0.789374 | 4 | 0.776529 | 0.529242 |
| Degradation of DVL | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.54735 |
| Degradation of DVL | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.515416 |
| Degradation of DVL | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.515416 |
| Degradation of DVL | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.515416 |
| Degradation of DVL | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.54735 |
| Degradation of DVL | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.54735 |
| Degradation of DVL | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.54735 |
| Degradation of DVL | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.5197 |
| Degradation of DVL | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.515416 |
| Degradation of DVL | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.54735 |
| Degradation of DVL | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.54735 |
| Degradation of DVL | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.572294 |
| Degradation of DVL | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.515416 |
| Degradation of DVL | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.718012 | 0.789374 | 4 | 0.776529 | 0.57244 |
| Degradation of GLI1 by the proteasome | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.309412 | 0.725115 | 4 | 0.72332 | 0.579609 |
| Degradation of GLI1 by the proteasome | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.54735 |
| Degradation of GLI1 by the proteasome | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.515416 |
| Degradation of GLI1 by the proteasome | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.547953 |
| Degradation of GLI1 by the proteasome | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.54735 |
| Degradation of GLI1 by the proteasome | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.515416 |
| Degradation of GLI1 by the proteasome | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.546881 |
| Degradation of GLI1 by the proteasome | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.515416 |
| Degradation of GLI1 by the proteasome | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.54735 |
| Degradation of GLI1 by the proteasome | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.515416 |
| Degradation of GLI1 by the proteasome | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.515416 |
| Degradation of GLI1 by the proteasome | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.515416 |
| Degradation of GLI1 by the proteasome | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.309412 | 0.725115 | 4 | 0.72332 | 0.568645 |
| Degradation of GLI1 by the proteasome | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.58858 |
| Degradation of GLI1 by the proteasome | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.57244 |
| Degradation of GLI1 by the proteasome | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.515416 |
| Degradation of GLI1 by the proteasome | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.54735 |
| Degradation of GLI1 by the proteasome | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.573417 |
| Degradation of GLI1 by the proteasome | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.565601 |
| Degradation of GLI1 by the proteasome | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.309412 | 0.725115 | 4 | 0.72332 | 0.515416 |
| Degradation of GLI1 by the proteasome | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.515416 |
| Degradation of GLI1 by the proteasome | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.54735 |
| Degradation of GLI1 by the proteasome | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.54735 |
| Degradation of GLI1 by the proteasome | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.54735 |
| Degradation of GLI1 by the proteasome | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.577481 |
| Degradation of GLI1 by the proteasome | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.515416 |
| Degradation of GLI1 by the proteasome | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.54735 |
| Degradation of GLI1 by the proteasome | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.5197 |
| Degradation of GLI1 by the proteasome | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.54735 |
| Degradation of GLI1 by the proteasome | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.515416 |
| Degradation of GLI1 by the proteasome | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.515416 |
| Degradation of GLI1 by the proteasome | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.515416 |
| Degradation of GLI1 by the proteasome | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.572294 |
| Degradation of GLI1 by the proteasome | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.54735 |
| Degradation of GLI1 by the proteasome | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.577847 |
| Degradation of GLI1 by the proteasome | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.309412 | 0.725115 | 4 | 0.72332 | 0.515416 |
| Degradation of GLI1 by the proteasome | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.309412 | 0.725115 | 4 | 0.72332 | 0.515416 |
| Degradation of GLI2 by the proteasome | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.57244 |
| Degradation of GLI2 by the proteasome | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.54735 |
| Degradation of GLI2 by the proteasome | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.515416 |
| Degradation of GLI2 by the proteasome | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.281422 | 0.692966 | 4 | 0.705801 | 0.515416 |
| Degradation of GLI2 by the proteasome | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.565601 |
| Degradation of GLI2 by the proteasome | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.54735 |
| Degradation of GLI2 by the proteasome | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.54735 |
| Degradation of GLI2 by the proteasome | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.573417 |
| Degradation of GLI2 by the proteasome | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.54735 |
| Degradation of GLI2 by the proteasome | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.515416 |
| Degradation of GLI2 by the proteasome | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.54735 |
| Degradation of GLI2 by the proteasome | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.515416 |
| Degradation of GLI2 by the proteasome | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.515416 |
| Degradation of GLI2 by the proteasome | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.515416 |
| Degradation of GLI2 by the proteasome | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.54735 |
| Degradation of GLI2 by the proteasome | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.54735 |
| Degradation of GLI2 by the proteasome | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.515416 |
| Degradation of GLI2 by the proteasome | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.281422 | 0.692966 | 4 | 0.705801 | 0.579609 |
| Degradation of GLI2 by the proteasome | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.547953 |
| Degradation of GLI2 by the proteasome | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.54735 |
| Degradation of GLI2 by the proteasome | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.515416 |
| Degradation of GLI2 by the proteasome | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.5197 |
| Degradation of GLI2 by the proteasome | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.515416 |
| Degradation of GLI2 by the proteasome | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.572294 |
| Degradation of GLI2 by the proteasome | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.515416 |
| Degradation of GLI2 by the proteasome | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.515416 |
| Degradation of GLI2 by the proteasome | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.577847 |
| Degradation of GLI2 by the proteasome | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.546881 |
| Degradation of GLI2 by the proteasome | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.515416 |
| Degradation of GLI2 by the proteasome | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.54735 |
| Degradation of GLI2 by the proteasome | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.515416 |
| Degradation of GLI2 by the proteasome | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.54735 |
| Degradation of GLI2 by the proteasome | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.281422 | 0.692966 | 4 | 0.705801 | 0.515416 |
| Degradation of GLI2 by the proteasome | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.515416 |
| Degradation of GLI2 by the proteasome | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.281422 | 0.692966 | 4 | 0.705801 | 0.568645 |
| Degradation of GLI2 by the proteasome | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.58858 |
| Degradation of GLI2 by the proteasome | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.281422 | 0.692966 | 4 | 0.705801 | 0.577481 |
| Depolymerisation of the Nuclear Lamina | LMNA | Lamin A, Lamin A/C, Prelamin-A/C, Prelamin-A/C [Cleaved into: Lamin-A/C, Prelamin-A/C [Cleaved into: Lamin-A/C], Uncharacterized protein | 8.228557 | 0.840417 | 3.395434 | 0.615601 | 0.5357 |
| Depolymerisation of the Nuclear Lamina | CDK1 | Cell division control protein 2, Cyclin dependent kinase 1, Cyclin-dependent kinase 1 | 8.228557 | 0.840417 | 3.395434 | 0.615601 | 0.514008 |
| Depolymerisation of the Nuclear Lamina | PRKCB | Protein kinase C, Protein kinase C beta type | 8.228557 | 0.840417 | 3.395434 | 0.615601 | 0.520598 |
| Depolymerisation of the Nuclear Lamina | PRKCA | Protein kinase C, Protein kinase C alpha type, Protein kinase C, alpha | 8.228557 | 0.840417 | 3.395434 | 0.615601 | 0.52202 |
| Depolymerisation of the Nuclear Lamina | CCNB1 | Cyclin B1, G2/mitotic-specific cyclin-B1, Uncharacterized protein | 8.228557 | 0.840417 | 3.395434 | 0.615601 | 0.513205 |
| Dopamine receptors | DRD1 | D(1A) dopamine receptor, Dopamine receptor D1, G_PROTEIN_RECEP_F1_2 domain-containing protein | 9.710875 | 0.909789 | 4 | 0.836368 | 0.557448 |
| Dopamine receptors | DRD3 | D(3) dopamine receptor, Dopamine receptor D3 | 9.710875 | 0.909789 | 4 | 0.836368 | 0.649772 |
| Dopamine receptors | DRD4 | D(4) dopamine receptor, Dopamine receptor D4, DRD4 | 9.710875 | 0.909789 | 4 | 0.836368 | 0.571927 |
| Dopamine receptors | DRD2 | D(2) dopamine receptor, Dopamine receptor 2 protein transcript variant 1 | 9.710875 | 0.909789 | 4 | 0.836368 | 0.651493 |
| Dopamine receptors | DRD5 | D(1B) dopamine receptor, Dopamine receptor D5, G_PROTEIN_RECEP_F1_2 domain-containing protein | 9.710875 | 0.909789 | 4 | 0.836368 | 0.551596 |
| Downregulation of ERBB2 signaling | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.216076 | 0.564224 | 3.043383 | 0.485465 | 0.579609 |
| Downregulation of ERBB2 signaling | ERBB2 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-2 | 9.216076 | 0.564224 | 3.043383 | 0.485465 | 0.589497 |
| Downregulation of ERBB2 signaling | HSP90AA1 | Heat shock protein 90 alpha family class A member 1, Heat shock protein HSP 90-alpha | 9.216076 | 0.564224 | 3.043383 | 0.485465 | 0.624325 |
| Downregulation of ERBB2 signaling | ERBB3 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-3 | 9.216076 | 0.564224 | 3.043383 | 0.485465 | 0.551662 |
| Downregulation of ERBB2 signaling | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 9.216076 | 0.564224 | 3.043383 | 0.485465 | 0.552041 |
| Downregulation of ERBB2 signaling | ERBB4 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-4 | 9.216076 | 0.564224 | 3.043383 | 0.485465 | 0.550399 |
| Downregulation of ERBB2 signaling | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 9.216076 | 0.564224 | 3.043383 | 0.485465 | 0.589663 |
| Downregulation of ERBB2 signaling | EGFR | Epidermal growth factor receptor, Receptor protein-tyrosine kinase | 9.216076 | 0.564224 | 3.043383 | 0.485465 | 0.625817 |
| Downregulation of ERBB2:ERBB3 signaling | ERBB2 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-2 | 8.695019 | 0.715782 | 2.73146 | 0.469099 | 0.589497 |
| Downregulation of ERBB2:ERBB3 signaling | ERBB3 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-3 | 8.695019 | 0.715782 | 2.73146 | 0.469099 | 0.551662 |
| Downregulation of ERBB2:ERBB3 signaling | EGFR | Epidermal growth factor receptor, Receptor protein-tyrosine kinase | 8.695019 | 0.715782 | 2.73146 | 0.469099 | 0.625817 |
| Downregulation of ERBB2:ERBB3 signaling | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 8.695019 | 0.715782 | 2.73146 | 0.469099 | 0.552041 |
| Downregulation of ERBB2:ERBB3 signaling | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 8.695019 | 0.715782 | 2.73146 | 0.469099 | 0.579609 |
| Downregulation of ERBB2:ERBB3 signaling | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 8.695019 | 0.715782 | 2.73146 | 0.469099 | 0.589663 |
| Downregulation of ERBB4 signaling | EGFR | Epidermal growth factor receptor, Receptor protein-tyrosine kinase | 9.413304 | 0.800107 | 2.354979 | 0.451082 | 0.625817 |
| Downregulation of ERBB4 signaling | ERBB4 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-4 | 9.413304 | 0.800107 | 2.354979 | 0.451082 | 0.550399 |
| Downregulation of ERBB4 signaling | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 9.413304 | 0.800107 | 2.354979 | 0.451082 | 0.594807 |
| Downregulation of ERBB4 signaling | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.413304 | 0.800107 | 2.354979 | 0.451082 | 0.579609 |
| Downstream TCR signaling | PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.787616 | 0.632983 | 4 | 0.701924 | 0.589385 |
| Downstream TCR signaling | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.5197 |
| Downstream TCR signaling | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.515416 |
| Downstream TCR signaling | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.54735 |
| Downstream TCR signaling | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.57244 |
| Downstream TCR signaling | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 9.787616 | 0.632983 | 4 | 0.701924 | 0.646924 |
| Downstream TCR signaling | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.787616 | 0.632983 | 4 | 0.701924 | 0.515416 |
| Downstream TCR signaling | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.54735 |
| Downstream TCR signaling | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.515416 |
| Downstream TCR signaling | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.515416 |
| Downstream TCR signaling | PRKCQ | Protein kinase C, Protein kinase C theta type | 9.787616 | 0.632983 | 4 | 0.701924 | 0.523352 |
| Downstream TCR signaling | UBE2V1 | Ubiquitin conjugating enzyme E2 V1, Ubiquitin-conjugating enzyme E2 variant 1 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.593364 |
| Downstream TCR signaling | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.515416 |
| Downstream TCR signaling | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.547953 |
| Downstream TCR signaling | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.565601 |
| Downstream TCR signaling | IKBKB | I-kappa-B kinase, Inhibitor of nuclear factor kappa-B kinase subunit beta | 9.787616 | 0.632983 | 4 | 0.701924 | 0.518113 |
| Downstream TCR signaling | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.54735 |
| Downstream TCR signaling | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.54735 |
| Downstream TCR signaling | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.515416 |
| Downstream TCR signaling | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.515416 |
| Downstream TCR signaling | RIPK2 | Receptor-interacting serine-threonine kinase 2, Receptor-interacting serine/threonine-protein kinase 2 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.530344 |
| Downstream TCR signaling | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.787616 | 0.632983 | 4 | 0.701924 | 0.660919 |
| Downstream TCR signaling | MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.519182 |
| Downstream TCR signaling | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.54735 |
| Downstream TCR signaling | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.515416 |
| Downstream TCR signaling | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.515416 |
| Downstream TCR signaling | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.546881 |
| Downstream TCR signaling | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.515416 |
| Downstream TCR signaling | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.54735 |
| Downstream TCR signaling | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.515416 |
| Downstream TCR signaling | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.577847 |
| Downstream TCR signaling | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.54735 |
| Downstream TCR signaling | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.515416 |
| Downstream TCR signaling | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.58858 |
| Downstream TCR signaling | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.573417 |
| Downstream TCR signaling | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.787616 | 0.632983 | 4 | 0.701924 | 0.579609 |
| Downstream TCR signaling | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.515416 |
| Downstream TCR signaling | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.54735 |
| Downstream TCR signaling | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.572294 |
| Downstream TCR signaling | LCK | Proto-oncogene tyrosine-protein kinase LCK, Tyrosine-protein kinase, Tyrosine-protein kinase Lck | 9.787616 | 0.632983 | 4 | 0.701924 | 0.543278 |
| Downstream TCR signaling | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.787616 | 0.632983 | 4 | 0.701924 | 0.568645 |
| Downstream TCR signaling | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.577481 |
| Downstream TCR signaling | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.54735 |
| Downstream TCR signaling | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.515416 |
| Downstream TCR signaling | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.787616 | 0.632983 | 4 | 0.701924 | 0.54735 |
| Downstream TCR signaling | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.787616 | 0.632983 | 4 | 0.701924 | 0.515416 |
| EML4 and NUDC in mitotic spindle formation | PPP2R5A | Serine/threonine protein phosphatase 2A regulatory subunit, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform | 9.013415 | 0.52377 | 4 | 0.607377 | 0.559184 |
| EML4 and NUDC in mitotic spindle formation | KIF2C | Kinesin-like protein, Kinesin-like protein KIF2C | 9.013415 | 0.52377 | 4 | 0.607377 | 0.611637 |
| EML4 and NUDC in mitotic spindle formation | INCENP | Inner centromere protein, Inner centromere protein, isoform A, Uncharacterized protein | 9.013415 | 0.52377 | 4 | 0.607377 | 0.527883 |
| EML4 and NUDC in mitotic spindle formation | NEK7 | NIMA related kinase 7, Protein kinase domain-containing protein, Serine/threonine-protein kinase Nek7 | 9.013415 | 0.52377 | 4 | 0.607377 | 0.513171 |
| EML4 and NUDC in mitotic spindle formation | PLK1 | Serine/threonine-protein kinase PLK, Serine/threonine-protein kinase PLK1 | 9.013415 | 0.52377 | 4 | 0.607377 | 0.504187 |
| EML4 and NUDC in mitotic spindle formation | AURKB | Aurora kinase, Aurora kinase B | 9.013415 | 0.52377 | 4 | 0.607377 | 0.524412 |
| EML4 and NUDC in mitotic spindle formation | RPS27 | 40S ribosomal protein S27 | 9.013415 | 0.52377 | 4 | 0.607377 | 0.549907 |
| EML4 and NUDC in mitotic spindle formation | CENPE | Centromere protein E, Centromere-associated protein E, Kinesin motor domain-containing protein | 9.013415 | 0.52377 | 4 | 0.607377 | 0.564477 |
| EML4 and NUDC in mitotic spindle formation | PPP1CC | Serine/threonine-protein phosphatase, Serine/threonine-protein phosphatase PP1-gamma catalytic subunit | 9.013415 | 0.52377 | 4 | 0.607377 | 0.503303 |
| Energy dependent regulation of mTOR by LKB1-AMPK | RPTOR | Regulatory associated protein of MTOR complex 1, Regulatory associated protein of MTOR, complex 1, Regulatory-associated protein of mTOR, Regulatory-associated protein of MTOR, complex 1, WD_REPEATS_REGION domain-containing protein | 8.487652 | 0.575126 | 2.684305 | 0.406626 | 0.543357 |
| Energy dependent regulation of mTOR by LKB1-AMPK | MTOR | Nucleoprotein TPR, Serine/threonine-protein kinase mTOR, Serine/threonine-protein kinase TOR | 8.487652 | 0.575126 | 2.684305 | 0.406626 | 0.619026 |
| Energy dependent regulation of mTOR by LKB1-AMPK | PRKAB1 | 5'-AMP-activated protein kinase subunit beta-1, 5'AMP-activated protein kinase beta-1 non-catalytic subunit, Protein kinase AMP-activated non-catalytic subunit beta 1, Protein kinase, AMP-activated, beta 1 non-catalytic subunit | 8.487652 | 0.575126 | 2.684305 | 0.406626 | 0.500481 |
| Energy dependent regulation of mTOR by LKB1-AMPK | STK11 | Non-specific serine/threonine protein kinase, Serine/threonine-protein kinase STK11 | 8.487652 | 0.575126 | 2.684305 | 0.406626 | 0.504599 |
| Energy dependent regulation of mTOR by LKB1-AMPK | PRKAG1 | 5'-AMP-activated protein kinase subunit gamma-1, Protein kinase, AMP-activated, gamma 1 non-catalytic subunit | 8.487652 | 0.575126 | 2.684305 | 0.406626 | 0.515113 |
| Energy dependent regulation of mTOR by LKB1-AMPK | PRKAG2 | 5'-AMP-activated protein kinase subunit gamma-2, 5'-AMP-activated protein kinase subunit gamma-2 isoform X8, Protein kinase AMP-activated non-catalytic subunit gamma 2, Protein kinase, AMP-activated, gamma 2 non-catalytic subunit, Uncharacterized protein | 8.487652 | 0.575126 | 2.684305 | 0.406626 | 0.510193 |
| Entry of Influenza Virion into Host Cell via Endocytosis | PB1 | Protein PB1-F2, RNA-directed RNA polymerase catalytic subunit | 7.145023 | 0.505388 | 0.508942 | ||
| EPH-ephrin mediated repulsion of cells | EPHB6 | EPH receptor B6, Ephrin type-B receptor 6, Receptor protein-tyrosine kinase | 9.128163 | 0.534246 | 4 | 0.618535 | 0.500457 |
| EPH-ephrin mediated repulsion of cells | APH1B | Aph-1 homolog B, gamma-secretase subunit, APH-1B', Gamma-secretase subunit APH-1B, Uncharacterized protein | 9.128163 | 0.534246 | 4 | 0.618535 | 0.60749 |
| EPH-ephrin mediated repulsion of cells | APH1A | Anterior pharynx defective 1 homolog A, Anterior pharynx defective 1a homolog, Aph-1 homolog A, gamma-secretase subunit, Gamma-secretase subunit APH-1A, Uncharacterized protein | 9.128163 | 0.534246 | 4 | 0.618535 | 0.60749 |
| EPH-ephrin mediated repulsion of cells | EPHA3 | Ephrin type-A receptor 3, Receptor protein-tyrosine kinase | 9.128163 | 0.534246 | 4 | 0.618535 | 0.500178 |
| EPH-ephrin mediated repulsion of cells | EPHB3 | Ephrin type-B receptor 3, Receptor protein-tyrosine kinase | 9.128163 | 0.534246 | 4 | 0.618535 | 0.502956 |
| EPH-ephrin mediated repulsion of cells | EPHA2 | Ephrin type-A receptor 2, Receptor protein-tyrosine kinase | 9.128163 | 0.534246 | 4 | 0.618535 | 0.530164 |
| EPH-ephrin mediated repulsion of cells | PSENEN | Gamma-secretase subunit PEN-2 | 9.128163 | 0.534246 | 4 | 0.618535 | 0.60725 |
| EPH-ephrin mediated repulsion of cells | EPHA4 | Ephrin type-A receptor 4, Receptor protein-tyrosine kinase | 9.128163 | 0.534246 | 4 | 0.618535 | 0.503164 |
| EPH-ephrin mediated repulsion of cells | EPHB2 | Ephrin type-B receptor 2, Receptor protein-tyrosine kinase | 9.128163 | 0.534246 | 4 | 0.618535 | 0.508582 |
| EPH-ephrin mediated repulsion of cells | YES1 | Tyrosine-protein kinase, Tyrosine-protein kinase yes, Tyrosine-protein kinase Yes | 9.128163 | 0.534246 | 4 | 0.618535 | 0.506641 |
| EPH-ephrin mediated repulsion of cells | PSEN2 | Presenilin, Presenilin-2 | 9.128163 | 0.534246 | 4 | 0.618535 | 0.607393 |
| EPH-ephrin mediated repulsion of cells | NCSTN | Nicastrin | 9.128163 | 0.534246 | 4 | 0.618535 | 0.60749 |
| EPH-ephrin mediated repulsion of cells | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 9.128163 | 0.534246 | 4 | 0.618535 | 0.594807 |
| EPH-ephrin mediated repulsion of cells | PSEN1 | Presenilin, Presenilin-1 | 9.128163 | 0.534246 | 4 | 0.618535 | 0.611222 |
| EPH-ephrin mediated repulsion of cells | MMP2 | 72 kDa gelatinase, 72 kDa type IV collagenase, Matrix metalloproteinase-2 | 9.128163 | 0.534246 | 4 | 0.618535 | 0.512398 |
| EPH-ephrin mediated repulsion of cells | EPHB4 | Ephrin type-B receptor 4, Receptor protein-tyrosine kinase | 9.128163 | 0.534246 | 4 | 0.618535 | 0.527016 |
| EPH-ephrin mediated repulsion of cells | EPHB1 | Ephrin type-B receptor 1, Receptor protein-tyrosine kinase | 9.128163 | 0.534246 | 4 | 0.618535 | 0.509548 |
| EPH-ephrin mediated repulsion of cells | EPHA8 | EPH receptor A8, Ephrin type-A receptor 8, Receptor protein-tyrosine kinase | 9.128163 | 0.534246 | 4 | 0.618535 | 0.518651 |
| EPH-ephrin mediated repulsion of cells | EPHA5 | Ephrin type-A receptor 5, Receptor protein-tyrosine kinase | 9.128163 | 0.534246 | 4 | 0.618535 | 0.515572 |
| EPH-Ephrin signaling | EPHB2 | Ephrin type-B receptor 2, Receptor protein-tyrosine kinase | 8.477081 | 0.452752 | 4 | 0.544199 | 0.508582 |
| EPH-Ephrin signaling | GRIN2A | Glutamate receptor, Glutamate receptor ionotropic, NMDA 2A | 8.477081 | 0.452752 | 4 | 0.544199 | 0.527756 |
| EPH-Ephrin signaling | EPHB6 | EPH receptor B6, Ephrin type-B receptor 6, Receptor protein-tyrosine kinase | 8.477081 | 0.452752 | 4 | 0.544199 | 0.500457 |
| EPH-Ephrin signaling | EPHA4 | Ephrin type-A receptor 4, Receptor protein-tyrosine kinase | 8.477081 | 0.452752 | 4 | 0.544199 | 0.503164 |
| EPH-Ephrin signaling | APH1A | Anterior pharynx defective 1 homolog A, Anterior pharynx defective 1a homolog, Aph-1 homolog A, gamma-secretase subunit, Gamma-secretase subunit APH-1A, Uncharacterized protein | 8.477081 | 0.452752 | 4 | 0.544199 | 0.60749 |
| EPH-Ephrin signaling | ROCK2 | Rho-associated protein kinase, Rho-associated protein kinase 2 | 8.477081 | 0.452752 | 4 | 0.544199 | 0.516942 |
| EPH-Ephrin signaling | PTK2 | Focal adhesion kinase 1, Non-specific protein-tyrosine kinase, Serine/threonine-protein kinase PTK2/STK2 | 8.477081 | 0.452752 | 4 | 0.544199 | 0.503121 |
| EPH-Ephrin signaling | EPHB4 | Ephrin type-B receptor 4, Receptor protein-tyrosine kinase | 8.477081 | 0.452752 | 4 | 0.544199 | 0.527016 |
| EPH-Ephrin signaling | YES1 | Tyrosine-protein kinase, Tyrosine-protein kinase yes, Tyrosine-protein kinase Yes | 8.477081 | 0.452752 | 4 | 0.544199 | 0.506641 |
| EPH-Ephrin signaling | EPHA2 | Ephrin type-A receptor 2, Receptor protein-tyrosine kinase | 8.477081 | 0.452752 | 4 | 0.544199 | 0.530164 |
| EPH-Ephrin signaling | PSEN1 | Presenilin, Presenilin-1 | 8.477081 | 0.452752 | 4 | 0.544199 | 0.611222 |
| EPH-Ephrin signaling | APH1B | Aph-1 homolog B, gamma-secretase subunit, APH-1B', Gamma-secretase subunit APH-1B, Uncharacterized protein | 8.477081 | 0.452752 | 4 | 0.544199 | 0.60749 |
| EPH-Ephrin signaling | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 8.477081 | 0.452752 | 4 | 0.544199 | 0.594807 |
| EPH-Ephrin signaling | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 8.477081 | 0.452752 | 4 | 0.544199 | 0.505988 |
| EPH-Ephrin signaling | GRIN2B | Glutamate receptor, Glutamate receptor ionotropic, NMDA 2B | 8.477081 | 0.452752 | 4 | 0.544199 | 0.56443 |
| EPH-Ephrin signaling | MMP2 | 72 kDa gelatinase, 72 kDa type IV collagenase, Matrix metalloproteinase-2 | 8.477081 | 0.452752 | 4 | 0.544199 | 0.512398 |
| EPH-Ephrin signaling | NCSTN | Nicastrin | 8.477081 | 0.452752 | 4 | 0.544199 | 0.60749 |
| EPH-Ephrin signaling | EPHA5 | Ephrin type-A receptor 5, Receptor protein-tyrosine kinase | 8.477081 | 0.452752 | 4 | 0.544199 | 0.515572 |
| EPH-Ephrin signaling | GRIN1 | Glutamate receptor, Glutamate receptor ionotropic, NMDA 1 | 8.477081 | 0.452752 | 4 | 0.544199 | 0.573676 |
| EPH-Ephrin signaling | PSENEN | Gamma-secretase subunit PEN-2 | 8.477081 | 0.452752 | 4 | 0.544199 | 0.60725 |
| EPH-Ephrin signaling | EPHA3 | Ephrin type-A receptor 3, Receptor protein-tyrosine kinase | 8.477081 | 0.452752 | 4 | 0.544199 | 0.500178 |
| EPH-Ephrin signaling | PSEN2 | Presenilin, Presenilin-2 | 8.477081 | 0.452752 | 4 | 0.544199 | 0.607393 |
| EPH-Ephrin signaling | EPHB1 | Ephrin type-B receptor 1, Receptor protein-tyrosine kinase | 8.477081 | 0.452752 | 4 | 0.544199 | 0.509548 |
| EPH-Ephrin signaling | EPHB3 | Ephrin type-B receptor 3, Receptor protein-tyrosine kinase | 8.477081 | 0.452752 | 4 | 0.544199 | 0.502956 |
| EPH-Ephrin signaling | EPHA8 | EPH receptor A8, Ephrin type-A receptor 8, Receptor protein-tyrosine kinase | 8.477081 | 0.452752 | 4 | 0.544199 | 0.518651 |
| EPH-Ephrin signaling | ACTR3 | Actin-like protein 3, Actin-related protein 3, ARP3 actin-related protein 3 homolog | 8.477081 | 0.452752 | 4 | 0.544199 | 0.520663 |
| ER-Phagosome pathway | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.54735 |
| ER-Phagosome pathway | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.57244 |
| ER-Phagosome pathway | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.515416 |
| ER-Phagosome pathway | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.515416 |
| ER-Phagosome pathway | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.546881 |
| ER-Phagosome pathway | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.573417 |
| ER-Phagosome pathway | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.515416 |
| ER-Phagosome pathway | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.54735 |
| ER-Phagosome pathway | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.565601 |
| ER-Phagosome pathway | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.515416 |
| ER-Phagosome pathway | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.515416 |
| ER-Phagosome pathway | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.54735 |
| ER-Phagosome pathway | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.58858 |
| ER-Phagosome pathway | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.515416 |
| ER-Phagosome pathway | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.54735 |
| ER-Phagosome pathway | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.547953 |
| ER-Phagosome pathway | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.5197 |
| ER-Phagosome pathway | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.572294 |
| ER-Phagosome pathway | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.515416 |
| ER-Phagosome pathway | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.54735 |
| ER-Phagosome pathway | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.599924 | 0.663922 | 4 | 0.707711 | 0.515416 |
| ER-Phagosome pathway | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.599924 | 0.663922 | 4 | 0.707711 | 0.515416 |
| ER-Phagosome pathway | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.54735 |
| ER-Phagosome pathway | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.577481 |
| ER-Phagosome pathway | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.54735 |
| ER-Phagosome pathway | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.515416 |
| ER-Phagosome pathway | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.515416 |
| ER-Phagosome pathway | BTK | Tyrosine-protein kinase, Tyrosine-protein kinase BTK | 9.599924 | 0.663922 | 4 | 0.707711 | 0.602174 |
| ER-Phagosome pathway | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.515416 |
| ER-Phagosome pathway | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.54735 |
| ER-Phagosome pathway | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.54735 |
| ER-Phagosome pathway | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.599924 | 0.663922 | 4 | 0.707711 | 0.568645 |
| ER-Phagosome pathway | IKBKB | I-kappa-B kinase, Inhibitor of nuclear factor kappa-B kinase subunit beta | 9.599924 | 0.663922 | 4 | 0.707711 | 0.518113 |
| ER-Phagosome pathway | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.599924 | 0.663922 | 4 | 0.707711 | 0.579609 |
| ER-Phagosome pathway | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.54735 |
| ER-Phagosome pathway | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.515416 |
| ER-Phagosome pathway | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.577847 |
| ER-Phagosome pathway | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.515416 |
| ER-Phagosome pathway | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.599924 | 0.663922 | 4 | 0.707711 | 0.515416 |
| Erythropoietin activates Phosphoinositide-3-kinase (PI3K) | PIK3CG | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.781822 | 0.690307 | 3.378149 | 0.616755 | 0.570021 |
| Erythropoietin activates Phosphoinositide-3-kinase (PI3K) | JAK2 | Janus kinase 2, Tyrosine-protein kinase, Tyrosine-protein kinase JAK2 | 9.781822 | 0.690307 | 3.378149 | 0.616755 | 0.573235 |
| Erythropoietin activates Phosphoinositide-3-kinase (PI3K) | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 9.781822 | 0.690307 | 3.378149 | 0.616755 | 0.646924 |
| Erythropoietin activates Phosphoinositide-3-kinase (PI3K) | PIK3CD | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.781822 | 0.690307 | 3.378149 | 0.616755 | 0.632077 |
| Erythropoietin activates Phosphoinositide-3-kinase (PI3K) | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.781822 | 0.690307 | 3.378149 | 0.616755 | 0.660919 |
| Erythropoietin activates Phosphoinositide-3-kinase (PI3K) | EPOR | Erythropoietin receptor | 9.781822 | 0.690307 | 3.378149 | 0.616755 | 0.508215 |
| Erythropoietin activates Phosphoinositide-3-kinase (PI3K) | PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.781822 | 0.690307 | 3.378149 | 0.616755 | 0.589385 |
| Eukaryotic Translation Elongation | RPS23 | 40S ribosomal protein S23, 40S ribosomal protein S23-A | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL26 | 60S ribosomal protein L26, GEO07453p1, KOW domain-containing protein, Ribosomal protein L26 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS3A | 40S ribosomal protein S3a | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL24 | 60S ribosomal protein L24, Ribosomal protein L24, TRASH domain-containing protein | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPLP0 | 60S acidic ribosomal protein P0 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL14 | 60S ribosomal protein L14 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL12 | 60S ribosomal protein L12 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL35 | 60S ribosomal protein L35 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS13 | 40S ribosomal protein S13 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL27 | 60S ribosomal protein L27 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS11 | 40S ribosomal protein S11 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL36 | 60S ribosomal protein L36 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS3 | 40S ribosomal protein S3, DNA-(apurinic or apyrimidinic site) lyase | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL7 | 60S ribosomal protein L7, MGC79754 protein, Ribosomal protein L7, Uncharacterized protein | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL3 | 60S ribosomal protein L3, LOC100145083 protein, Ribosomal protein L3, Uncharacterized protein | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS10 | 40S ribosomal protein S10, Ribosomal protein S10, S10_plectin domain-containing protein | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL7A | 60S ribosomal protein L7-A, 60S ribosomal protein L7a | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS25 | 40S ribosomal protein S25 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS21 | 40S ribosomal protein S21 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS20 | 40S ribosomal protein S20 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL15 | 60S ribosomal protein L15, Ribosomal protein L15 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL5 | 60S ribosomal protein L5, Ribosomal protein L5 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL22 | 60S ribosomal protein L22, 60S ribosomal protein L22 1, Ribosomal protein L22, Uncharacterized protein | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL38 | 60S ribosomal protein L38 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL19 | 60S ribosomal protein L19, Ribosomal protein L19 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS8 | 40S ribosomal protein S8 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPSA | 40S ribosomal protein SA | 5.091402 | 0.903382 | 4 | 0.59485 | 0.521531 |
| Eukaryotic Translation Elongation | RPS2 | 40S ribosomal protein S2 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL10 | 60S ribosomal protein L10, MGC89303 protein | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL28 | 60S ribosomal protein L28 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS7 | 40S ribosomal protein S7 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS16 | 40S ribosomal protein S16, Ribosomal protein S16, Uncharacterized protein | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS26 | 40S ribosomal protein S26 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL27A | 60S ribosomal protein L27-A, 60S ribosomal protein L27a | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS6 | 40S ribosomal protein S6 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.566528 |
| Eukaryotic Translation Elongation | RPS17 | 40S ribosomal protein S17 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL39 | 60S ribosomal protein L39, LOC100135132 protein, Ribosomal protein L39 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS19 | 40S ribosomal protein S19, Uncharacterized protein | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL34 | 60S ribosomal protein L34 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL11 | 60S ribosomal protein L11 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL13 | 60S ribosomal protein L13 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL17 | 60S ribosomal protein L17 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS4Y1 | 40S ribosomal protein S4, 40S ribosomal protein S4, Y isoform 1 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS29 | 40S ribosomal protein S29 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL4 | 60S ribosomal protein L4, Ribos_L4_asso_C domain-containing protein, Ribosomal protein L4 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS27 | 40S ribosomal protein S27 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.549907 |
| Eukaryotic Translation Elongation | RPL8 | 60S ribosomal protein L8 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS28 | 40S ribosomal protein S28 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL18A | 60S ribosomal protein L18-A, 60S ribosomal protein L18a | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS24 | 40S ribosomal protein S24 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS18 | 40S ribosomal protein S18 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL30 | 60S ribosomal protein L30 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPLP1 | 60S acidic ribosomal protein P1, Ribosomal protein, large, P1 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS9 | 40S ribosomal protein S9 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL35A | 60S ribosomal protein L35-A, 60S ribosomal protein L35a | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL37 | 60S ribosomal protein L37, Ribosomal protein L37 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL13A | 60S ribosomal protein L13a | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL29 | 60S ribosomal protein L29 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL31 | 60S ribosomal protein L31 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS15 | 40S ribosomal protein S15 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL21 | 60S ribosomal protein L21 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL36A | 60S ribosomal protein L36-A, 60S ribosomal protein L36a, IP17351p, ribosomal protein L36a, Uncharacterized protein | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | FAU | 40S ribosomal protein S30 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS12 | 40S ribosomal protein S12 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL41 | 60S ribosomal protein L41 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL37A | 60S ribosomal protein L37-A, 60S ribosomal protein L37a, MGC89854 protein, Probable 60S ribosomal protein L37-A, Ribosomal protein L37a | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS4X | 40S ribosomal protein S4, 40S ribosomal protein S4, X isoform | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL23A | 60S ribosomal protein L23-A, 60S ribosomal protein L23a, FI01658p, MGC89958 protein, Ribosomal protein L23a, Ribosomal_L23eN domain-containing protein | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS15A | 40S ribosomal protein S15a | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS14 | 40S ribosomal protein S14, Ribosomal protein S14, Uncharacterized protein | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL32 | 60S ribosomal protein L32, Hypothetical LOC496894, Ribosomal protein L32, Uncharacterized protein | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL10A | 60S ribosomal protein L10a, Ribosomal protein | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL18 | 60S ribosomal protein L18, Ribosomal protein L18 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL23 | 60S ribosomal protein L23 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPL6 | 60S ribosomal protein L6 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Elongation | RPS5 | 40S ribosomal protein S5, 40S ribosomal protein S5 [Cleaved into: 40S ribosomal protein S5, N-terminally processed], 40S ribosomal protein S5-A, Ribosomal protein S5 | 5.091402 | 0.903382 | 4 | 0.59485 | 0.579609 |
| Eukaryotic Translation Termination | RPS26 | 40S ribosomal protein S26 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS27 | 40S ribosomal protein S27 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.549907 |
| Eukaryotic Translation Termination | RPL18 | 60S ribosomal protein L18, Ribosomal protein L18 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL39 | 60S ribosomal protein L39, LOC100135132 protein, Ribosomal protein L39 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL27 | 60S ribosomal protein L27 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS5 | 40S ribosomal protein S5, 40S ribosomal protein S5 [Cleaved into: 40S ribosomal protein S5, N-terminally processed], 40S ribosomal protein S5-A, Ribosomal protein S5 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL5 | 60S ribosomal protein L5, Ribosomal protein L5 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL10 | 60S ribosomal protein L10, MGC89303 protein | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL22 | 60S ribosomal protein L22, 60S ribosomal protein L22 1, Ribosomal protein L22, Uncharacterized protein | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL11 | 60S ribosomal protein L11 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL36A | 60S ribosomal protein L36-A, 60S ribosomal protein L36a, IP17351p, ribosomal protein L36a, Uncharacterized protein | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL15 | 60S ribosomal protein L15, Ribosomal protein L15 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS3 | 40S ribosomal protein S3, DNA-(apurinic or apyrimidinic site) lyase | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS28 | 40S ribosomal protein S28 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL32 | 60S ribosomal protein L32, Hypothetical LOC496894, Ribosomal protein L32, Uncharacterized protein | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL8 | 60S ribosomal protein L8 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL12 | 60S ribosomal protein L12 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS8 | 40S ribosomal protein S8 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL7 | 60S ribosomal protein L7, MGC79754 protein, Ribosomal protein L7, Uncharacterized protein | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL23 | 60S ribosomal protein L23 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS25 | 40S ribosomal protein S25 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS18 | 40S ribosomal protein S18 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL3 | 60S ribosomal protein L3, LOC100145083 protein, Ribosomal protein L3, Uncharacterized protein | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL13A | 60S ribosomal protein L13a | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS23 | 40S ribosomal protein S23, 40S ribosomal protein S23-A | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL31 | 60S ribosomal protein L31 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPLP1 | 60S acidic ribosomal protein P1, Ribosomal protein, large, P1 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL30 | 60S ribosomal protein L30 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL29 | 60S ribosomal protein L29 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL37A | 60S ribosomal protein L37-A, 60S ribosomal protein L37a, MGC89854 protein, Probable 60S ribosomal protein L37-A, Ribosomal protein L37a | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL14 | 60S ribosomal protein L14 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS29 | 40S ribosomal protein S29 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPLP0 | 60S acidic ribosomal protein P0 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL21 | 60S ribosomal protein L21 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS12 | 40S ribosomal protein S12 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS16 | 40S ribosomal protein S16, Ribosomal protein S16, Uncharacterized protein | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS7 | 40S ribosomal protein S7 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPSA | 40S ribosomal protein SA | 11.669631 | 0.904343 | 4 | 0.934696 | 0.521531 |
| Eukaryotic Translation Termination | RPL41 | 60S ribosomal protein L41 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS24 | 40S ribosomal protein S24 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS20 | 40S ribosomal protein S20 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL19 | 60S ribosomal protein L19, Ribosomal protein L19 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL23A | 60S ribosomal protein L23-A, 60S ribosomal protein L23a, FI01658p, MGC89958 protein, Ribosomal protein L23a, Ribosomal_L23eN domain-containing protein | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS3A | 40S ribosomal protein S3a | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS15A | 40S ribosomal protein S15a | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS6 | 40S ribosomal protein S6 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.566528 |
| Eukaryotic Translation Termination | RPS11 | 40S ribosomal protein S11 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL38 | 60S ribosomal protein L38 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL37 | 60S ribosomal protein L37, Ribosomal protein L37 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS21 | 40S ribosomal protein S21 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL35 | 60S ribosomal protein L35 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS19 | 40S ribosomal protein S19, Uncharacterized protein | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS4X | 40S ribosomal protein S4, 40S ribosomal protein S4, X isoform | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL27A | 60S ribosomal protein L27-A, 60S ribosomal protein L27a | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL26 | 60S ribosomal protein L26, GEO07453p1, KOW domain-containing protein, Ribosomal protein L26 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS15 | 40S ribosomal protein S15 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL7A | 60S ribosomal protein L7-A, 60S ribosomal protein L7a | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS14 | 40S ribosomal protein S14, Ribosomal protein S14, Uncharacterized protein | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS9 | 40S ribosomal protein S9 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL28 | 60S ribosomal protein L28 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS13 | 40S ribosomal protein S13 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL6 | 60S ribosomal protein L6 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | FAU | 40S ribosomal protein S30 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL24 | 60S ribosomal protein L24, Ribosomal protein L24, TRASH domain-containing protein | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL18A | 60S ribosomal protein L18-A, 60S ribosomal protein L18a | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL35A | 60S ribosomal protein L35-A, 60S ribosomal protein L35a | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS10 | 40S ribosomal protein S10, Ribosomal protein S10, S10_plectin domain-containing protein | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL4 | 60S ribosomal protein L4, Ribos_L4_asso_C domain-containing protein, Ribosomal protein L4 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS17 | 40S ribosomal protein S17 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL17 | 60S ribosomal protein L17 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL34 | 60S ribosomal protein L34 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS2 | 40S ribosomal protein S2 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL10A | 60S ribosomal protein L10a, Ribosomal protein | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL13 | 60S ribosomal protein L13 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPL36 | 60S ribosomal protein L36 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Eukaryotic Translation Termination | RPS4Y1 | 40S ribosomal protein S4, 40S ribosomal protein S4, Y isoform 1 | 11.669631 | 0.904343 | 4 | 0.934696 | 0.579609 |
| Extra-nuclear estrogen signaling | PRKCZ | Protein kinase C, Protein kinase C zeta type | 9.535892 | 0.459983 | 4 | 0.602438 | 0.513845 |
| Extra-nuclear estrogen signaling | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 9.535892 | 0.459983 | 4 | 0.602438 | 0.505988 |
| Extra-nuclear estrogen signaling | MMP3 | Stromelysin-1, ZnMc domain-containing protein | 9.535892 | 0.459983 | 4 | 0.602438 | 0.50644 |
| Extra-nuclear estrogen signaling | HSP90AA1 | Heat shock protein 90 alpha family class A member 1, Heat shock protein HSP 90-alpha | 9.535892 | 0.459983 | 4 | 0.602438 | 0.624325 |
| Extra-nuclear estrogen signaling | ESR2 | Estrogen receptor 2, Estrogen receptor beta | 9.535892 | 0.459983 | 4 | 0.602438 | 0.53484 |
| Extra-nuclear estrogen signaling | MMP2 | 72 kDa gelatinase, 72 kDa type IV collagenase, Matrix metalloproteinase-2 | 9.535892 | 0.459983 | 4 | 0.602438 | 0.512398 |
| Extra-nuclear estrogen signaling | EGFR | Epidermal growth factor receptor, Receptor protein-tyrosine kinase | 9.535892 | 0.459983 | 4 | 0.602438 | 0.625817 |
| Extra-nuclear estrogen signaling | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.535892 | 0.459983 | 4 | 0.602438 | 0.660919 |
| Extra-nuclear estrogen signaling | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 9.535892 | 0.459983 | 4 | 0.602438 | 0.552041 |
| Extra-nuclear estrogen signaling | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 9.535892 | 0.459983 | 4 | 0.602438 | 0.646924 |
| Extra-nuclear estrogen signaling | SHC1 | SHC-transforming protein 1 | 9.535892 | 0.459983 | 4 | 0.602438 | 0.521858 |
| Extra-nuclear estrogen signaling | CCND1 | Cyclin D1, G1/S-specific cyclin-D1 | 9.535892 | 0.459983 | 4 | 0.602438 | 0.525654 |
| Extra-nuclear estrogen signaling | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 9.535892 | 0.459983 | 4 | 0.602438 | 0.594807 |
| Extra-nuclear estrogen signaling | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 9.535892 | 0.459983 | 4 | 0.602438 | 0.541254 |
| Extra-nuclear estrogen signaling | GNAI2 | G protein subunit alpha i2, Guanine nucleotide binding protein, Guanine nucleotide-binding protein G(i) subunit alpha-2 | 9.535892 | 0.459983 | 4 | 0.602438 | 0.512918 |
| Extra-nuclear estrogen signaling | IGF1R | Insulin-like growth factor 1 receptor, Tyrosine-protein kinase receptor | 9.535892 | 0.459983 | 4 | 0.602438 | 0.595333 |
| Extra-nuclear estrogen signaling | PTK2 | Focal adhesion kinase 1, Non-specific protein-tyrosine kinase, Serine/threonine-protein kinase PTK2/STK2 | 9.535892 | 0.459983 | 4 | 0.602438 | 0.503121 |
| Extra-nuclear estrogen signaling | ESR1 | Estrogen receptor | 9.535892 | 0.459983 | 4 | 0.602438 | 0.561602 |
| Extra-nuclear estrogen signaling | SHC | DShc protein | 9.535892 | 0.459983 | 4 | 0.602438 | 0.605 |
| Extra-nuclear estrogen signaling | BCL2 | Apoptosis regulator Bcl-2 | 9.535892 | 0.459983 | 4 | 0.602438 | 1 |
| Extra-nuclear estrogen signaling | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 9.535892 | 0.459983 | 4 | 0.602438 | 0.589663 |
| Extrinsic Pathway of Fibrin Clot Formation | F10 | Coagulation factor VII, Coagulation factor X, MGC107878 protein, Uncharacterized protein | 9.247442 | 0.838497 | 3.34255 | 0.649135 | 0.588553 |
| Extrinsic Pathway of Fibrin Clot Formation | F9 | Christmas factor, Coagulation factor IX | 9.247442 | 0.838497 | 3.34255 | 0.649135 | 0.632191 |
| Extrinsic Pathway of Fibrin Clot Formation | F3 | Coagulation factor III, Tissue factor | 9.247442 | 0.838497 | 3.34255 | 0.649135 | 0.51249 |
| Extrinsic Pathway of Fibrin Clot Formation | TFPI | Tissue factor pathway inhibitor, Uncharacterized protein | 9.247442 | 0.838497 | 3.34255 | 0.649135 | 0.886316 |
| Extrinsic Pathway of Fibrin Clot Formation | F7 | Coagulation factor VII | 9.247442 | 0.838497 | 3.34255 | 0.649135 | 0.519515 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.515416 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.577847 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.54735 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.515416 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.515416 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.515416 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.515416 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.515416 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.515416 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.54735 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.58858 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.565601 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.572294 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.54735 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.515416 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.546881 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.763119 | 0.808014 | 4 | 0.788176 | 0.579609 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.515416 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.5197 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.577481 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.515416 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.763119 | 0.808014 | 4 | 0.788176 | 0.515416 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.54735 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.763119 | 0.808014 | 4 | 0.788176 | 0.515416 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.54735 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.54735 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.515416 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.54735 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.54735 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.763119 | 0.808014 | 4 | 0.788176 | 0.568645 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.515416 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.547953 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.573417 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | AURKA | Aurora kinase, Aurora kinase A | 9.763119 | 0.808014 | 4 | 0.788176 | 0.54457 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.515416 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.54735 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | AURKB | Aurora kinase, Aurora kinase B | 9.763119 | 0.808014 | 4 | 0.788176 | 0.524412 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.54735 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.763119 | 0.808014 | 4 | 0.788176 | 0.57244 |
| Fc epsilon receptor (FCERI) signaling | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.515416 |
| Fc epsilon receptor (FCERI) signaling | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 8.636516 | 0.523063 | 4 | 0.58758 | 0.505988 |
| Fc epsilon receptor (FCERI) signaling | SHC | DShc protein | 8.636516 | 0.523063 | 4 | 0.58758 | 0.605 |
| Fc epsilon receptor (FCERI) signaling | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.54735 |
| Fc epsilon receptor (FCERI) signaling | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.54735 |
| Fc epsilon receptor (FCERI) signaling | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.54735 |
| Fc epsilon receptor (FCERI) signaling | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.515416 |
| Fc epsilon receptor (FCERI) signaling | SYK | Tyrosine-protein kinase, Tyrosine-protein kinase SYK | 8.636516 | 0.523063 | 4 | 0.58758 | 0.557587 |
| Fc epsilon receptor (FCERI) signaling | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.515416 |
| Fc epsilon receptor (FCERI) signaling | IKBKB | I-kappa-B kinase, Inhibitor of nuclear factor kappa-B kinase subunit beta | 8.636516 | 0.523063 | 4 | 0.58758 | 0.518113 |
| Fc epsilon receptor (FCERI) signaling | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 8.636516 | 0.523063 | 4 | 0.58758 | 0.515416 |
| Fc epsilon receptor (FCERI) signaling | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.515416 |
| Fc epsilon receptor (FCERI) signaling | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.515416 |
| Fc epsilon receptor (FCERI) signaling | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.577847 |
| Fc epsilon receptor (FCERI) signaling | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.515416 |
| Fc epsilon receptor (FCERI) signaling | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 8.636516 | 0.523063 | 4 | 0.58758 | 0.660919 |
| Fc epsilon receptor (FCERI) signaling | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.5197 |
| Fc epsilon receptor (FCERI) signaling | MAPK8 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 8 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.503967 |
| Fc epsilon receptor (FCERI) signaling | PRKCQ | Protein kinase C, Protein kinase C theta type | 8.636516 | 0.523063 | 4 | 0.58758 | 0.523352 |
| Fc epsilon receptor (FCERI) signaling | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 8.636516 | 0.523063 | 4 | 0.58758 | 0.568645 |
| Fc epsilon receptor (FCERI) signaling | MAPK10 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 10 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.506109 |
| Fc epsilon receptor (FCERI) signaling | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.573417 |
| Fc epsilon receptor (FCERI) signaling | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 8.636516 | 0.523063 | 4 | 0.58758 | 0.515416 |
| Fc epsilon receptor (FCERI) signaling | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.54735 |
| Fc epsilon receptor (FCERI) signaling | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.54735 |
| Fc epsilon receptor (FCERI) signaling | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.572294 |
| Fc epsilon receptor (FCERI) signaling | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.54735 |
| Fc epsilon receptor (FCERI) signaling | PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 8.636516 | 0.523063 | 4 | 0.58758 | 0.589385 |
| Fc epsilon receptor (FCERI) signaling | MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.519182 |
| Fc epsilon receptor (FCERI) signaling | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.54735 |
| Fc epsilon receptor (FCERI) signaling | BTK | Tyrosine-protein kinase, Tyrosine-protein kinase BTK | 8.636516 | 0.523063 | 4 | 0.58758 | 0.602174 |
| Fc epsilon receptor (FCERI) signaling | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.541254 |
| Fc epsilon receptor (FCERI) signaling | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.57244 |
| Fc epsilon receptor (FCERI) signaling | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.515416 |
| Fc epsilon receptor (FCERI) signaling | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.515416 |
| Fc epsilon receptor (FCERI) signaling | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.54735 |
| Fc epsilon receptor (FCERI) signaling | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 8.636516 | 0.523063 | 4 | 0.58758 | 0.646924 |
| Fc epsilon receptor (FCERI) signaling | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.547953 |
| Fc epsilon receptor (FCERI) signaling | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.546881 |
| Fc epsilon receptor (FCERI) signaling | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.54735 |
| Fc epsilon receptor (FCERI) signaling | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.58858 |
| Fc epsilon receptor (FCERI) signaling | SHC1 | SHC-transforming protein 1 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.521858 |
| Fc epsilon receptor (FCERI) signaling | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.515416 |
| Fc epsilon receptor (FCERI) signaling | UBE2V1 | Ubiquitin conjugating enzyme E2 V1, Ubiquitin-conjugating enzyme E2 variant 1 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.593364 |
| Fc epsilon receptor (FCERI) signaling | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.577481 |
| Fc epsilon receptor (FCERI) signaling | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.515416 |
| Fc epsilon receptor (FCERI) signaling | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.565601 |
| Fc epsilon receptor (FCERI) signaling | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.515416 |
| Fc epsilon receptor (FCERI) signaling | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.515416 |
| Fc epsilon receptor (FCERI) signaling | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.54735 |
| Fc epsilon receptor (FCERI) signaling | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 8.636516 | 0.523063 | 4 | 0.58758 | 0.579609 |
| Fc epsilon receptor (FCERI) signaling | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 8.636516 | 0.523063 | 4 | 0.58758 | 0.515416 |
| FCERI mediated NF-kB activation | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.54735 |
| FCERI mediated NF-kB activation | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.515416 |
| FCERI mediated NF-kB activation | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.54735 |
| FCERI mediated NF-kB activation | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.547953 |
| FCERI mediated NF-kB activation | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.565601 |
| FCERI mediated NF-kB activation | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.577847 |
| FCERI mediated NF-kB activation | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.548571 | 0.640495 | 4 | 0.693348 | 0.515416 |
| FCERI mediated NF-kB activation | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.515416 |
| FCERI mediated NF-kB activation | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.54735 |
| FCERI mediated NF-kB activation | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.515416 |
| FCERI mediated NF-kB activation | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.546881 |
| FCERI mediated NF-kB activation | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.515416 |
| FCERI mediated NF-kB activation | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.515416 |
| FCERI mediated NF-kB activation | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.572294 |
| FCERI mediated NF-kB activation | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.515416 |
| FCERI mediated NF-kB activation | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.548571 | 0.640495 | 4 | 0.693348 | 0.515416 |
| FCERI mediated NF-kB activation | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.548571 | 0.640495 | 4 | 0.693348 | 0.568645 |
| FCERI mediated NF-kB activation | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.54735 |
| FCERI mediated NF-kB activation | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.54735 |
| FCERI mediated NF-kB activation | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.54735 |
| FCERI mediated NF-kB activation | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.54735 |
| FCERI mediated NF-kB activation | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.548571 | 0.640495 | 4 | 0.693348 | 0.579609 |
| FCERI mediated NF-kB activation | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.58858 |
| FCERI mediated NF-kB activation | UBE2V1 | Ubiquitin conjugating enzyme E2 V1, Ubiquitin-conjugating enzyme E2 variant 1 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.593364 |
| FCERI mediated NF-kB activation | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.54735 |
| FCERI mediated NF-kB activation | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.54735 |
| FCERI mediated NF-kB activation | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.54735 |
| FCERI mediated NF-kB activation | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.515416 |
| FCERI mediated NF-kB activation | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.515416 |
| FCERI mediated NF-kB activation | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.577481 |
| FCERI mediated NF-kB activation | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.57244 |
| FCERI mediated NF-kB activation | PRKCQ | Protein kinase C, Protein kinase C theta type | 9.548571 | 0.640495 | 4 | 0.693348 | 0.523352 |
| FCERI mediated NF-kB activation | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.515416 |
| FCERI mediated NF-kB activation | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.515416 |
| FCERI mediated NF-kB activation | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.515416 |
| FCERI mediated NF-kB activation | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.515416 |
| FCERI mediated NF-kB activation | MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.519182 |
| FCERI mediated NF-kB activation | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.515416 |
| FCERI mediated NF-kB activation | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.5197 |
| FCERI mediated NF-kB activation | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.548571 | 0.640495 | 4 | 0.693348 | 0.573417 |
| FCERI mediated NF-kB activation | IKBKB | I-kappa-B kinase, Inhibitor of nuclear factor kappa-B kinase subunit beta | 9.548571 | 0.640495 | 4 | 0.693348 | 0.518113 |
| FCGR3A-mediated IL10 synthesis | ADCY9 | Adenylate cyclase 9, Adenylate cyclase type 9 | 8.54185 | 0.488795 | 3.629709 | 0.513207 | 0.550534 |
| FCGR3A-mediated IL10 synthesis | YES1 | Tyrosine-protein kinase, Tyrosine-protein kinase yes, Tyrosine-protein kinase Yes | 8.54185 | 0.488795 | 3.629709 | 0.513207 | 0.506641 |
| FCGR3A-mediated IL10 synthesis | HCK | Tyrosine-protein kinase, Tyrosine-protein kinase HCK | 8.54185 | 0.488795 | 3.629709 | 0.513207 | 0.515497 |
| FCGR3A-mediated IL10 synthesis | PRKAR2A | cAMP-dependent protein kinase type II-alpha regulatory subunit, Protein kinase cAMP-dependent type II regulatory subunit alpha, Protein kinase, cAMP-dependent, regulatory subunit type II alpha, Uncharacterized protein | 8.54185 | 0.488795 | 3.629709 | 0.513207 | 0.527065 |
| FCGR3A-mediated IL10 synthesis | ADCY7 | Adenylate cyclase type 7 | 8.54185 | 0.488795 | 3.629709 | 0.513207 | 0.550655 |
| FCGR3A-mediated IL10 synthesis | SYK | Tyrosine-protein kinase, Tyrosine-protein kinase SYK | 8.54185 | 0.488795 | 3.629709 | 0.513207 | 0.557587 |
| FCGR3A-mediated IL10 synthesis | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 8.54185 | 0.488795 | 3.629709 | 0.513207 | 0.594807 |
| FLT3 Signaling | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 8.251472 | 0.474888 | 4 | 0.543628 | 0.505988 |
| FLT3 Signaling | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 8.251472 | 0.474888 | 4 | 0.543628 | 0.552041 |
| FLT3 Signaling | LCK | Proto-oncogene tyrosine-protein kinase LCK, Tyrosine-protein kinase, Tyrosine-protein kinase Lck | 8.251472 | 0.474888 | 4 | 0.543628 | 0.543278 |
| FLT3 Signaling | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 8.251472 | 0.474888 | 4 | 0.543628 | 0.579609 |
| FLT3 Signaling | FLT3 | Receptor protein-tyrosine kinase, Receptor-type tyrosine-protein kinase FLT3 | 8.251472 | 0.474888 | 4 | 0.543628 | 0.529563 |
| FLT3 Signaling | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 8.251472 | 0.474888 | 4 | 0.543628 | 0.646924 |
| FLT3 Signaling | SYK | Tyrosine-protein kinase, Tyrosine-protein kinase SYK | 8.251472 | 0.474888 | 4 | 0.543628 | 0.557587 |
| FLT3 Signaling | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 8.251472 | 0.474888 | 4 | 0.543628 | 0.589663 |
| FLT3 Signaling | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 8.251472 | 0.474888 | 4 | 0.543628 | 0.660919 |
| FLT3 Signaling | HCK | Tyrosine-protein kinase, Tyrosine-protein kinase HCK | 8.251472 | 0.474888 | 4 | 0.543628 | 0.515497 |
| FLT3 Signaling | ABL2 | Tyrosine-protein kinase, Tyrosine-protein kinase ABL2 | 8.251472 | 0.474888 | 4 | 0.543628 | 0.52908 |
| Formation of a pool of free 40S subunits | RPS29 | 40S ribosomal protein S29 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS3 | 40S ribosomal protein S3, DNA-(apurinic or apyrimidinic site) lyase | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS11 | 40S ribosomal protein S11 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS17 | 40S ribosomal protein S17 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS23 | 40S ribosomal protein S23, 40S ribosomal protein S23-A | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL30 | 60S ribosomal protein L30 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS14 | 40S ribosomal protein S14, Ribosomal protein S14, Uncharacterized protein | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS15A | 40S ribosomal protein S15a | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS2 | 40S ribosomal protein S2 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS26 | 40S ribosomal protein S26 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS10 | 40S ribosomal protein S10, Ribosomal protein S10, S10_plectin domain-containing protein | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL5 | 60S ribosomal protein L5, Ribosomal protein L5 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPLP0 | 60S acidic ribosomal protein P0 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL9 | 60S ribosomal protein L9 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL23 | 60S ribosomal protein L23 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL34 | 60S ribosomal protein L34 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS4Y1 | 40S ribosomal protein S4, 40S ribosomal protein S4, Y isoform 1 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL18 | 60S ribosomal protein L18, Ribosomal protein L18 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL22 | 60S ribosomal protein L22, 60S ribosomal protein L22 1, Ribosomal protein L22, Uncharacterized protein | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS18 | 40S ribosomal protein S18 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL38 | 60S ribosomal protein L38 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL35A | 60S ribosomal protein L35-A, 60S ribosomal protein L35a | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS12 | 40S ribosomal protein S12 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL12 | 60S ribosomal protein L12 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL28 | 60S ribosomal protein L28 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS6 | 40S ribosomal protein S6 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.566528 |
| Formation of a pool of free 40S subunits | RPL23A | 60S ribosomal protein L23-A, 60S ribosomal protein L23a, FI01658p, MGC89958 protein, Ribosomal protein L23a, Ribosomal_L23eN domain-containing protein | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS16 | 40S ribosomal protein S16, Ribosomal protein S16, Uncharacterized protein | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL24 | 60S ribosomal protein L24, Ribosomal protein L24, TRASH domain-containing protein | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL39 | 60S ribosomal protein L39, LOC100135132 protein, Ribosomal protein L39 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPSA | 40S ribosomal protein SA | 11.645676 | 0.914537 | 4 | 0.938557 | 0.521531 |
| Formation of a pool of free 40S subunits | RPS3A | 40S ribosomal protein S3a | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL10 | 60S ribosomal protein L10, MGC89303 protein | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS7 | 40S ribosomal protein S7 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL37A | 60S ribosomal protein L37-A, 60S ribosomal protein L37a, MGC89854 protein, Probable 60S ribosomal protein L37-A, Ribosomal protein L37a | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS9 | 40S ribosomal protein S9 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL41 | 60S ribosomal protein L41 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS4X | 40S ribosomal protein S4, 40S ribosomal protein S4, X isoform | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS5 | 40S ribosomal protein S5, 40S ribosomal protein S5 [Cleaved into: 40S ribosomal protein S5, N-terminally processed], 40S ribosomal protein S5-A, Ribosomal protein S5 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS8 | 40S ribosomal protein S8 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL29 | 60S ribosomal protein L29 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL37 | 60S ribosomal protein L37, Ribosomal protein L37 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS20 | 40S ribosomal protein S20 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS28 | 40S ribosomal protein S28 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL27A | 60S ribosomal protein L27-A, 60S ribosomal protein L27a | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL18A | 60S ribosomal protein L18-A, 60S ribosomal protein L18a | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPLP1 | 60S acidic ribosomal protein P1, Ribosomal protein, large, P1 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL32 | 60S ribosomal protein L32, Hypothetical LOC496894, Ribosomal protein L32, Uncharacterized protein | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL36 | 60S ribosomal protein L36 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL13A | 60S ribosomal protein L13a | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL26 | 60S ribosomal protein L26, GEO07453p1, KOW domain-containing protein, Ribosomal protein L26 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL7A | 60S ribosomal protein L7-A, 60S ribosomal protein L7a | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL14 | 60S ribosomal protein L14 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL35 | 60S ribosomal protein L35 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL15 | 60S ribosomal protein L15, Ribosomal protein L15 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL17 | 60S ribosomal protein L17 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS15 | 40S ribosomal protein S15 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL19 | 60S ribosomal protein L19, Ribosomal protein L19 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL8 | 60S ribosomal protein L8 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS21 | 40S ribosomal protein S21 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL36A | 60S ribosomal protein L36-A, 60S ribosomal protein L36a, IP17351p, ribosomal protein L36a, Uncharacterized protein | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS13 | 40S ribosomal protein S13 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL3 | 60S ribosomal protein L3, LOC100145083 protein, Ribosomal protein L3, Uncharacterized protein | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS19 | 40S ribosomal protein S19, Uncharacterized protein | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL4 | 60S ribosomal protein L4, Ribos_L4_asso_C domain-containing protein, Ribosomal protein L4 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL13 | 60S ribosomal protein L13 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL6 | 60S ribosomal protein L6 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS27 | 40S ribosomal protein S27 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.549907 |
| Formation of a pool of free 40S subunits | RPS24 | 40S ribosomal protein S24 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL21 | 60S ribosomal protein L21 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL7 | 60S ribosomal protein L7, MGC79754 protein, Ribosomal protein L7, Uncharacterized protein | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL10A | 60S ribosomal protein L10a, Ribosomal protein | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL27 | 60S ribosomal protein L27 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL31 | 60S ribosomal protein L31 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPS25 | 40S ribosomal protein S25 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | FAU | 40S ribosomal protein S30 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of a pool of free 40S subunits | RPL11 | 60S ribosomal protein L11 | 11.645676 | 0.914537 | 4 | 0.938557 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS16 | 40S ribosomal protein S16, Ribosomal protein S16, Uncharacterized protein | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS14 | 40S ribosomal protein S14, Ribosomal protein S14, Uncharacterized protein | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS18 | 40S ribosomal protein S18 | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS5 | 40S ribosomal protein S5, 40S ribosomal protein S5 [Cleaved into: 40S ribosomal protein S5, N-terminally processed], 40S ribosomal protein S5-A, Ribosomal protein S5 | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS15 | 40S ribosomal protein S15 | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS3 | 40S ribosomal protein S3, DNA-(apurinic or apyrimidinic site) lyase | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS4X | 40S ribosomal protein S4, 40S ribosomal protein S4, X isoform | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS25 | 40S ribosomal protein S25 | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS29 | 40S ribosomal protein S29 | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS13 | 40S ribosomal protein S13 | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPSA | 40S ribosomal protein SA | 10.89176 | 0.869183 | 4 | 0.876986 | 0.521531 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS11 | 40S ribosomal protein S11 | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS21 | 40S ribosomal protein S21 | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS12 | 40S ribosomal protein S12 | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS10 | 40S ribosomal protein S10, Ribosomal protein S10, S10_plectin domain-containing protein | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS3A | 40S ribosomal protein S3a | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS23 | 40S ribosomal protein S23, 40S ribosomal protein S23-A | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS2 | 40S ribosomal protein S2 | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | FAU | 40S ribosomal protein S30 | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS9 | 40S ribosomal protein S9 | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS20 | 40S ribosomal protein S20 | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS27 | 40S ribosomal protein S27 | 10.89176 | 0.869183 | 4 | 0.876986 | 0.549907 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS15A | 40S ribosomal protein S15a | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS26 | 40S ribosomal protein S26 | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS28 | 40S ribosomal protein S28 | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS17 | 40S ribosomal protein S17 | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS6 | 40S ribosomal protein S6 | 10.89176 | 0.869183 | 4 | 0.876986 | 0.566528 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS4Y1 | 40S ribosomal protein S4, 40S ribosomal protein S4, Y isoform 1 | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS24 | 40S ribosomal protein S24 | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS8 | 40S ribosomal protein S8 | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS19 | 40S ribosomal protein S19, Uncharacterized protein | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Formation of the ternary complex, and subsequently, the 43S complex | RPS7 | 40S ribosomal protein S7 | 10.89176 | 0.869183 | 4 | 0.876986 | 0.579609 |
| Frs2-mediated activation | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 8.949797 | 0.713383 | 3.37958 | 0.590499 | 0.541254 |
| Frs2-mediated activation | BRAF | Non-specific serine/threonine protein kinase, Serine/threonine-protein kinase B-raf | 8.949797 | 0.713383 | 3.37958 | 0.590499 | 0.590074 |
| Frs2-mediated activation | RAP1A | RAP1A, member of RAS oncogene family, Ras-related protein Rap-1A, Uncharacterized protein | 8.949797 | 0.713383 | 3.37958 | 0.590499 | 0.530823 |
| Frs2-mediated activation | MAP2K1 | Dual specificity mitogen-activated protein kinase kinase 1, MAP2K1 protein, Mitogen-activated protein kinase kinase 1, Protein kinase domain-containing protein | 8.949797 | 0.713383 | 3.37958 | 0.590499 | 0.545374 |
| Frs2-mediated activation | NTRK1 | High affinity nerve growth factor receptor, Tyrosine-protein kinase receptor | 8.949797 | 0.713383 | 3.37958 | 0.590499 | 0.527885 |
| Frs2-mediated activation | YWHAB | 14-3-3 protein beta/alpha, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta | 8.949797 | 0.713383 | 3.37958 | 0.590499 | 0.513843 |
| Fusion of the Influenza Virion to the Host Cell Endosome | PB1 | Protein PB1-F2, RNA-directed RNA polymerase catalytic subunit | 7.405237 | 0.528279 | 0.508942 | ||
| G alpha (q) signalling events | PRKCQ | Protein kinase C, Protein kinase C theta type | 9.752014 | 0.347427 | 4 | 0.557309 | 0.523352 |
| G alpha (q) signalling events | ADRA1B | Alpha-1B adrenergic receptor | 9.752014 | 0.347427 | 4 | 0.557309 | 0.601659 |
| G alpha (q) signalling events | F2R | CWC22 homolog, spliceosome-associated protein, Proteinase-activated receptor 1 | 9.752014 | 0.347427 | 4 | 0.557309 | 0.58059 |
| G alpha (q) signalling events | GNRHR | Gonadotropin-releasing hormone receptor | 9.752014 | 0.347427 | 4 | 0.557309 | 0.689059 |
| G alpha (q) signalling events | APP | ABPP, Amyloid-beta A4 protein, Amyloid-beta precursor protein | 9.752014 | 0.347427 | 4 | 0.557309 | 0.506343 |
| G alpha (q) signalling events | F2RL3 | F2R like thrombin or trypsin receptor 3, F2R-like thrombin or trypsin receptor 3, G_PROTEIN_RECEP_F1_2 domain-containing protein, Proteinase-activated receptor 4 | 9.752014 | 0.347427 | 4 | 0.557309 | 0.537847 |
| G alpha (q) signalling events | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 9.752014 | 0.347427 | 4 | 0.557309 | 0.541254 |
| G alpha (q) signalling events | KNG1 | Bradykinin, Kininogen 1, Kininogen-1, Kininogen-1 [Cleaved into: Kininogen-1 heavy chain; Bradykinin; Kininogen-1 light chain], Uncharacterized protein | 9.752014 | 0.347427 | 4 | 0.557309 | 0.566192 |
| G alpha (q) signalling events | NMUR1 | Neuromedin U receptor 1, Neuromedin U receptor type 1, Neuromedin-U receptor 1 | 9.752014 | 0.347427 | 4 | 0.557309 | 0.62859 |
| G alpha (q) signalling events | NMUR2 | Neuromedin U receptor 2, Neuromedin U receptor type 2, Neuromedin-U receptor 2 | 9.752014 | 0.347427 | 4 | 0.557309 | 0.612605 |
| G alpha (q) signalling events | MMP3 | Stromelysin-1, ZnMc domain-containing protein | 9.752014 | 0.347427 | 4 | 0.557309 | 0.50644 |
| G alpha (q) signalling events | TACR3 | Neuromedin-K receptor, NK-3 receptor | 9.752014 | 0.347427 | 4 | 0.557309 | 0.602621 |
| G alpha (q) signalling events | PRKCA | Protein kinase C, Protein kinase C alpha type, Protein kinase C, alpha | 9.752014 | 0.347427 | 4 | 0.557309 | 0.52202 |
| G alpha (q) signalling events | CYSLTR2 | CYSLTR2 protein, Cysteinyl leukotriene receptor 2, G_PROTEIN_RECEP_F1_2 domain-containing protein | 9.752014 | 0.347427 | 4 | 0.557309 | 0.566351 |
| G alpha (q) signalling events | AGTR1 | Agtr1.2 protein, Angiotensin II receptor type 1, Type-1 angiotensin II receptor, Type-1A angiotensin II receptor | 9.752014 | 0.347427 | 4 | 0.557309 | 0.648107 |
| G alpha (q) signalling events | F2 | Activation peptide fragment 1, Prothrombin | 9.752014 | 0.347427 | 4 | 0.557309 | 0.5402 |
| G alpha (q) signalling events | TACR2 | NK-2 receptor, Substance-K receptor | 9.752014 | 0.347427 | 4 | 0.557309 | 0.601846 |
| G alpha (q) signalling events | EGFR | Epidermal growth factor receptor, Receptor protein-tyrosine kinase | 9.752014 | 0.347427 | 4 | 0.557309 | 0.625817 |
| G alpha (q) signalling events | MCHR1 | G-protein coupled receptor 24, Melanin-concentrating hormone receptor 1 | 9.752014 | 0.347427 | 4 | 0.557309 | 0.678762 |
| G alpha (q) signalling events | LTB4R2 | Leukotriene B4 receptor 2 | 9.752014 | 0.347427 | 4 | 0.557309 | 0.627297 |
| G alpha (q) signalling events | PRKCE | Protein kinase C, Protein kinase C epsilon type | 9.752014 | 0.347427 | 4 | 0.557309 | 0.512211 |
| G alpha (q) signalling events | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 9.752014 | 0.347427 | 4 | 0.557309 | 0.646924 |
| G alpha (q) signalling events | PRKCD | Protein kinase C, Protein kinase C delta type | 9.752014 | 0.347427 | 4 | 0.557309 | 0.520528 |
| G alpha (q) signalling events | CYSLTR1 | Cysteinyl leukotriene receptor 1 | 9.752014 | 0.347427 | 4 | 0.557309 | 0.552802 |
| G alpha (q) signalling events | P2RY1 | P2Y purinoceptor 1 | 9.752014 | 0.347427 | 4 | 0.557309 | 0.550301 |
| G alpha (q) signalling events | HTR2B | 5-hydroxytryptamine receptor 2B | 9.752014 | 0.347427 | 4 | 0.557309 | 0.563255 |
| G alpha (q) signalling events | BTK | Tyrosine-protein kinase, Tyrosine-protein kinase BTK | 9.752014 | 0.347427 | 4 | 0.557309 | 0.602174 |
| G alpha (q) signalling events | ADRA1A | Alpha-1A adrenergic receptor | 9.752014 | 0.347427 | 4 | 0.557309 | 0.61112 |
| G alpha (q) signalling events | OXTR | Isotocin receptor-like 2, Oxytocin receptor, Oxytocin-like receptor | 9.752014 | 0.347427 | 4 | 0.557309 | 0.526619 |
| G alpha (q) signalling events | BDKRB1 | B1 bradykinin receptor, Bradykinin receptor B1 | 9.752014 | 0.347427 | 4 | 0.557309 | 0.547577 |
| G alpha (q) signalling events | ADRA1D | Alpha-1D adrenergic receptor, Procollagen C-endopeptidase enhancer | 9.752014 | 0.347427 | 4 | 0.557309 | 0.602889 |
| G alpha (q) signalling events | TACR1 | NK-1 receptor, Substance-P receptor | 9.752014 | 0.347427 | 4 | 0.557309 | 0.738091 |
| G alpha (q) signalling events | CCKBR | Cholecystokinin-2 receptor, Gastrin/cholecystokinin type B receptor | 9.752014 | 0.347427 | 4 | 0.557309 | 0.593267 |
| G alpha (q) signalling events | BDKRB2 | B1 bradykinin receptor, B2 bradykinin receptor | 9.752014 | 0.347427 | 4 | 0.557309 | 0.518514 |
| G alpha (q) signalling events | KISS1R | G protein-coupled receptor 54, KiSS-1 receptor, KISS1 receptor | 9.752014 | 0.347427 | 4 | 0.557309 | 0.63445 |
| G alpha (q) signalling events | HTR2A | 5-hydroxytryptamine receptor 2A | 9.752014 | 0.347427 | 4 | 0.557309 | 0.628347 |
| G alpha (q) signalling events | MCHR2 | Melanin concentrating hormone receptor 2, Melanin concentrating hormone receptor 2a, Melanin-concentrating hormone receptor 2 | 9.752014 | 0.347427 | 4 | 0.557309 | 0.520996 |
| G alpha (q) signalling events | HCRTR2 | Hypocretin receptor type 2, Orexin receptor type 2 | 9.752014 | 0.347427 | 4 | 0.557309 | 0.558726 |
| G alpha (q) signalling events | GHSR | GH-releasing peptide receptor, Growth hormone secretagogue receptor type 1 | 9.752014 | 0.347427 | 4 | 0.557309 | 0.542668 |
| G alpha (q) signalling events | PRKCH | Protein kinase C, Protein kinase C eta type | 9.752014 | 0.347427 | 4 | 0.557309 | 0.506005 |
| G alpha (q) signalling events | AVPR1A | Arginine vasopressin receptor 1A, G_PROTEIN_RECEP_F1_2 domain-containing protein, Vasopressin V1a receptor | 9.752014 | 0.347427 | 4 | 0.557309 | 0.54747 |
| G alpha (q) signalling events | FPR1 | FK506-binding protein 1, fMet-Leu-Phe receptor, Formyl peptide receptor 1 | 9.752014 | 0.347427 | 4 | 0.557309 | 0.511829 |
| G alpha (q) signalling events | UTS2R | G_PROTEIN_RECEP_F1_2 domain-containing protein, Urotensin II receptor, Urotensin-2 receptor | 9.752014 | 0.347427 | 4 | 0.557309 | 0.594282 |
| G alpha (q) signalling events | PTAFR | Platelet-activating factor receptor | 9.752014 | 0.347427 | 4 | 0.557309 | 0.612734 |
| G alpha (q) signalling events | LTB4R | Leukotriene B4 receptor, Leukotriene B4 receptor 1 | 9.752014 | 0.347427 | 4 | 0.557309 | 0.604494 |
| G alpha (q) signalling events | AVPR1B | G_PROTEIN_RECEP_F1_2 domain-containing protein, Vasopressin V1b receptor | 9.752014 | 0.347427 | 4 | 0.557309 | 0.514485 |
| G alpha (q) signalling events | CCR2 | C-C chemokine receptor type 2, C-C chemokine receptor type 2 isoform B, C-C motif chemokine receptor 2, Chemokine, G_PROTEIN_RECEP_F1_2 domain-containing protein | 9.752014 | 0.347427 | 4 | 0.557309 | 0.562183 |
| G alpha (q) signalling events | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 9.752014 | 0.347427 | 4 | 0.557309 | 0.505988 |
| G alpha (q) signalling events | CASR | Calcium-sensing receptor, Extracellular calcium-sensing receptor, G_PROTEIN_RECEP_F3_4 domain-containing protein | 9.752014 | 0.347427 | 4 | 0.557309 | 0.606556 |
| G alpha (q) signalling events | CHRM3 | Muscarinic acetylcholine receptor, Muscarinic acetylcholine receptor M3 | 9.752014 | 0.347427 | 4 | 0.557309 | 0.510046 |
| G alpha (q) signalling events | HTR2C | 5-hydroxytryptamine receptor 2C | 9.752014 | 0.347427 | 4 | 0.557309 | 0.576739 |
| G alpha (q) signalling events | HCRTR1 | Hypocretin receptor 1, Orexin receptor type 1 | 9.752014 | 0.347427 | 4 | 0.557309 | 0.541349 |
| G alpha (q) signalling events | CHRM1 | Muscarinic acetylcholine receptor, Muscarinic acetylcholine receptor M1 | 9.752014 | 0.347427 | 4 | 0.557309 | 0.532293 |
| G alpha (q) signalling events | EDNRB | Endothelin receptor non-selective type, Endothelin receptor type B | 9.752014 | 0.347427 | 4 | 0.557309 | 0.518386 |
| G alpha (q) signalling events | AGTR1B | Angiotensin II receptor, type 1b | 9.752014 | 0.347427 | 4 | 0.557309 | 0.614033 |
| G alpha (q) signalling events | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.752014 | 0.347427 | 4 | 0.557309 | 0.660919 |
| G alpha (q) signalling events | HRH1 | Histamine H1 receptor | 9.752014 | 0.347427 | 4 | 0.557309 | 0.518086 |
| G alpha (q) signalling events | CCKAR | Cholecystokinin receptor type A, Cholecystokinin-2 receptor | 9.752014 | 0.347427 | 4 | 0.557309 | 0.582813 |
| G alpha (s) signalling events | HTR7 | 5-hydroxytryptamine receptor 7, 5-hydroxytryptamine receptor 7 transcript variant 1 | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.601446 |
| G alpha (s) signalling events | CYSLTR1 | Cysteinyl leukotriene receptor 1 | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.552802 |
| G alpha (s) signalling events | FPR1 | FK506-binding protein 1, fMet-Leu-Phe receptor, Formyl peptide receptor 1 | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.511829 |
| G alpha (s) signalling events | MC5R | Melanocortin receptor 5, mRNA cap guanine-N7 methyltransferase | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.501364 |
| G alpha (s) signalling events | CRHR2 | Corticotropin releasing hormone receptor 2, Corticotropin-releasing factor receptor 2, Corticotropin-releasing hormone receptor 2, Uncharacterized protein | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.569524 |
| G alpha (s) signalling events | PDE10A | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A, Histone acetyltransferase, Phosphodiesterase | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.660672 |
| G alpha (s) signalling events | ADM2 | Adrenomedullin 2, Protein ADM2 | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.537164 |
| G alpha (s) signalling events | HTR6 | 5-hydroxytryptamine, 5-hydroxytryptamine receptor 6, G_PROTEIN_RECEP_F1_2 domain-containing protein | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.508125 |
| G alpha (s) signalling events | ADCY9 | Adenylate cyclase 9, Adenylate cyclase type 9 | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.550534 |
| G alpha (s) signalling events | VIPR2 | Vasoactive intestinal peptide receptor 2, Vasoactive intestinal polypeptide receptor 2 | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.5519 |
| G alpha (s) signalling events | AVPR2 | AVPR V2, Vasopressin V2 receptor | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.569629 |
| G alpha (s) signalling events | GPBAR1 | G protein-coupled bile acid receptor 1, G-protein coupled bile acid receptor 1 | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.520207 |
| G alpha (s) signalling events | MC4R | Melanocortin receptor 4 | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.598533 |
| G alpha (s) signalling events | PDE4A | cAMP-specific 3',5'-cyclic phosphodiesterase 4A, Phosphodiesterase | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.54461 |
| G alpha (s) signalling events | CRHR1 | Corticotropin-releasing factor receptor 1 | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.722459 |
| G alpha (s) signalling events | ADCYAP1 | GLUCAGON domain-containing protein, PACAP-related peptide, Pituitary adenylate cyclase-activating polypeptide | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.549868 |
| G alpha (s) signalling events | MC1R | Melanocyte-stimulating hormone receptor | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.526117 |
| G alpha (s) signalling events | GLP1R | Glucagon-like peptide 1 receptor | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.586361 |
| G alpha (s) signalling events | CYSLTR2 | CYSLTR2 protein, Cysteinyl leukotriene receptor 2, G_PROTEIN_RECEP_F1_2 domain-containing protein | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.566351 |
| G alpha (s) signalling events | CALCRL | Calcitonin gene-related peptide type 1 receptor | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.56686 |
| G alpha (s) signalling events | GLP2R | Glucagon like peptide 2 receptor, Glucagon-like peptide 2 receptor | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.677329 |
| G alpha (s) signalling events | GNAI2 | G protein subunit alpha i2, Guanine nucleotide binding protein, Guanine nucleotide-binding protein G(i) subunit alpha-2 | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.512918 |
| G alpha (s) signalling events | ADCY7 | Adenylate cyclase type 7 | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.550655 |
| G alpha (s) signalling events | CALCA | Calcitonin, Calcitonin [Cleaved into: Calcitonin; Katacalcin, Calcitonin gene-related peptide 1, Calcitonin/calcitonin-related polypeptide, alpha | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.535129 |
| G alpha (s) signalling events | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.594807 |
| G alpha (s) signalling events | DRD5 | D(1B) dopamine receptor, Dopamine receptor D5, G_PROTEIN_RECEP_F1_2 domain-containing protein | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.551596 |
| G alpha (s) signalling events | PDE4C | cAMP-specific 3',5'-cyclic phosphodiesterase 4C, Phosphodiesterase | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.526451 |
| G alpha (s) signalling events | RAMP1 | Receptor activity modifying protein 1, Receptor activity-modifying protein 1, Zgc:91818 | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.582824 |
| G alpha (s) signalling events | DRD1 | D(1A) dopamine receptor, Dopamine receptor D1, G_PROTEIN_RECEP_F1_2 domain-containing protein | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.557448 |
| G alpha (s) signalling events | ADCYAP1R1 | ADCYAP receptor type I, Pituitary adenylate cyclase-activating polypeptide type I receptor, Type I adenylate cyclase activating polypeptide receptor, Uncharacterized protein | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.594673 |
| G alpha (s) signalling events | PDE4D | cAMP-specific 3',5'-cyclic phosphodiesterase 4D, Phosphodiesterase | 9.29322 | 0.353249 | 3.271 | 0.438765 | 0.535211 |
| G alpha (z) signalling events | ADRA2A | Adrenoceptor alpha 2A, Alpha-2A adrenergic receptor | 9.387855 | 0.575049 | 4 | 0.652334 | 0.530746 |
| G alpha (z) signalling events | ADRA2C | Adrenoceptor alpha 2C, Alpha-2C adrenergic receptor | 9.387855 | 0.575049 | 4 | 0.652334 | 0.528655 |
| G alpha (z) signalling events | PRKCD | Protein kinase C, Protein kinase C delta type | 9.387855 | 0.575049 | 4 | 0.652334 | 0.520528 |
| G alpha (z) signalling events | GNAI2 | G protein subunit alpha i2, Guanine nucleotide binding protein, Guanine nucleotide-binding protein G(i) subunit alpha-2 | 9.387855 | 0.575049 | 4 | 0.652334 | 0.512918 |
| G alpha (z) signalling events | ADCY9 | Adenylate cyclase 9, Adenylate cyclase type 9 | 9.387855 | 0.575049 | 4 | 0.652334 | 0.550534 |
| G alpha (z) signalling events | PRKCB | Protein kinase C, Protein kinase C beta type | 9.387855 | 0.575049 | 4 | 0.652334 | 0.520598 |
| G alpha (z) signalling events | ADCY7 | Adenylate cyclase type 7 | 9.387855 | 0.575049 | 4 | 0.652334 | 0.550655 |
| G alpha (z) signalling events | PRKCA | Protein kinase C, Protein kinase C alpha type, Protein kinase C, alpha | 9.387855 | 0.575049 | 4 | 0.652334 | 0.52202 |
| G alpha (z) signalling events | PRKCQ | Protein kinase C, Protein kinase C theta type | 9.387855 | 0.575049 | 4 | 0.652334 | 0.523352 |
| G alpha (z) signalling events | PRKCH | Protein kinase C, Protein kinase C eta type | 9.387855 | 0.575049 | 4 | 0.652334 | 0.506005 |
| G alpha (z) signalling events | PRKCE | Protein kinase C, Protein kinase C epsilon type | 9.387855 | 0.575049 | 4 | 0.652334 | 0.512211 |
| G alpha (z) signalling events | ADRA2B | Adrenoceptor alpha 2B, Alpha-2B adrenergic receptor, G_PROTEIN_RECEP_F1_2 domain-containing protein | 9.387855 | 0.575049 | 4 | 0.652334 | 0.524078 |
| G alpha (z) signalling events | PRKCG | Protein kinase C, Protein kinase C gamma type, Sperm-associated antigen 9 | 9.387855 | 0.575049 | 4 | 0.652334 | 0.500344 |
| G2/M Checkpoints | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.54735 |
| G2/M Checkpoints | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.577481 |
| G2/M Checkpoints | PDE10A | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A, Histone acetyltransferase, Phosphodiesterase | 9.727017 | 0.525579 | 4 | 0.645096 | 0.660672 |
| G2/M Checkpoints | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.515416 |
| G2/M Checkpoints | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.5197 |
| G2/M Checkpoints | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.727017 | 0.525579 | 4 | 0.645096 | 0.579609 |
| G2/M Checkpoints | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.573417 |
| G2/M Checkpoints | MCM4 | DNA helicase, DNA replication licensing factor mcm4, DNA replication licensing factor MCM4 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.515269 |
| G2/M Checkpoints | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.54735 |
| G2/M Checkpoints | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.54735 |
| G2/M Checkpoints | ATR | Non-specific serine/threonine protein kinase, Serine/threonine-protein kinase ATR | 9.727017 | 0.525579 | 4 | 0.645096 | 0.518847 |
| G2/M Checkpoints | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.515416 |
| G2/M Checkpoints | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.515416 |
| G2/M Checkpoints | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.54735 |
| G2/M Checkpoints | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.515416 |
| G2/M Checkpoints | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.58858 |
| G2/M Checkpoints | TP53 | Cellular tumor antigen p53 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.525937 |
| G2/M Checkpoints | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.727017 | 0.525579 | 4 | 0.645096 | 0.568645 |
| G2/M Checkpoints | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.546881 |
| G2/M Checkpoints | YWHAB | 14-3-3 protein beta/alpha, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta | 9.727017 | 0.525579 | 4 | 0.645096 | 0.513843 |
| G2/M Checkpoints | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.54735 |
| G2/M Checkpoints | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.54735 |
| G2/M Checkpoints | YWHAZ | 14_3_3 domain-containing protein, 14-3-3 protein zeta, 14-3-3 protein zeta/delta, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta | 9.727017 | 0.525579 | 4 | 0.645096 | 0.602005 |
| G2/M Checkpoints | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.515416 |
| G2/M Checkpoints | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.54735 |
| G2/M Checkpoints | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.515416 |
| G2/M Checkpoints | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.572294 |
| G2/M Checkpoints | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.57244 |
| G2/M Checkpoints | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.54735 |
| G2/M Checkpoints | CRK | 14_3_3 domain-containing protein, Adapter molecule crk, Adapter molecule Crk, CRK proto-oncogene, adaptor protein, Uncharacterized protein, V-crk avian sarcoma virus CT10 oncogene homolog | 9.727017 | 0.525579 | 4 | 0.645096 | 0.513369 |
| G2/M Checkpoints | CCNB1 | Cyclin B1, G2/mitotic-specific cyclin-B1, Uncharacterized protein | 9.727017 | 0.525579 | 4 | 0.645096 | 0.513205 |
| G2/M Checkpoints | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.54735 |
| G2/M Checkpoints | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.515416 |
| G2/M Checkpoints | BLM | Bloom syndrome protein, Bloom syndrome protein homolog, DNA helicase | 9.727017 | 0.525579 | 4 | 0.645096 | 0.509426 |
| G2/M Checkpoints | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.577847 |
| G2/M Checkpoints | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.727017 | 0.525579 | 4 | 0.645096 | 0.515416 |
| G2/M Checkpoints | CDK1 | Cell division control protein 2, Cyclin dependent kinase 1, Cyclin-dependent kinase 1 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.514008 |
| G2/M Checkpoints | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.515416 |
| G2/M Checkpoints | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.727017 | 0.525579 | 4 | 0.645096 | 0.515416 |
| G2/M Checkpoints | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.565601 |
| G2/M Checkpoints | PTK2 | Focal adhesion kinase 1, Non-specific protein-tyrosine kinase, Serine/threonine-protein kinase PTK2/STK2 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.503121 |
| G2/M Checkpoints | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.515416 |
| G2/M Checkpoints | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.54735 |
| G2/M Checkpoints | ATRIP | ATR interacting protein, ATR-interacting protein, Uncharacterized protein | 9.727017 | 0.525579 | 4 | 0.645096 | 0.54377 |
| G2/M Checkpoints | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.515416 |
| G2/M Checkpoints | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.515416 |
| G2/M Checkpoints | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.515416 |
| G2/M Checkpoints | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.515416 |
| G2/M Checkpoints | CDK2 | Cyclin dependent kinase 2, Cyclin-dependent kinase 2, Protein kinase domain-containing protein, Uncharacterized protein | 9.727017 | 0.525579 | 4 | 0.645096 | 0.532011 |
| G2/M Checkpoints | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.727017 | 0.525579 | 4 | 0.645096 | 0.547953 |
| GABA receptor activation | GABRA1 | GABA(A) receptor subunit alpha-1, Gamma-aminobutyric acid receptor subunit alpha-1 | 8.262772 | 0.49141 | 4 | 0.552472 | 0.508013 |
| GABA receptor activation | GABRA3 | Gamma-aminobutyric acid, Gamma-aminobutyric acid receptor subunit alpha-3, Gamma-aminobutyric acid type A receptor alpha3 subunit, Gamma-aminobutyric acid type A receptor subunit alpha3, Uncharacterized protein | 8.262772 | 0.49141 | 4 | 0.552472 | 0.516173 |
| GABA receptor activation | GNAI2 | G protein subunit alpha i2, Guanine nucleotide binding protein, Guanine nucleotide-binding protein G(i) subunit alpha-2 | 8.262772 | 0.49141 | 4 | 0.552472 | 0.512918 |
| GABA receptor activation | KCNJ5 | Cardiac inward rectifier, G protein-activated inward rectifier potassium channel 4, Potassium inwardly rectifying channel subfamily J member 5 | 8.262772 | 0.49141 | 4 | 0.552472 | 0.565449 |
| GABA receptor activation | GABRG2 | GABA(A) receptor subunit gamma-2, Gamma-aminobutyric acid, Gamma-aminobutyric acid receptor subunit gamma-2, Gamma-aminobutyric acid type A receptor subunit gamma2 | 8.262772 | 0.49141 | 4 | 0.552472 | 0.522585 |
| GABA receptor activation | ADCY7 | Adenylate cyclase type 7 | 8.262772 | 0.49141 | 4 | 0.552472 | 0.550655 |
| GABA receptor activation | GABRA5 | Gamma-aminobutyric acid, Gamma-aminobutyric acid receptor subunit alpha-5, Gamma-aminobutyric acid type A receptor alpha5 subunit, Gamma-aminobutyric acid type A receptor subunit alpha5, Uncharacterized protein | 8.262772 | 0.49141 | 4 | 0.552472 | 0.519175 |
| GABA receptor activation | ADCY9 | Adenylate cyclase 9, Adenylate cyclase type 9 | 8.262772 | 0.49141 | 4 | 0.552472 | 0.550534 |
| GABA receptor activation | GABRA2 | GABA(A) receptor subunit alpha-2, Gamma-aminobutyric acid receptor subunit alpha-2 | 8.262772 | 0.49141 | 4 | 0.552472 | 0.519767 |
| GABA receptor activation | GABBR2 | G_PROTEIN_RECEP_F3_4 domain-containing protein, Gabbr2 protein, Gamma-aminobutyric acid, Gamma-aminobutyric acid type B receptor subunit 2 | 8.262772 | 0.49141 | 4 | 0.552472 | 0.542461 |
| GABA receptor activation | KCNJ3 | G protein-activated inward rectifier potassium channel 1 | 8.262772 | 0.49141 | 4 | 0.552472 | 0.56031 |
| GABA receptor activation | GABBR1 | Gamma-aminobutyric acid, Gamma-aminobutyric acid type B receptor subunit 1 | 8.262772 | 0.49141 | 4 | 0.552472 | 0.528076 |
| GABA receptor activation | GABRA6 | Gamma-aminobutyric acid receptor subunit alpha-6, Gamma-aminobutyric acid type A receptor alpha6 subunit, Gamma-aminobutyric acid type A receptor subunit alpha6 | 8.262772 | 0.49141 | 4 | 0.552472 | 0.50028 |
| GABA receptor activation | GABRB3 | Gabrb3 protein, Gamma-aminobutyric acid receptor subunit beta-3, Gamma-aminobutyric acid type A receptor beta3 subunit, Gamma-aminobutyric acid type A receptor subunit beta3 | 8.262772 | 0.49141 | 4 | 0.552472 | 0.53034 |
| Gastrin-CREB signalling pathway via PKC and MAPK | EGFR | Epidermal growth factor receptor, Receptor protein-tyrosine kinase | 8.333876 | 0.545064 | 2.827883 | 0.412141 | 0.625817 |
| Gastrin-CREB signalling pathway via PKC and MAPK | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 8.333876 | 0.545064 | 2.827883 | 0.412141 | 0.541254 |
| Gastrin-CREB signalling pathway via PKC and MAPK | CCKBR | Cholecystokinin-2 receptor, Gastrin/cholecystokinin type B receptor | 8.333876 | 0.545064 | 2.827883 | 0.412141 | 0.593267 |
| Gastrin-CREB signalling pathway via PKC and MAPK | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 8.333876 | 0.545064 | 2.827883 | 0.412141 | 0.505988 |
| Gastrin-CREB signalling pathway via PKC and MAPK | PRKCA | Protein kinase C, Protein kinase C alpha type, Protein kinase C, alpha | 8.333876 | 0.545064 | 2.827883 | 0.412141 | 0.52202 |
| Gastrin-CREB signalling pathway via PKC and MAPK | MMP3 | Stromelysin-1, ZnMc domain-containing protein | 8.333876 | 0.545064 | 2.827883 | 0.412141 | 0.50644 |
| Generic Transcription Pathway | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.54735 |
| Generic Transcription Pathway | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.57244 |
| Generic Transcription Pathway | ATRIP | ATR interacting protein, ATR-interacting protein, Uncharacterized protein | 7.880884 | 0.348754 | 4 | 0.461443 | 0.54377 |
| Generic Transcription Pathway | HDAC7 | Hist_deacetyl domain-containing protein, Histone deacetylase, Histone deacetylase 7 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.522826 |
| Generic Transcription Pathway | KMT2A | Histone-lysine N-methyltransferase, Histone-lysine N-methyltransferase 2A | 7.880884 | 0.348754 | 4 | 0.461443 | 0.518598 |
| Generic Transcription Pathway | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 7.880884 | 0.348754 | 4 | 0.461443 | 0.552041 |
| Generic Transcription Pathway | ERBB2 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-2 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.589497 |
| Generic Transcription Pathway | NOTCH3 | Neurogenic locus notch homolog protein 3 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.555427 |
| Generic Transcription Pathway | MAPK14 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 14 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.591941 |
| Generic Transcription Pathway | CDK4 | Cyclin dependent kinase 4, Cyclin-dependent kinase 4, Cyclin-dependent kinase 4, isoform A | 7.880884 | 0.348754 | 4 | 0.461443 | 0.511296 |
| Generic Transcription Pathway | EZH2 | [Histone H3]-lysine(27) N-trimethyltransferase, Histone-lysine N-methyltransferase EZH2 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.53644 |
| Generic Transcription Pathway | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.515416 |
| Generic Transcription Pathway | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 7.880884 | 0.348754 | 4 | 0.461443 | 0.515416 |
| Generic Transcription Pathway | VDRA | Vitamin D3 receptor A | 7.880884 | 0.348754 | 4 | 0.461443 | 0.529536 |
| Generic Transcription Pathway | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.54735 |
| Generic Transcription Pathway | CCNT1 | Cyclin-T1 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.643075 |
| Generic Transcription Pathway | GRIN2A | Glutamate receptor, Glutamate receptor ionotropic, NMDA 2A | 7.880884 | 0.348754 | 4 | 0.461443 | 0.527756 |
| Generic Transcription Pathway | OPRM1 | Mu-type opioid receptor | 7.880884 | 0.348754 | 4 | 0.461443 | 0.614279 |
| Generic Transcription Pathway | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.565601 |
| Generic Transcription Pathway | REP | Rab proteins geranylgeranyltransferase component A, Replicase polyprotein 1ab | 7.880884 | 0.348754 | 4 | 0.461443 | 0.746505 |
| Generic Transcription Pathway | CCNA2 | Cyclin-A2 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.527977 |
| Generic Transcription Pathway | ABL1 | Tyrosine-protein kinase, Tyrosine-protein kinase ABL1 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.590046 |
| Generic Transcription Pathway | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.515416 |
| Generic Transcription Pathway | NR3C1 | Glucocorticoid receptor | 7.880884 | 0.348754 | 4 | 0.461443 | 0.59851 |
| Generic Transcription Pathway | CDK6 | Cyclin dependent kinase 6, Cyclin-dependent kinase, Cyclin-dependent kinase 6 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.509686 |
| Generic Transcription Pathway | HDAC6 | Histone deacetylase 6, Histone deacetylase 6, isoform G | 7.880884 | 0.348754 | 4 | 0.461443 | 0.531859 |
| Generic Transcription Pathway | CSF1R | Macrophage colony-stimulating factor 1 receptor, Receptor protein-tyrosine kinase | 7.880884 | 0.348754 | 4 | 0.461443 | 0.517773 |
| Generic Transcription Pathway | NR1H3 | Nuclear receptor subfamily 1 group H member 3, Nuclear receptor subfamily 1, group H, member 3, Oxysterols receptor LXR-alpha, Uncharacterized protein | 7.880884 | 0.348754 | 4 | 0.461443 | 0.501369 |
| Generic Transcription Pathway | PGR | Nuclear receptor subfamily 3 group C member 3, Progesterone receptor | 7.880884 | 0.348754 | 4 | 0.461443 | 0.5833 |
| Generic Transcription Pathway | BLM | Bloom syndrome protein, Bloom syndrome protein homolog, DNA helicase | 7.880884 | 0.348754 | 4 | 0.461443 | 0.509426 |
| Generic Transcription Pathway | CCNE2 | Cyclin E2, Cyclin N-terminal domain-containing protein, G1/S-specific cyclin-E2 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.503983 |
| Generic Transcription Pathway | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.54735 |
| Generic Transcription Pathway | RORC | Nuclear receptor ROR-gamma, RAR related orphan receptor C, RAR-related orphan receptor C, Uncharacterized protein | 7.880884 | 0.348754 | 4 | 0.461443 | 0.556066 |
| Generic Transcription Pathway | ESR2 | Estrogen receptor 2, Estrogen receptor beta | 7.880884 | 0.348754 | 4 | 0.461443 | 0.53484 |
| Generic Transcription Pathway | ESR1 | Estrogen receptor | 7.880884 | 0.348754 | 4 | 0.461443 | 0.561602 |
| Generic Transcription Pathway | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.515416 |
| Generic Transcription Pathway | CCND3 | Cyclin D3, Cyclin N-terminal domain-containing protein, G1/S-specific cyclin-D3 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.562307 |
| Generic Transcription Pathway | CCNE1 | Cyclin E1, Cyclin N-terminal domain-containing protein, G1/S-specific cyclin-E1 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.526741 |
| Generic Transcription Pathway | HDAC8 | Histone deacetylase, Histone deacetylase 8, WD repeat and SOCS box containing 2 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.535733 |
| Generic Transcription Pathway | CCND1 | Cyclin D1, G1/S-specific cyclin-D1 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.525654 |
| Generic Transcription Pathway | YWHAZ | 14_3_3 domain-containing protein, 14-3-3 protein zeta, 14-3-3 protein zeta/delta, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta | 7.880884 | 0.348754 | 4 | 0.461443 | 0.602005 |
| Generic Transcription Pathway | MDM4 | Double minute 4 protein, Protein Mdm4 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.510741 |
| Generic Transcription Pathway | HIPK1 | Homeodomain interacting protein kinase 1, Homeodomain-interacting protein kinase 1 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.512719 |
| Generic Transcription Pathway | HDAC10 | Histone deacetylase 10, Polyamine deacetylase HDAC10 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.525881 |
| Generic Transcription Pathway | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 7.880884 | 0.348754 | 4 | 0.461443 | 0.515416 |
| Generic Transcription Pathway | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.541254 |
| Generic Transcription Pathway | YAP1 | Transcriptional coactivator YAP1, Yes1 associated transcriptional regulator | 7.880884 | 0.348754 | 4 | 0.461443 | 0.628833 |
| Generic Transcription Pathway | CSNK2A2 | Casein kinase 2 alpha 2, Casein kinase II subunit alpha' | 7.880884 | 0.348754 | 4 | 0.461443 | 0.53622 |
| Generic Transcription Pathway | PLK2 | Serine/threonine-protein kinase PLK, Serine/threonine-protein kinase PLK2 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.533485 |
| Generic Transcription Pathway | CDK1 | Cell division control protein 2, Cyclin dependent kinase 1, Cyclin-dependent kinase 1 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.514008 |
| Generic Transcription Pathway | TCF7L2 | HMG box domain-containing protein, Transcription factor 7 like 2, Transcription factor 7-like 2 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.688556 |
| Generic Transcription Pathway | MTOR | Nucleoprotein TPR, Serine/threonine-protein kinase mTOR, Serine/threonine-protein kinase TOR | 7.880884 | 0.348754 | 4 | 0.461443 | 0.619026 |
| Generic Transcription Pathway | CDK2 | Cyclin dependent kinase 2, Cyclin-dependent kinase 2, Protein kinase domain-containing protein, Uncharacterized protein | 7.880884 | 0.348754 | 4 | 0.461443 | 0.532011 |
| Generic Transcription Pathway | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.54735 |
| Generic Transcription Pathway | ATR | Non-specific serine/threonine protein kinase, Serine/threonine-protein kinase ATR | 7.880884 | 0.348754 | 4 | 0.461443 | 0.518847 |
| Generic Transcription Pathway | MEN1 | Actin like 6A, Menin | 7.880884 | 0.348754 | 4 | 0.461443 | 0.517021 |
| Generic Transcription Pathway | RPTOR | Regulatory associated protein of MTOR complex 1, Regulatory associated protein of MTOR, complex 1, Regulatory-associated protein of mTOR, Regulatory-associated protein of MTOR, complex 1, WD_REPEATS_REGION domain-containing protein | 7.880884 | 0.348754 | 4 | 0.461443 | 0.543357 |
| Generic Transcription Pathway | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.515416 |
| Generic Transcription Pathway | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.54735 |
| Generic Transcription Pathway | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.589663 |
| Generic Transcription Pathway | PRKAB1 | 5'-AMP-activated protein kinase subunit beta-1, 5'AMP-activated protein kinase beta-1 non-catalytic subunit, Protein kinase AMP-activated non-catalytic subunit beta 1, Protein kinase, AMP-activated, beta 1 non-catalytic subunit | 7.880884 | 0.348754 | 4 | 0.461443 | 0.500481 |
| Generic Transcription Pathway | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.515416 |
| Generic Transcription Pathway | POLG | Bromodomain and PHD finger containing 1, Genome polyprotein [Cleaved into: Core protein precursor | 7.880884 | 0.348754 | 4 | 0.461443 | 0.681085 |
| Generic Transcription Pathway | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.572294 |
| Generic Transcription Pathway | EGFR | Epidermal growth factor receptor, Receptor protein-tyrosine kinase | 7.880884 | 0.348754 | 4 | 0.461443 | 0.625817 |
| Generic Transcription Pathway | NOTCH1 | Neurogenic locus notch homolog protein 1 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.591875 |
| Generic Transcription Pathway | TAOK2 | TAO kinase 2 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.511145 |
| Generic Transcription Pathway | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.515416 |
| Generic Transcription Pathway | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.58858 |
| Generic Transcription Pathway | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.5197 |
| Generic Transcription Pathway | ITGAL | Integrin alpha-L, Integrin subunit alpha L | 7.880884 | 0.348754 | 4 | 0.461443 | 0.590214 |
| Generic Transcription Pathway | HDAC4 | Histone deacetylase, Histone deacetylase 4 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.528897 |
| Generic Transcription Pathway | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 7.880884 | 0.348754 | 4 | 0.461443 | 0.579609 |
| Generic Transcription Pathway | KIT | Mast/stem cell growth factor receptor, Mast/stem cell growth factor receptor Kit | 7.880884 | 0.348754 | 4 | 0.461443 | 0.562353 |
| Generic Transcription Pathway | CTSL | Cathepsin L1 isoform 1 preproprotein, Procathepsin L | 7.880884 | 0.348754 | 4 | 0.461443 | 0.51103 |
| Generic Transcription Pathway | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.573417 |
| Generic Transcription Pathway | EHMT2 | Euchromatic histone lysine methyltransferase 2, Euchromatic histone-lysine N-methyltransferase 2, Histone-lysine N-methyltransferase EHMT2 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.520106 |
| Generic Transcription Pathway | BBC3 | Bcl-2-binding component 3, isoforms 1/2 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.520237 |
| Generic Transcription Pathway | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.547953 |
| Generic Transcription Pathway | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 7.880884 | 0.348754 | 4 | 0.461443 | 0.594807 |
| Generic Transcription Pathway | CDK9 | CDK9, Cyclin dependent kinase 9, Cyclin-dependent kinase 9, Probable cyclin-dependent kinase 9 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.528649 |
| Generic Transcription Pathway | HDAC2 | Histone deacetylase 2 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.52963 |
| Generic Transcription Pathway | NR1H2 | Nuclear receptor subfamily 1 group H member 2, Oxysterols receptor LXR-beta | 7.880884 | 0.348754 | 4 | 0.461443 | 0.525711 |
| Generic Transcription Pathway | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.515416 |
| Generic Transcription Pathway | VDR | 1,25-dihydroxyvitamin D3 receptor, Vitamin D receptor, Vitamin D3 receptor | 7.880884 | 0.348754 | 4 | 0.461443 | 0.518981 |
| Generic Transcription Pathway | ZNF664 | Zinc finger protein 664 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.502655 |
| Generic Transcription Pathway | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.515416 |
| Generic Transcription Pathway | PRKCQ | Protein kinase C, Protein kinase C theta type | 7.880884 | 0.348754 | 4 | 0.461443 | 0.523352 |
| Generic Transcription Pathway | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.515416 |
| Generic Transcription Pathway | CBX6 | Chromo domain-containing protein, Chromobox 6, Chromobox protein homolog 6 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.575593 |
| Generic Transcription Pathway | MET | Hepatocyte growth factor receptor, HGF/SF receptor | 7.880884 | 0.348754 | 4 | 0.461443 | 0.5914 |
| Generic Transcription Pathway | TP53RK | EKC/KEOPS complex subunit Tp53rk, EKC/KEOPS complex subunit TP53RK, Non-specific serine/threonine protein kinase | 7.880884 | 0.348754 | 4 | 0.461443 | 0.504969 |
| Generic Transcription Pathway | GRIN2B | Glutamate receptor, Glutamate receptor ionotropic, NMDA 2B | 7.880884 | 0.348754 | 4 | 0.461443 | 0.56443 |
| Generic Transcription Pathway | PRKAG2 | 5'-AMP-activated protein kinase subunit gamma-2, 5'-AMP-activated protein kinase subunit gamma-2 isoform X8, Protein kinase AMP-activated non-catalytic subunit gamma 2, Protein kinase, AMP-activated, gamma 2 non-catalytic subunit, Uncharacterized protein | 7.880884 | 0.348754 | 4 | 0.461443 | 0.510193 |
| Generic Transcription Pathway | HDAC3 | Histone deacetylase, Histone deacetylase 3 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.528637 |
| Generic Transcription Pathway | YES1 | Tyrosine-protein kinase, Tyrosine-protein kinase yes, Tyrosine-protein kinase Yes | 7.880884 | 0.348754 | 4 | 0.461443 | 0.506641 |
| Generic Transcription Pathway | OPRK1 | Kappa-type opioid receptor, Opioid receptor kappa 1, Opioid receptor, kappa 1 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.590275 |
| Generic Transcription Pathway | CHD4 | Chromodomain-helicase-DNA-binding protein 4, DNA helicase | 7.880884 | 0.348754 | 4 | 0.461443 | 0.566824 |
| Generic Transcription Pathway | YWHAB | 14-3-3 protein beta/alpha, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta | 7.880884 | 0.348754 | 4 | 0.461443 | 0.513843 |
| Generic Transcription Pathway | CSNK2A1 | Casein kinase 2, alpha 1 polypeptide, Casein kinase II subunit alpha, Protein kinase domain-containing protein | 7.880884 | 0.348754 | 4 | 0.461443 | 0.538737 |
| Generic Transcription Pathway | CTNNB1 | Catenin, Catenin beta-1, Uncharacterized protein | 7.880884 | 0.348754 | 4 | 0.461443 | 0.657916 |
| Generic Transcription Pathway | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.515416 |
| Generic Transcription Pathway | CTSK | Cathepsin K | 7.880884 | 0.348754 | 4 | 0.461443 | 0.511213 |
| Generic Transcription Pathway | MMP13 | Collagenase 3 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.515873 |
| Generic Transcription Pathway | PDE10A | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A, Histone acetyltransferase, Phosphodiesterase | 7.880884 | 0.348754 | 4 | 0.461443 | 0.660672 |
| Generic Transcription Pathway | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.54735 |
| Generic Transcription Pathway | HDAC11 | Hdac11 protein, Hist_deacetyl domain-containing protein, Histone deacetylase 11, Histone deacetylase 11, isoform A | 7.880884 | 0.348754 | 4 | 0.461443 | 0.524904 |
| Generic Transcription Pathway | MAPK11 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 11 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.567539 |
| Generic Transcription Pathway | AURKB | Aurora kinase, Aurora kinase B | 7.880884 | 0.348754 | 4 | 0.461443 | 0.524412 |
| Generic Transcription Pathway | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.54735 |
| Generic Transcription Pathway | TEAD1 | TEA domain transcription factor 1, TEA domain-containing protein, Transcriptional enhancer factor TEF-1 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.632738 |
| Generic Transcription Pathway | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.515416 |
| Generic Transcription Pathway | MDM2 | E3 ubiquitin-protein ligase Mdm2 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.659771 |
| Generic Transcription Pathway | PRKCB | Protein kinase C, Protein kinase C beta type | 7.880884 | 0.348754 | 4 | 0.461443 | 0.520598 |
| Generic Transcription Pathway | CCNC | Cyclin-C | 7.880884 | 0.348754 | 4 | 0.461443 | 0.503295 |
| Generic Transcription Pathway | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.54735 |
| Generic Transcription Pathway | TP53 | Cellular tumor antigen p53 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.525937 |
| Generic Transcription Pathway | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.577481 |
| Generic Transcription Pathway | HDAC9 | Histone deacetylase, Histone deacetylase 9 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.525623 |
| Generic Transcription Pathway | CCNB1 | Cyclin B1, G2/mitotic-specific cyclin-B1, Uncharacterized protein | 7.880884 | 0.348754 | 4 | 0.461443 | 0.513205 |
| Generic Transcription Pathway | HDAC5 | Histone deacetylase, Histone deacetylase 5 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.523224 |
| Generic Transcription Pathway | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.577847 |
| Generic Transcription Pathway | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.515416 |
| Generic Transcription Pathway | STK11 | Non-specific serine/threonine protein kinase, Serine/threonine-protein kinase STK11 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.504599 |
| Generic Transcription Pathway | CRK | 14_3_3 domain-containing protein, Adapter molecule crk, Adapter molecule Crk, CRK proto-oncogene, adaptor protein, Uncharacterized protein, V-crk avian sarcoma virus CT10 oncogene homolog | 7.880884 | 0.348754 | 4 | 0.461443 | 0.513369 |
| Generic Transcription Pathway | RBPJ | Recombination signal binding protein for immunoglobulin kappa J region, Recombination signal-binding protein for immunoglobulin kappa J region, Recombining binding protein suppressor of hairless, Uncharacterized protein | 7.880884 | 0.348754 | 4 | 0.461443 | 0.647854 |
| Generic Transcription Pathway | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.54735 |
| Generic Transcription Pathway | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.515416 |
| Generic Transcription Pathway | SMAD3 | Mothers against decapentaplegic homolog, Mothers against decapentaplegic homolog 3 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.502216 |
| Generic Transcription Pathway | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.546881 |
| Generic Transcription Pathway | HDAC1 | Histone deacetylase 1, Histone deacetylase HDAC1 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.540105 |
| Generic Transcription Pathway | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.54735 |
| Generic Transcription Pathway | AURKA | Aurora kinase, Aurora kinase A | 7.880884 | 0.348754 | 4 | 0.461443 | 0.54457 |
| Generic Transcription Pathway | CCNA1 | Cyclin A1, Cyclin-A1, Cyclin-A2 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.506777 |
| Generic Transcription Pathway | PPARGC1A | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | 7.880884 | 0.348754 | 4 | 0.461443 | 0.539093 |
| Generic Transcription Pathway | PTPN1 | Protein-tyrosine-phosphatase, Tyrosine-protein phosphatase non-receptor type, Tyrosine-protein phosphatase non-receptor type 1 | 7.880884 | 0.348754 | 4 | 0.461443 | 0.510671 |
| Generic Transcription Pathway | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 7.880884 | 0.348754 | 4 | 0.461443 | 0.568645 |
| Generic Transcription Pathway | CSNK2B | Casein kinase II subunit beta | 7.880884 | 0.348754 | 4 | 0.461443 | 0.537688 |
| Generic Transcription Pathway | PRKAG1 | 5'-AMP-activated protein kinase subunit gamma-1, Protein kinase, AMP-activated, gamma 1 non-catalytic subunit | 7.880884 | 0.348754 | 4 | 0.461443 | 0.515113 |
| GLI3 is processed to GLI3R by the proteasome | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.515416 |
| GLI3 is processed to GLI3R by the proteasome | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.54735 |
| GLI3 is processed to GLI3R by the proteasome | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.572294 |
| GLI3 is processed to GLI3R by the proteasome | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.573417 |
| GLI3 is processed to GLI3R by the proteasome | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.515416 |
| GLI3 is processed to GLI3R by the proteasome | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.515416 |
| GLI3 is processed to GLI3R by the proteasome | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.515416 |
| GLI3 is processed to GLI3R by the proteasome | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.515416 |
| GLI3 is processed to GLI3R by the proteasome | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.515416 |
| GLI3 is processed to GLI3R by the proteasome | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.401417 | 0.70503 | 4 | 0.718024 | 0.515416 |
| GLI3 is processed to GLI3R by the proteasome | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.565601 |
| GLI3 is processed to GLI3R by the proteasome | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.54735 |
| GLI3 is processed to GLI3R by the proteasome | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.401417 | 0.70503 | 4 | 0.718024 | 0.579609 |
| GLI3 is processed to GLI3R by the proteasome | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.515416 |
| GLI3 is processed to GLI3R by the proteasome | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.515416 |
| GLI3 is processed to GLI3R by the proteasome | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.54735 |
| GLI3 is processed to GLI3R by the proteasome | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.577481 |
| GLI3 is processed to GLI3R by the proteasome | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.515416 |
| GLI3 is processed to GLI3R by the proteasome | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.515416 |
| GLI3 is processed to GLI3R by the proteasome | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.515416 |
| GLI3 is processed to GLI3R by the proteasome | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.54735 |
| GLI3 is processed to GLI3R by the proteasome | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.515416 |
| GLI3 is processed to GLI3R by the proteasome | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.54735 |
| GLI3 is processed to GLI3R by the proteasome | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.54735 |
| GLI3 is processed to GLI3R by the proteasome | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.546881 |
| GLI3 is processed to GLI3R by the proteasome | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.515416 |
| GLI3 is processed to GLI3R by the proteasome | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.57244 |
| GLI3 is processed to GLI3R by the proteasome | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.547953 |
| GLI3 is processed to GLI3R by the proteasome | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.401417 | 0.70503 | 4 | 0.718024 | 0.515416 |
| GLI3 is processed to GLI3R by the proteasome | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.401417 | 0.70503 | 4 | 0.718024 | 0.568645 |
| GLI3 is processed to GLI3R by the proteasome | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.58858 |
| GLI3 is processed to GLI3R by the proteasome | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.5197 |
| GLI3 is processed to GLI3R by the proteasome | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.54735 |
| GLI3 is processed to GLI3R by the proteasome | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.54735 |
| GLI3 is processed to GLI3R by the proteasome | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.54735 |
| GLI3 is processed to GLI3R by the proteasome | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.577847 |
| GLI3 is processed to GLI3R by the proteasome | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.401417 | 0.70503 | 4 | 0.718024 | 0.54735 |
| Glucagon signaling in metabolic regulation | CYSLTR1 | Cysteinyl leukotriene receptor 1 | 8.686529 | 0.531045 | 2.820143 | 0.418898 | 0.552802 |
| Glucagon signaling in metabolic regulation | CYSLTR2 | CYSLTR2 protein, Cysteinyl leukotriene receptor 2, G_PROTEIN_RECEP_F1_2 domain-containing protein | 8.686529 | 0.531045 | 2.820143 | 0.418898 | 0.566351 |
| Glucagon signaling in metabolic regulation | ADCY7 | Adenylate cyclase type 7 | 8.686529 | 0.531045 | 2.820143 | 0.418898 | 0.550655 |
| Glucagon signaling in metabolic regulation | PRKAR2A | cAMP-dependent protein kinase type II-alpha regulatory subunit, Protein kinase cAMP-dependent type II regulatory subunit alpha, Protein kinase, cAMP-dependent, regulatory subunit type II alpha, Uncharacterized protein | 8.686529 | 0.531045 | 2.820143 | 0.418898 | 0.527065 |
| Glucagon signaling in metabolic regulation | ADCY9 | Adenylate cyclase 9, Adenylate cyclase type 9 | 8.686529 | 0.531045 | 2.820143 | 0.418898 | 0.550534 |
| GP1b-IX-V activation signalling | YWHAZ | 14_3_3 domain-containing protein, 14-3-3 protein zeta, 14-3-3 protein zeta/delta, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta | 10.669639 | 0.797653 | 4 | 0.829762 | 0.602005 |
| GP1b-IX-V activation signalling | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 10.669639 | 0.797653 | 4 | 0.829762 | 0.646924 |
| GP1b-IX-V activation signalling | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 10.669639 | 0.797653 | 4 | 0.829762 | 0.594807 |
| GP1b-IX-V activation signalling | VWF | von Willebrand factor | 10.669639 | 0.797653 | 4 | 0.829762 | 0.548181 |
| GP1b-IX-V activation signalling | RAF1 | Non-specific serine/threonine protein kinase, RAF proto-oncogene serine/threonine-protein kinase | 10.669639 | 0.797653 | 4 | 0.829762 | 0.573556 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL23 | 60S ribosomal protein L23 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS21 | 40S ribosomal protein S21 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS28 | 40S ribosomal protein S28 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL26 | 60S ribosomal protein L26, GEO07453p1, KOW domain-containing protein, Ribosomal protein L26 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL6 | 60S ribosomal protein L6 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS9 | 40S ribosomal protein S9 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS3 | 40S ribosomal protein S3, DNA-(apurinic or apyrimidinic site) lyase | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL27A | 60S ribosomal protein L27-A, 60S ribosomal protein L27a | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL21 | 60S ribosomal protein L21 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS14 | 40S ribosomal protein S14, Ribosomal protein S14, Uncharacterized protein | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS23 | 40S ribosomal protein S23, 40S ribosomal protein S23-A | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS25 | 40S ribosomal protein S25 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL18A | 60S ribosomal protein L18-A, 60S ribosomal protein L18a | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL36 | 60S ribosomal protein L36 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS2 | 40S ribosomal protein S2 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS3A | 40S ribosomal protein S3a | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS12 | 40S ribosomal protein S12 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL35 | 60S ribosomal protein L35 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL18 | 60S ribosomal protein L18, Ribosomal protein L18 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS19 | 40S ribosomal protein S19, Uncharacterized protein | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL9 | 60S ribosomal protein L9 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL12 | 60S ribosomal protein L12 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL10A | 60S ribosomal protein L10a, Ribosomal protein | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS17 | 40S ribosomal protein S17 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS4Y1 | 40S ribosomal protein S4, 40S ribosomal protein S4, Y isoform 1 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS13 | 40S ribosomal protein S13 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL39 | 60S ribosomal protein L39, LOC100135132 protein, Ribosomal protein L39 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL3 | 60S ribosomal protein L3, LOC100145083 protein, Ribosomal protein L3, Uncharacterized protein | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL14 | 60S ribosomal protein L14 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS7 | 40S ribosomal protein S7 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS26 | 40S ribosomal protein S26 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPSA | 40S ribosomal protein SA | 11.273966 | 0.870578 | 4 | 0.897401 | 0.521531 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS15A | 40S ribosomal protein S15a | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL32 | 60S ribosomal protein L32, Hypothetical LOC496894, Ribosomal protein L32, Uncharacterized protein | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPLP0 | 60S acidic ribosomal protein P0 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS20 | 40S ribosomal protein S20 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL29 | 60S ribosomal protein L29 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL15 | 60S ribosomal protein L15, Ribosomal protein L15 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL36A | 60S ribosomal protein L36-A, 60S ribosomal protein L36a, IP17351p, ribosomal protein L36a, Uncharacterized protein | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS29 | 40S ribosomal protein S29 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL11 | 60S ribosomal protein L11 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS24 | 40S ribosomal protein S24 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL4 | 60S ribosomal protein L4, Ribos_L4_asso_C domain-containing protein, Ribosomal protein L4 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL35A | 60S ribosomal protein L35-A, 60S ribosomal protein L35a | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS15 | 40S ribosomal protein S15 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPLP1 | 60S acidic ribosomal protein P1, Ribosomal protein, large, P1 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL19 | 60S ribosomal protein L19, Ribosomal protein L19 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL41 | 60S ribosomal protein L41 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL8 | 60S ribosomal protein L8 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS27 | 40S ribosomal protein S27 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.549907 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS18 | 40S ribosomal protein S18 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL17 | 60S ribosomal protein L17 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS8 | 40S ribosomal protein S8 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS4X | 40S ribosomal protein S4, 40S ribosomal protein S4, X isoform | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL30 | 60S ribosomal protein L30 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS11 | 40S ribosomal protein S11 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL31 | 60S ribosomal protein L31 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS6 | 40S ribosomal protein S6 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.566528 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL23A | 60S ribosomal protein L23-A, 60S ribosomal protein L23a, FI01658p, MGC89958 protein, Ribosomal protein L23a, Ribosomal_L23eN domain-containing protein | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL5 | 60S ribosomal protein L5, Ribosomal protein L5 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL22 | 60S ribosomal protein L22, 60S ribosomal protein L22 1, Ribosomal protein L22, Uncharacterized protein | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL27 | 60S ribosomal protein L27 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS5 | 40S ribosomal protein S5, 40S ribosomal protein S5 [Cleaved into: 40S ribosomal protein S5, N-terminally processed], 40S ribosomal protein S5-A, Ribosomal protein S5 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL13A | 60S ribosomal protein L13a | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL24 | 60S ribosomal protein L24, Ribosomal protein L24, TRASH domain-containing protein | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL37 | 60S ribosomal protein L37, Ribosomal protein L37 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL34 | 60S ribosomal protein L34 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL38 | 60S ribosomal protein L38 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS16 | 40S ribosomal protein S16, Ribosomal protein S16, Uncharacterized protein | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL7 | 60S ribosomal protein L7, MGC79754 protein, Ribosomal protein L7, Uncharacterized protein | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL37A | 60S ribosomal protein L37-A, 60S ribosomal protein L37a, MGC89854 protein, Probable 60S ribosomal protein L37-A, Ribosomal protein L37a | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL28 | 60S ribosomal protein L28 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL13 | 60S ribosomal protein L13 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL7A | 60S ribosomal protein L7-A, 60S ribosomal protein L7a | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | FAU | 40S ribosomal protein S30 | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPS10 | 40S ribosomal protein S10, Ribosomal protein S10, S10_plectin domain-containing protein | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | RPL10 | 60S ribosomal protein L10, MGC89303 protein | 11.273966 | 0.870578 | 4 | 0.897401 | 0.579609 |
| HCMV Early Events | EZH2 | [Histone H3]-lysine(27) N-trimethyltransferase, Histone-lysine N-methyltransferase EZH2 | 8.485709 | 0.480759 | 3.59969 | 0.50274 | 0.53644 |
| HCMV Early Events | EGFR | Epidermal growth factor receptor, Receptor protein-tyrosine kinase | 8.485709 | 0.480759 | 3.59969 | 0.50274 | 0.625817 |
| HCMV Early Events | HDAC3 | Histone deacetylase, Histone deacetylase 3 | 8.485709 | 0.480759 | 3.59969 | 0.50274 | 0.528637 |
| Hedgehog 'off' state | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.515416 |
| Hedgehog 'off' state | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.515416 |
| Hedgehog 'off' state | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.111032 | 0.621961 | 4 | 0.661509 | 0.515416 |
| Hedgehog 'off' state | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.515416 |
| Hedgehog 'off' state | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.515416 |
| Hedgehog 'off' state | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.54735 |
| Hedgehog 'off' state | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.54735 |
| Hedgehog 'off' state | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.515416 |
| Hedgehog 'off' state | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.573417 |
| Hedgehog 'off' state | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.54735 |
| Hedgehog 'off' state | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.577481 |
| Hedgehog 'off' state | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.515416 |
| Hedgehog 'off' state | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.515416 |
| Hedgehog 'off' state | SMO | Protein smoothened, Smoothened homolog, Smoothened, frizzled class receptor | 9.111032 | 0.621961 | 4 | 0.661509 | 0.547562 |
| Hedgehog 'off' state | PRKAR2A | cAMP-dependent protein kinase type II-alpha regulatory subunit, Protein kinase cAMP-dependent type II regulatory subunit alpha, Protein kinase, cAMP-dependent, regulatory subunit type II alpha, Uncharacterized protein | 9.111032 | 0.621961 | 4 | 0.661509 | 0.527065 |
| Hedgehog 'off' state | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.515416 |
| Hedgehog 'off' state | ADCY7 | Adenylate cyclase type 7 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.550655 |
| Hedgehog 'off' state | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.54735 |
| Hedgehog 'off' state | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.565601 |
| Hedgehog 'off' state | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.54735 |
| Hedgehog 'off' state | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.111032 | 0.621961 | 4 | 0.661509 | 0.515416 |
| Hedgehog 'off' state | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.515416 |
| Hedgehog 'off' state | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.515416 |
| Hedgehog 'off' state | ADCY9 | Adenylate cyclase 9, Adenylate cyclase type 9 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.550534 |
| Hedgehog 'off' state | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.572294 |
| Hedgehog 'off' state | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.5197 |
| Hedgehog 'off' state | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.54735 |
| Hedgehog 'off' state | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.58858 |
| Hedgehog 'off' state | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.515416 |
| Hedgehog 'off' state | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.546881 |
| Hedgehog 'off' state | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.54735 |
| Hedgehog 'off' state | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.515416 |
| Hedgehog 'off' state | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.57244 |
| Hedgehog 'off' state | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.547953 |
| Hedgehog 'off' state | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.111032 | 0.621961 | 4 | 0.661509 | 0.568645 |
| Hedgehog 'off' state | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.54735 |
| Hedgehog 'off' state | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.111032 | 0.621961 | 4 | 0.661509 | 0.579609 |
| Hedgehog 'off' state | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.54735 |
| Hedgehog 'off' state | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.54735 |
| Hedgehog 'off' state | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.577847 |
| Hedgehog 'off' state | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.111032 | 0.621961 | 4 | 0.661509 | 0.515416 |
| Hedgehog 'on' state | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.5197 |
| Hedgehog 'on' state | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.456334 | 0.716756 | 4 | 0.72672 | 0.515416 |
| Hedgehog 'on' state | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.54735 |
| Hedgehog 'on' state | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.54735 |
| Hedgehog 'on' state | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.54735 |
| Hedgehog 'on' state | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.577481 |
| Hedgehog 'on' state | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.547953 |
| Hedgehog 'on' state | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.57244 |
| Hedgehog 'on' state | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.54735 |
| Hedgehog 'on' state | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.546881 |
| Hedgehog 'on' state | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.54735 |
| Hedgehog 'on' state | ULK3 | Serine/threonine-protein kinase ULK3, Unc-51-like kinase 3 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.512263 |
| Hedgehog 'on' state | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.54735 |
| Hedgehog 'on' state | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.456334 | 0.716756 | 4 | 0.72672 | 0.515416 |
| Hedgehog 'on' state | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.515416 |
| Hedgehog 'on' state | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.456334 | 0.716756 | 4 | 0.72672 | 0.579609 |
| Hedgehog 'on' state | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.565601 |
| Hedgehog 'on' state | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.54735 |
| Hedgehog 'on' state | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.54735 |
| Hedgehog 'on' state | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.515416 |
| Hedgehog 'on' state | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.577847 |
| Hedgehog 'on' state | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.54735 |
| Hedgehog 'on' state | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.573417 |
| Hedgehog 'on' state | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.58858 |
| Hedgehog 'on' state | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.515416 |
| Hedgehog 'on' state | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.515416 |
| Hedgehog 'on' state | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.515416 |
| Hedgehog 'on' state | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.515416 |
| Hedgehog 'on' state | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.456334 | 0.716756 | 4 | 0.72672 | 0.568645 |
| Hedgehog 'on' state | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.515416 |
| Hedgehog 'on' state | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.54735 |
| Hedgehog 'on' state | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.515416 |
| Hedgehog 'on' state | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.515416 |
| Hedgehog 'on' state | SMO | Protein smoothened, Smoothened homolog, Smoothened, frizzled class receptor | 9.456334 | 0.716756 | 4 | 0.72672 | 0.547562 |
| Hedgehog 'on' state | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.515416 |
| Hedgehog 'on' state | SHH | Hedgehog protein, Sonic hedgehog protein | 9.456334 | 0.716756 | 4 | 0.72672 | 0.510466 |
| Hedgehog 'on' state | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.515416 |
| Hedgehog 'on' state | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.515416 |
| Hedgehog 'on' state | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.572294 |
| Hedgehog 'on' state | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.456334 | 0.716756 | 4 | 0.72672 | 0.515416 |
| Hedgehog ligand biogenesis | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.54735 |
| Hedgehog ligand biogenesis | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.54735 |
| Hedgehog ligand biogenesis | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.515416 |
| Hedgehog ligand biogenesis | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.60391 | 0.753492 | 4 | 0.752702 | 0.579609 |
| Hedgehog ligand biogenesis | SHH | Hedgehog protein, Sonic hedgehog protein | 9.60391 | 0.753492 | 4 | 0.752702 | 0.510466 |
| Hedgehog ligand biogenesis | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.515416 |
| Hedgehog ligand biogenesis | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.57244 |
| Hedgehog ligand biogenesis | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.547953 |
| Hedgehog ligand biogenesis | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.573417 |
| Hedgehog ligand biogenesis | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.54735 |
| Hedgehog ligand biogenesis | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.58858 |
| Hedgehog ligand biogenesis | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.60391 | 0.753492 | 4 | 0.752702 | 0.568645 |
| Hedgehog ligand biogenesis | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.515416 |
| Hedgehog ligand biogenesis | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.60391 | 0.753492 | 4 | 0.752702 | 0.515416 |
| Hedgehog ligand biogenesis | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.515416 |
| Hedgehog ligand biogenesis | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.515416 |
| Hedgehog ligand biogenesis | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.515416 |
| Hedgehog ligand biogenesis | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.54735 |
| Hedgehog ligand biogenesis | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.515416 |
| Hedgehog ligand biogenesis | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.546881 |
| Hedgehog ligand biogenesis | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.54735 |
| Hedgehog ligand biogenesis | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.577847 |
| Hedgehog ligand biogenesis | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.515416 |
| Hedgehog ligand biogenesis | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.577481 |
| Hedgehog ligand biogenesis | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.515416 |
| Hedgehog ligand biogenesis | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.565601 |
| Hedgehog ligand biogenesis | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.54735 |
| Hedgehog ligand biogenesis | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.515416 |
| Hedgehog ligand biogenesis | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.515416 |
| Hedgehog ligand biogenesis | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.515416 |
| Hedgehog ligand biogenesis | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.60391 | 0.753492 | 4 | 0.752702 | 0.515416 |
| Hedgehog ligand biogenesis | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.54735 |
| Hedgehog ligand biogenesis | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.54735 |
| Hedgehog ligand biogenesis | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.54735 |
| Hedgehog ligand biogenesis | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.515416 |
| Hedgehog ligand biogenesis | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.54735 |
| Hedgehog ligand biogenesis | VCP | 15S Mg(2+)-ATPase p97 subunit, Transitional endoplasmic reticulum ATPase | 9.60391 | 0.753492 | 4 | 0.752702 | 0.526633 |
| Hedgehog ligand biogenesis | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.5197 |
| Hedgehog ligand biogenesis | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.60391 | 0.753492 | 4 | 0.752702 | 0.572294 |
| HSP90 chaperone cycle for steroid hormone receptors (SHR) | PGR | Nuclear receptor subfamily 3 group C member 3, Progesterone receptor | 8.989027 | 0.512427 | 4 | 0.600448 | 0.5833 |
| HSP90 chaperone cycle for steroid hormone receptors (SHR) | DNAJA1 | DnaJ, DnaJ heat shock protein family, DnaJ homolog subfamily A member 1, DnaJ homolog subfamily A member 1 homolog, DnaJ homolog subfamily A member 1 isoform 1, Uncharacterized protein | 8.989027 | 0.512427 | 4 | 0.600448 | 0.515668 |
| HSP90 chaperone cycle for steroid hormone receptors (SHR) | HSP90AA1 | Heat shock protein 90 alpha family class A member 1, Heat shock protein HSP 90-alpha | 8.989027 | 0.512427 | 4 | 0.600448 | 0.624325 |
| HSP90 chaperone cycle for steroid hormone receptors (SHR) | NR3C1 | Glucocorticoid receptor | 8.989027 | 0.512427 | 4 | 0.600448 | 0.59851 |
| HSP90 chaperone cycle for steroid hormone receptors (SHR) | HSP90AB1 | Heat shock protein 90 alpha family class B member 1, Heat shock protein 90kDa alpha, Heat shock protein HSP 90-beta, Heat shock protein HSP 90-beta isoform a | 8.989027 | 0.512427 | 4 | 0.600448 | 0.569354 |
| Influenza Infection | CANX | Calnexin, Uncharacterized protein | 7.405237 | 0.826895 | 4 | 0.675975 | 0.530822 |
| Influenza Infection | RPS27 | 40S ribosomal protein S27 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.549907 |
| Influenza Infection | RPL28 | 60S ribosomal protein L28 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS13 | 40S ribosomal protein S13 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL23 | 60S ribosomal protein L23 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS15 | 40S ribosomal protein S15 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL41 | 60S ribosomal protein L41 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS12 | 40S ribosomal protein S12 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL24 | 60S ribosomal protein L24, Ribosomal protein L24, TRASH domain-containing protein | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | SLC25A6 | ADP/ATP translocase 3, Solute carrier family 25 member 6, Uncharacterized protein | 7.405237 | 0.826895 | 4 | 0.675975 | 0.546978 |
| Influenza Infection | RPL10A | 60S ribosomal protein L10a, Ribosomal protein | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS15A | 40S ribosomal protein S15a | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS4X | 40S ribosomal protein S4, 40S ribosomal protein S4, X isoform | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS14 | 40S ribosomal protein S14, Ribosomal protein S14, Uncharacterized protein | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPSA | 40S ribosomal protein SA | 7.405237 | 0.826895 | 4 | 0.675975 | 0.521531 |
| Influenza Infection | RPL35 | 60S ribosomal protein L35 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL34 | 60S ribosomal protein L34 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL17 | 60S ribosomal protein L17 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL32 | 60S ribosomal protein L32, Hypothetical LOC496894, Ribosomal protein L32, Uncharacterized protein | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL11 | 60S ribosomal protein L11 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL12 | 60S ribosomal protein L12 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS19 | 40S ribosomal protein S19, Uncharacterized protein | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL37 | 60S ribosomal protein L37, Ribosomal protein L37 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL29 | 60S ribosomal protein L29 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPLP1 | 60S acidic ribosomal protein P1, Ribosomal protein, large, P1 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS9 | 40S ribosomal protein S9 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL15 | 60S ribosomal protein L15, Ribosomal protein L15 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS8 | 40S ribosomal protein S8 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL7A | 60S ribosomal protein L7-A, 60S ribosomal protein L7a | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS17 | 40S ribosomal protein S17 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL10 | 60S ribosomal protein L10, MGC89303 protein | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS29 | 40S ribosomal protein S29 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL8 | 60S ribosomal protein L8 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS3 | 40S ribosomal protein S3, DNA-(apurinic or apyrimidinic site) lyase | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL14 | 60S ribosomal protein L14 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL31 | 60S ribosomal protein L31 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL19 | 60S ribosomal protein L19, Ribosomal protein L19 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS18 | 40S ribosomal protein S18 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS24 | 40S ribosomal protein S24 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | PB1 | Protein PB1-F2, RNA-directed RNA polymerase catalytic subunit | 7.405237 | 0.826895 | 4 | 0.675975 | 0.508942 |
| Influenza Infection | RPS26 | 40S ribosomal protein S26 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL5 | 60S ribosomal protein L5, Ribosomal protein L5 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL7 | 60S ribosomal protein L7, MGC79754 protein, Ribosomal protein L7, Uncharacterized protein | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS6 | 40S ribosomal protein S6 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.566528 |
| Influenza Infection | RPL39 | 60S ribosomal protein L39, LOC100135132 protein, Ribosomal protein L39 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL36 | 60S ribosomal protein L36 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL27 | 60S ribosomal protein L27 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS10 | 40S ribosomal protein S10, Ribosomal protein S10, S10_plectin domain-containing protein | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL37A | 60S ribosomal protein L37-A, 60S ribosomal protein L37a, MGC89854 protein, Probable 60S ribosomal protein L37-A, Ribosomal protein L37a | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS2 | 40S ribosomal protein S2 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS20 | 40S ribosomal protein S20 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS21 | 40S ribosomal protein S21 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL27A | 60S ribosomal protein L27-A, 60S ribosomal protein L27a | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS7 | 40S ribosomal protein S7 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL4 | 60S ribosomal protein L4, Ribos_L4_asso_C domain-containing protein, Ribosomal protein L4 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS4Y1 | 40S ribosomal protein S4, 40S ribosomal protein S4, Y isoform 1 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS11 | 40S ribosomal protein S11 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL26 | 60S ribosomal protein L26, GEO07453p1, KOW domain-containing protein, Ribosomal protein L26 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS28 | 40S ribosomal protein S28 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS5 | 40S ribosomal protein S5, 40S ribosomal protein S5 [Cleaved into: 40S ribosomal protein S5, N-terminally processed], 40S ribosomal protein S5-A, Ribosomal protein S5 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL13 | 60S ribosomal protein L13 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL13A | 60S ribosomal protein L13a | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL38 | 60S ribosomal protein L38 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS16 | 40S ribosomal protein S16, Ribosomal protein S16, Uncharacterized protein | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | FAU | 40S ribosomal protein S30 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL36A | 60S ribosomal protein L36-A, 60S ribosomal protein L36a, IP17351p, ribosomal protein L36a, Uncharacterized protein | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL3 | 60S ribosomal protein L3, LOC100145083 protein, Ribosomal protein L3, Uncharacterized protein | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | HSP90AA1 | Heat shock protein 90 alpha family class A member 1, Heat shock protein HSP 90-alpha | 7.405237 | 0.826895 | 4 | 0.675975 | 0.624325 |
| Influenza Infection | RPS23 | 40S ribosomal protein S23, 40S ribosomal protein S23-A | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL22 | 60S ribosomal protein L22, 60S ribosomal protein L22 1, Ribosomal protein L22, Uncharacterized protein | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL6 | 60S ribosomal protein L6 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL21 | 60S ribosomal protein L21 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL30 | 60S ribosomal protein L30 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL18A | 60S ribosomal protein L18-A, 60S ribosomal protein L18a | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS3A | 40S ribosomal protein S3a | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPLP0 | 60S acidic ribosomal protein P0 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL18 | 60S ribosomal protein L18, Ribosomal protein L18 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPS25 | 40S ribosomal protein S25 | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL23A | 60S ribosomal protein L23-A, 60S ribosomal protein L23a, FI01658p, MGC89958 protein, Ribosomal protein L23a, Ribosomal_L23eN domain-containing protein | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| Influenza Infection | RPL35A | 60S ribosomal protein L35-A, 60S ribosomal protein L35a | 7.405237 | 0.826895 | 4 | 0.675975 | 0.579609 |
| InlA-mediated entry of Listeria monocytogenes into host cells | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.210632 | 0.795438 | 2.280101 | 0.429449 | 0.579609 |
| InlA-mediated entry of Listeria monocytogenes into host cells | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 9.210632 | 0.795438 | 2.280101 | 0.429449 | 0.594807 |
| InlA-mediated entry of Listeria monocytogenes into host cells | CTNNB1 | Catenin, Catenin beta-1, Uncharacterized protein | 9.210632 | 0.795438 | 2.280101 | 0.429449 | 0.657916 |
| Insulin receptor signalling cascade | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 9.371883 | 0.471471 | 3.329185 | 0.499145 | 0.589663 |
| Insulin receptor signalling cascade | SHC1 | SHC-transforming protein 1 | 9.371883 | 0.471471 | 3.329185 | 0.499145 | 0.521858 |
| Insulin receptor signalling cascade | FLT3 | Receptor protein-tyrosine kinase, Receptor-type tyrosine-protein kinase FLT3 | 9.371883 | 0.471471 | 3.329185 | 0.499145 | 0.529563 |
| Insulin receptor signalling cascade | FGFR3 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 3 | 9.371883 | 0.471471 | 3.329185 | 0.499145 | 0.560785 |
| Insulin receptor signalling cascade | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 9.371883 | 0.471471 | 3.329185 | 0.499145 | 0.552041 |
| Insulin receptor signalling cascade | FGF1 | Fibroblast growth factor 1, Multifunctional fusion protein [Includes: Fibroblast growth factor 1, Putative fibroblast growth factor 1 | 9.371883 | 0.471471 | 3.329185 | 0.499145 | 0.509844 |
| Insulin receptor signalling cascade | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 9.371883 | 0.471471 | 3.329185 | 0.499145 | 0.541254 |
| Insulin receptor signalling cascade | INSR | Insulin receptor, Tyrosine-protein kinase receptor | 9.371883 | 0.471471 | 3.329185 | 0.499145 | 0.563226 |
| Insulin receptor signalling cascade | FGFR4 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 4 | 9.371883 | 0.471471 | 3.329185 | 0.499145 | 0.555253 |
| Insulin receptor signalling cascade | FGFR2 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 2 | 9.371883 | 0.471471 | 3.329185 | 0.499145 | 0.577012 |
| Insulin receptor signalling cascade | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 9.371883 | 0.471471 | 3.329185 | 0.499145 | 0.505988 |
| Insulin receptor signalling cascade | SHC | DShc protein | 9.371883 | 0.471471 | 3.329185 | 0.499145 | 0.605 |
| Insulin receptor signalling cascade | PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.371883 | 0.471471 | 3.329185 | 0.499145 | 0.589385 |
| Insulin receptor signalling cascade | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 9.371883 | 0.471471 | 3.329185 | 0.499145 | 0.646924 |
| Insulin receptor signalling cascade | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.371883 | 0.471471 | 3.329185 | 0.499145 | 0.660919 |
| Insulin receptor signalling cascade | FGFR1 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 1 | 9.371883 | 0.471471 | 3.329185 | 0.499145 | 0.55821 |
| Integration of viral DNA into host genomic DNA | GAG-POL | Gag-Pol polyprotein | 7.548022 | 0.54084 | 0.64835 | ||
| Integrin signaling | SHC | DShc protein | 9.055285 | 0.541957 | 3.169992 | 0.490264 | 0.605 |
| Integrin signaling | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 9.055285 | 0.541957 | 3.169992 | 0.490264 | 0.594807 |
| Integrin signaling | CRK | 14_3_3 domain-containing protein, Adapter molecule crk, Adapter molecule Crk, CRK proto-oncogene, adaptor protein, Uncharacterized protein, V-crk avian sarcoma virus CT10 oncogene homolog | 9.055285 | 0.541957 | 3.169992 | 0.490264 | 0.513369 |
| Integrin signaling | SYK | Tyrosine-protein kinase, Tyrosine-protein kinase SYK | 9.055285 | 0.541957 | 3.169992 | 0.490264 | 0.557587 |
| Integrin signaling | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 9.055285 | 0.541957 | 3.169992 | 0.490264 | 0.552041 |
| Integrin signaling | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 9.055285 | 0.541957 | 3.169992 | 0.490264 | 0.589663 |
| Integrin signaling | FN1 | Fibronectin | 9.055285 | 0.541957 | 3.169992 | 0.490264 | 0.578096 |
| Integrin signaling | SHC1 | SHC-transforming protein 1 | 9.055285 | 0.541957 | 3.169992 | 0.490264 | 0.521858 |
| Integrin signaling | PTPN1 | Protein-tyrosine-phosphatase, Tyrosine-protein phosphatase non-receptor type, Tyrosine-protein phosphatase non-receptor type 1 | 9.055285 | 0.541957 | 3.169992 | 0.490264 | 0.510671 |
| Integrin signaling | VWF | von Willebrand factor | 9.055285 | 0.541957 | 3.169992 | 0.490264 | 0.548181 |
| Integrin signaling | PTK2 | Focal adhesion kinase 1, Non-specific protein-tyrosine kinase, Serine/threonine-protein kinase PTK2/STK2 | 9.055285 | 0.541957 | 3.169992 | 0.490264 | 0.503121 |
| Integrin signaling | RAP1A | RAP1A, member of RAS oncogene family, Ras-related protein Rap-1A, Uncharacterized protein | 9.055285 | 0.541957 | 3.169992 | 0.490264 | 0.530823 |
| Interleukin-1 signaling | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.515416 |
| Interleukin-1 signaling | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.310883 | 0.559449 | 4 | 0.640563 | 0.515416 |
| Interleukin-1 signaling | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.54735 |
| Interleukin-1 signaling | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.515416 |
| Interleukin-1 signaling | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.57244 |
| Interleukin-1 signaling | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.54735 |
| Interleukin-1 signaling | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.515416 |
| Interleukin-1 signaling | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.310883 | 0.559449 | 4 | 0.640563 | 0.579609 |
| Interleukin-1 signaling | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.577481 |
| Interleukin-1 signaling | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.573417 |
| Interleukin-1 signaling | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.515416 |
| Interleukin-1 signaling | UBE2V1 | Ubiquitin conjugating enzyme E2 V1, Ubiquitin-conjugating enzyme E2 variant 1 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.593364 |
| Interleukin-1 signaling | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.515416 |
| Interleukin-1 signaling | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.54735 |
| Interleukin-1 signaling | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.54735 |
| Interleukin-1 signaling | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.54735 |
| Interleukin-1 signaling | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.565601 |
| Interleukin-1 signaling | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.546881 |
| Interleukin-1 signaling | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.54735 |
| Interleukin-1 signaling | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.572294 |
| Interleukin-1 signaling | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.577847 |
| Interleukin-1 signaling | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.515416 |
| Interleukin-1 signaling | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.515416 |
| Interleukin-1 signaling | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.515416 |
| Interleukin-1 signaling | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.547953 |
| Interleukin-1 signaling | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.54735 |
| Interleukin-1 signaling | MAP2K1 | Dual specificity mitogen-activated protein kinase kinase 1, MAP2K1 protein, Mitogen-activated protein kinase kinase 1, Protein kinase domain-containing protein | 9.310883 | 0.559449 | 4 | 0.640563 | 0.545374 |
| Interleukin-1 signaling | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.310883 | 0.559449 | 4 | 0.640563 | 0.568645 |
| Interleukin-1 signaling | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.515416 |
| Interleukin-1 signaling | MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.519182 |
| Interleukin-1 signaling | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.54735 |
| Interleukin-1 signaling | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.515416 |
| Interleukin-1 signaling | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.515416 |
| Interleukin-1 signaling | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.54735 |
| Interleukin-1 signaling | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.515416 |
| Interleukin-1 signaling | APP | ABPP, Amyloid-beta A4 protein, Amyloid-beta precursor protein | 9.310883 | 0.559449 | 4 | 0.640563 | 0.506343 |
| Interleukin-1 signaling | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.5197 |
| Interleukin-1 signaling | RIPK2 | Receptor-interacting serine-threonine kinase 2, Receptor-interacting serine/threonine-protein kinase 2 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.530344 |
| Interleukin-1 signaling | IRAK4 | Interleukin-1 receptor-associated kinase 4 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.50686 |
| Interleukin-1 signaling | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.515416 |
| Interleukin-1 signaling | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.58858 |
| Interleukin-1 signaling | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.54735 |
| Interleukin-1 signaling | IKBKB | I-kappa-B kinase, Inhibitor of nuclear factor kappa-B kinase subunit beta | 9.310883 | 0.559449 | 4 | 0.640563 | 0.518113 |
| Interleukin-1 signaling | MDM2 | E3 ubiquitin-protein ligase Mdm2 | 9.310883 | 0.559449 | 4 | 0.640563 | 0.659771 |
| Interleukin-1 signaling | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.310883 | 0.559449 | 4 | 0.640563 | 0.515416 |
| Interleukin-3, Interleukin-5 and GM-CSF signaling | JAK1 | Solute carrier family 39, Tyrosine-protein kinase, Tyrosine-protein kinase JAK1 | 9.357212 | 0.468499 | 3.28043 | 0.489996 | 0.551488 |
| Interleukin-3, Interleukin-5 and GM-CSF signaling | CRK | 14_3_3 domain-containing protein, Adapter molecule crk, Adapter molecule Crk, CRK proto-oncogene, adaptor protein, Uncharacterized protein, V-crk avian sarcoma virus CT10 oncogene homolog | 9.357212 | 0.468499 | 3.28043 | 0.489996 | 0.513369 |
| Interleukin-3, Interleukin-5 and GM-CSF signaling | PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.357212 | 0.468499 | 3.28043 | 0.489996 | 0.589385 |
| Interleukin-3, Interleukin-5 and GM-CSF signaling | SYK | Tyrosine-protein kinase, Tyrosine-protein kinase SYK | 9.357212 | 0.468499 | 3.28043 | 0.489996 | 0.557587 |
| Interleukin-3, Interleukin-5 and GM-CSF signaling | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.357212 | 0.468499 | 3.28043 | 0.489996 | 0.579609 |
| Interleukin-3, Interleukin-5 and GM-CSF signaling | JAK2 | Janus kinase 2, Tyrosine-protein kinase, Tyrosine-protein kinase JAK2 | 9.357212 | 0.468499 | 3.28043 | 0.489996 | 0.573235 |
| Interleukin-3, Interleukin-5 and GM-CSF signaling | HCK | Tyrosine-protein kinase, Tyrosine-protein kinase HCK | 9.357212 | 0.468499 | 3.28043 | 0.489996 | 0.515497 |
| Interleukin-3, Interleukin-5 and GM-CSF signaling | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 9.357212 | 0.468499 | 3.28043 | 0.489996 | 0.646924 |
| Interleukin-3, Interleukin-5 and GM-CSF signaling | PIK3CD | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.357212 | 0.468499 | 3.28043 | 0.489996 | 0.632077 |
| Interleukin-3, Interleukin-5 and GM-CSF signaling | YES1 | Tyrosine-protein kinase, Tyrosine-protein kinase yes, Tyrosine-protein kinase Yes | 9.357212 | 0.468499 | 3.28043 | 0.489996 | 0.506641 |
| Interleukin-3, Interleukin-5 and GM-CSF signaling | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.357212 | 0.468499 | 3.28043 | 0.489996 | 0.660919 |
| Interleukin-3, Interleukin-5 and GM-CSF signaling | JAK3 | Tyrosine-protein kinase, Tyrosine-protein kinase JAK3 | 9.357212 | 0.468499 | 3.28043 | 0.489996 | 0.537649 |
| Interleukin-3, Interleukin-5 and GM-CSF signaling | YWHAZ | 14_3_3 domain-containing protein, 14-3-3 protein zeta, 14-3-3 protein zeta/delta, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta | 9.357212 | 0.468499 | 3.28043 | 0.489996 | 0.602005 |
| Interleukin-3, Interleukin-5 and GM-CSF signaling | SHC1 | SHC-transforming protein 1 | 9.357212 | 0.468499 | 3.28043 | 0.489996 | 0.521858 |
| Intrinsic Pathway of Fibrin Clot Formation | F11 | Coagulation factor XI | 9.662353 | 0.656089 | 3.235661 | 0.571915 | 0.56599 |
| Intrinsic Pathway of Fibrin Clot Formation | KLKB1 | Kallikrein B1, Plasma kallikrein, Uncharacterized protein | 9.662353 | 0.656089 | 3.235661 | 0.571915 | 0.594535 |
| Intrinsic Pathway of Fibrin Clot Formation | F9 | Christmas factor, Coagulation factor IX | 9.662353 | 0.656089 | 3.235661 | 0.571915 | 0.632191 |
| Intrinsic Pathway of Fibrin Clot Formation | VWF | von Willebrand factor | 9.662353 | 0.656089 | 3.235661 | 0.571915 | 0.548181 |
| Intrinsic Pathway of Fibrin Clot Formation | F2 | Activation peptide fragment 1, Prothrombin | 9.662353 | 0.656089 | 3.235661 | 0.571915 | 0.5402 |
| Intrinsic Pathway of Fibrin Clot Formation | F12 | Coagulation factor XII, Uncharacterized protein | 9.662353 | 0.656089 | 3.235661 | 0.571915 | 0.516654 |
| Intrinsic Pathway of Fibrin Clot Formation | KNG1 | Bradykinin, Kininogen 1, Kininogen-1, Kininogen-1 [Cleaved into: Kininogen-1 heavy chain; Bradykinin; Kininogen-1 light chain], Uncharacterized protein | 9.662353 | 0.656089 | 3.235661 | 0.571915 | 0.566192 |
| Intrinsic Pathway of Fibrin Clot Formation | PRCP | Lysosomal Pro-X carboxypeptidase, Prolylcarboxypeptidase, Uncharacterized protein | 9.662353 | 0.656089 | 3.235661 | 0.571915 | 0.583031 |
| Intrinsic Pathway of Fibrin Clot Formation | F10 | Coagulation factor VII, Coagulation factor X, MGC107878 protein, Uncharacterized protein | 9.662353 | 0.656089 | 3.235661 | 0.571915 | 0.588553 |
| Ion transport by P-type ATPases | ATP12A | Potassium-transporting ATPase alpha chain 2, Sodium/potassium-transporting ATPase subunit alpha | 9.053456 | 0.562069 | 3.165884 | 0.497513 | 0.510281 |
| Ion transport by P-type ATPases | ATP4A | ATPase H+/K+-transporting subunit alpha, Potassium-transporting ATPase alpha chain 1, Sodium/potassium-transporting ATPase subunit alpha | 9.053456 | 0.562069 | 3.165884 | 0.497513 | 0.564197 |
| Ion transport by P-type ATPases | ATP4B | Potassium-transporting ATPase subunit beta, Sodium/potassium-transporting ATPase subunit beta | 9.053456 | 0.562069 | 3.165884 | 0.497513 | 0.567762 |
| IRS-mediated signalling | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 10.006764 | 0.474241 | 2.587363 | 0.410006 | 0.505988 |
| IRS-mediated signalling | FLT3 | Receptor protein-tyrosine kinase, Receptor-type tyrosine-protein kinase FLT3 | 10.006764 | 0.474241 | 2.587363 | 0.410006 | 0.529563 |
| IRS-mediated signalling | FGF1 | Fibroblast growth factor 1, Multifunctional fusion protein [Includes: Fibroblast growth factor 1, Putative fibroblast growth factor 1 | 10.006764 | 0.474241 | 2.587363 | 0.410006 | 0.509844 |
| IRS-mediated signalling | FGFR4 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 4 | 10.006764 | 0.474241 | 2.587363 | 0.410006 | 0.555253 |
| IRS-mediated signalling | FGFR2 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 2 | 10.006764 | 0.474241 | 2.587363 | 0.410006 | 0.577012 |
| IRS-mediated signalling | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 10.006764 | 0.474241 | 2.587363 | 0.410006 | 0.646924 |
| IRS-mediated signalling | PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 10.006764 | 0.474241 | 2.587363 | 0.410006 | 0.589385 |
| IRS-mediated signalling | FGFR1 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 1 | 10.006764 | 0.474241 | 2.587363 | 0.410006 | 0.55821 |
| IRS-mediated signalling | FGFR3 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 3 | 10.006764 | 0.474241 | 2.587363 | 0.410006 | 0.560785 |
| IRS-mediated signalling | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 10.006764 | 0.474241 | 2.587363 | 0.410006 | 0.589663 |
| IRS-mediated signalling | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 10.006764 | 0.474241 | 2.587363 | 0.410006 | 0.660919 |
| IRS-mediated signalling | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 10.006764 | 0.474241 | 2.587363 | 0.410006 | 0.552041 |
| JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 | MAPK10 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 10 | 8.502858 | 0.568547 | 3.167014 | 0.477765 | 0.506109 |
| JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 8.502858 | 0.568547 | 3.167014 | 0.477765 | 0.579609 |
| JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 | MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | 8.502858 | 0.568547 | 3.167014 | 0.477765 | 0.519182 |
| JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 | MAPK8 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 8 | 8.502858 | 0.568547 | 3.167014 | 0.477765 | 0.503967 |
| JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 | UBE2V1 | Ubiquitin conjugating enzyme E2 V1, Ubiquitin-conjugating enzyme E2 variant 1 | 8.502858 | 0.568547 | 3.167014 | 0.477765 | 0.593364 |
| JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 | MDM2 | E3 ubiquitin-protein ligase Mdm2 | 8.502858 | 0.568547 | 3.167014 | 0.477765 | 0.659771 |
| JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 | RIPK2 | Receptor-interacting serine-threonine kinase 2, Receptor-interacting serine/threonine-protein kinase 2 | 8.502858 | 0.568547 | 3.167014 | 0.477765 | 0.530344 |
| Kinesins | KIFC1 | Kinesin-like protein, Kinesin-like protein KIFC1 | 8.664513 | 0.459442 | 3.393643 | 0.472746 | 0.517415 |
| Kinesins | KIF11 | Kinesin family member 11, Kinesin motor domain-containing protein, Kinesin-like protein KIF11 | 8.664513 | 0.459442 | 3.393643 | 0.472746 | 0.527775 |
| Kinesins | KIF2C | Kinesin-like protein, Kinesin-like protein KIF2C | 8.664513 | 0.459442 | 3.393643 | 0.472746 | 0.611637 |
| Kinesins | CENPE | Centromere protein E, Centromere-associated protein E, Kinesin motor domain-containing protein | 8.664513 | 0.459442 | 3.393643 | 0.472746 | 0.564477 |
| KSRP (KHSRP) binds and destabilizes mRNA | MAPK11 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 11 | 8.673083 | 0.801383 | 3.357304 | 0.611555 | 0.567539 |
| KSRP (KHSRP) binds and destabilizes mRNA | MAPK14 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 14 | 8.673083 | 0.801383 | 3.357304 | 0.611555 | 0.591941 |
| KSRP (KHSRP) binds and destabilizes mRNA | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 8.673083 | 0.801383 | 3.357304 | 0.611555 | 0.552041 |
| KSRP (KHSRP) binds and destabilizes mRNA | YWHAZ | 14_3_3 domain-containing protein, 14-3-3 protein zeta, 14-3-3 protein zeta/delta, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta | 8.673083 | 0.801383 | 3.357304 | 0.611555 | 0.602005 |
| KSRP (KHSRP) binds and destabilizes mRNA | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 8.673083 | 0.801383 | 3.357304 | 0.611555 | 0.589663 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL39 | 60S ribosomal protein L39, LOC100135132 protein, Ribosomal protein L39 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS26 | 40S ribosomal protein S26 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL7 | 60S ribosomal protein L7, MGC79754 protein, Ribosomal protein L7, Uncharacterized protein | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL38 | 60S ribosomal protein L38 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPLP0 | 60S acidic ribosomal protein P0 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL9 | 60S ribosomal protein L9 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL10A | 60S ribosomal protein L10a, Ribosomal protein | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS20 | 40S ribosomal protein S20 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS18 | 40S ribosomal protein S18 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL4 | 60S ribosomal protein L4, Ribos_L4_asso_C domain-containing protein, Ribosomal protein L4 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL13A | 60S ribosomal protein L13a | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS28 | 40S ribosomal protein S28 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL29 | 60S ribosomal protein L29 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL32 | 60S ribosomal protein L32, Hypothetical LOC496894, Ribosomal protein L32, Uncharacterized protein | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS3 | 40S ribosomal protein S3, DNA-(apurinic or apyrimidinic site) lyase | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL23 | 60S ribosomal protein L23 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL11 | 60S ribosomal protein L11 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL28 | 60S ribosomal protein L28 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPLP1 | 60S acidic ribosomal protein P1, Ribosomal protein, large, P1 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL7A | 60S ribosomal protein L7-A, 60S ribosomal protein L7a | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS21 | 40S ribosomal protein S21 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL8 | 60S ribosomal protein L8 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS12 | 40S ribosomal protein S12 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL6 | 60S ribosomal protein L6 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS14 | 40S ribosomal protein S14, Ribosomal protein S14, Uncharacterized protein | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS7 | 40S ribosomal protein S7 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS24 | 40S ribosomal protein S24 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL10 | 60S ribosomal protein L10, MGC89303 protein | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL14 | 60S ribosomal protein L14 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS11 | 40S ribosomal protein S11 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS25 | 40S ribosomal protein S25 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL22 | 60S ribosomal protein L22, 60S ribosomal protein L22 1, Ribosomal protein L22, Uncharacterized protein | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS13 | 40S ribosomal protein S13 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL5 | 60S ribosomal protein L5, Ribosomal protein L5 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS19 | 40S ribosomal protein S19, Uncharacterized protein | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS4X | 40S ribosomal protein S4, 40S ribosomal protein S4, X isoform | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL12 | 60S ribosomal protein L12 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS4Y1 | 40S ribosomal protein S4, 40S ribosomal protein S4, Y isoform 1 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS8 | 40S ribosomal protein S8 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL15 | 60S ribosomal protein L15, Ribosomal protein L15 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL3 | 60S ribosomal protein L3, LOC100145083 protein, Ribosomal protein L3, Uncharacterized protein | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL35A | 60S ribosomal protein L35-A, 60S ribosomal protein L35a | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL23A | 60S ribosomal protein L23-A, 60S ribosomal protein L23a, FI01658p, MGC89958 protein, Ribosomal protein L23a, Ribosomal_L23eN domain-containing protein | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS15A | 40S ribosomal protein S15a | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS2 | 40S ribosomal protein S2 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS16 | 40S ribosomal protein S16, Ribosomal protein S16, Uncharacterized protein | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL41 | 60S ribosomal protein L41 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL35 | 60S ribosomal protein L35 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS29 | 40S ribosomal protein S29 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS10 | 40S ribosomal protein S10, Ribosomal protein S10, S10_plectin domain-containing protein | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL21 | 60S ribosomal protein L21 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL37A | 60S ribosomal protein L37-A, 60S ribosomal protein L37a, MGC89854 protein, Probable 60S ribosomal protein L37-A, Ribosomal protein L37a | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL19 | 60S ribosomal protein L19, Ribosomal protein L19 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPSA | 40S ribosomal protein SA | 11.235091 | 0.874662 | 4 | 0.897438 | 0.521531 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL17 | 60S ribosomal protein L17 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL18A | 60S ribosomal protein L18-A, 60S ribosomal protein L18a | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS9 | 40S ribosomal protein S9 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL37 | 60S ribosomal protein L37, Ribosomal protein L37 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL18 | 60S ribosomal protein L18, Ribosomal protein L18 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL36 | 60S ribosomal protein L36 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL27 | 60S ribosomal protein L27 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS27 | 40S ribosomal protein S27 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.549907 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS5 | 40S ribosomal protein S5, 40S ribosomal protein S5 [Cleaved into: 40S ribosomal protein S5, N-terminally processed], 40S ribosomal protein S5-A, Ribosomal protein S5 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL26 | 60S ribosomal protein L26, GEO07453p1, KOW domain-containing protein, Ribosomal protein L26 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL27A | 60S ribosomal protein L27-A, 60S ribosomal protein L27a | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS17 | 40S ribosomal protein S17 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | FAU | 40S ribosomal protein S30 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS23 | 40S ribosomal protein S23, 40S ribosomal protein S23-A | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL31 | 60S ribosomal protein L31 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS15 | 40S ribosomal protein S15 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS6 | 40S ribosomal protein S6 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.566528 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL34 | 60S ribosomal protein L34 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL13 | 60S ribosomal protein L13 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL36A | 60S ribosomal protein L36-A, 60S ribosomal protein L36a, IP17351p, ribosomal protein L36a, Uncharacterized protein | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL30 | 60S ribosomal protein L30 | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPL24 | 60S ribosomal protein L24, Ribosomal protein L24, TRASH domain-containing protein | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L13a-mediated translational silencing of Ceruloplasmin expression | RPS3A | 40S ribosomal protein S3a | 11.235091 | 0.874662 | 4 | 0.897438 | 0.579609 |
| L1CAM interactions | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 6.889992 | 0.419729 | 4 | 0.445811 | 0.541254 |
| L1CAM interactions | CSNK2A2 | Casein kinase 2 alpha 2, Casein kinase II subunit alpha' | 6.889992 | 0.419729 | 4 | 0.445811 | 0.53622 |
| L1CAM interactions | SCN4A | Sodium channel protein, Sodium channel protein type 4 subunit alpha | 6.889992 | 0.419729 | 4 | 0.445811 | 0.695739 |
| L1CAM interactions | EPHB2 | Ephrin type-B receptor 2, Receptor protein-tyrosine kinase | 6.889992 | 0.419729 | 4 | 0.445811 | 0.508582 |
| L1CAM interactions | CSNK2B | Casein kinase II subunit beta | 6.889992 | 0.419729 | 4 | 0.445811 | 0.537688 |
| L1CAM interactions | CSNK2A1 | Casein kinase 2, alpha 1 polypeptide, Casein kinase II subunit alpha, Protein kinase domain-containing protein | 6.889992 | 0.419729 | 4 | 0.445811 | 0.538737 |
| L1CAM interactions | FGFR1 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 1 | 6.889992 | 0.419729 | 4 | 0.445811 | 0.55821 |
| L1CAM interactions | SCN2A | Sodium channel protein, Sodium channel protein type 2 subunit alpha | 6.889992 | 0.419729 | 4 | 0.445811 | 0.538824 |
| L1CAM interactions | MAP2K1 | Dual specificity mitogen-activated protein kinase kinase 1, MAP2K1 protein, Mitogen-activated protein kinase kinase 1, Protein kinase domain-containing protein | 6.889992 | 0.419729 | 4 | 0.445811 | 0.545374 |
| L1CAM interactions | EGFR | Epidermal growth factor receptor, Receptor protein-tyrosine kinase | 6.889992 | 0.419729 | 4 | 0.445811 | 0.625817 |
| L1CAM interactions | SCN9A | Sodium channel protein, Sodium channel protein type 9 subunit alpha | 6.889992 | 0.419729 | 4 | 0.445811 | 0.589531 |
| L1CAM interactions | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 6.889992 | 0.419729 | 4 | 0.445811 | 0.594807 |
| Leukotriene receptors | CYSLTR1 | Cysteinyl leukotriene receptor 1 | 8.513215 | 0.786826 | 2.674433 | 0.476785 | 0.552802 |
| Leukotriene receptors | CYSLTR2 | CYSLTR2 protein, Cysteinyl leukotriene receptor 2, G_PROTEIN_RECEP_F1_2 domain-containing protein | 8.513215 | 0.786826 | 2.674433 | 0.476785 | 0.566351 |
| Leukotriene receptors | LTB4R | Leukotriene B4 receptor, Leukotriene B4 receptor 1 | 8.513215 | 0.786826 | 2.674433 | 0.476785 | 0.604494 |
| Leukotriene receptors | LTB4R2 | Leukotriene B4 receptor 2 | 8.513215 | 0.786826 | 2.674433 | 0.476785 | 0.627297 |
| LGI-ADAM interactions | CACNG8 | Voltage-dependent calcium channel gamma-8 subunit | 8.368379 | 0.850197 | 3.624135 | 0.668031 | 0.579696 |
| LGI-ADAM interactions | CACNG4 | Voltage-dependent calcium channel gamma-4 subunit | 8.368379 | 0.850197 | 3.624135 | 0.668031 | 0.598629 |
| LGI-ADAM interactions | CACNG2 | Voltage-dependent calcium channel gamma-2 subunit | 8.368379 | 0.850197 | 3.624135 | 0.668031 | 0.593521 |
| LGI-ADAM interactions | CACNG3 | Calcium voltage-gated channel auxiliary subunit gamma 3, Voltage-dependent calcium channel gamma-3 subunit | 8.368379 | 0.850197 | 3.624135 | 0.668031 | 0.598629 |
| Macroautophagy | PRKAB1 | 5'-AMP-activated protein kinase subunit beta-1, 5'AMP-activated protein kinase beta-1 non-catalytic subunit, Protein kinase AMP-activated non-catalytic subunit beta 1, Protein kinase, AMP-activated, beta 1 non-catalytic subunit | 8.745663 | 0.523873 | 4 | 0.593616 | 0.500481 |
| Macroautophagy | CSNK2A1 | Casein kinase 2, alpha 1 polypeptide, Casein kinase II subunit alpha, Protein kinase domain-containing protein | 8.745663 | 0.523873 | 4 | 0.593616 | 0.538737 |
| Macroautophagy | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 8.745663 | 0.523873 | 4 | 0.593616 | 0.594807 |
| Macroautophagy | VCP | 15S Mg(2+)-ATPase p97 subunit, Transitional endoplasmic reticulum ATPase | 8.745663 | 0.523873 | 4 | 0.593616 | 0.526633 |
| Macroautophagy | ULK1 | Non-specific serine/threonine protein kinase, Serine/threonine-protein kinase, Serine/threonine-protein kinase ULK1 | 8.745663 | 0.523873 | 4 | 0.593616 | 0.511522 |
| Macroautophagy | PRKAG1 | 5'-AMP-activated protein kinase subunit gamma-1, Protein kinase, AMP-activated, gamma 1 non-catalytic subunit | 8.745663 | 0.523873 | 4 | 0.593616 | 0.515113 |
| Macroautophagy | CSNK2B | Casein kinase II subunit beta | 8.745663 | 0.523873 | 4 | 0.593616 | 0.537688 |
| Macroautophagy | PRKAG2 | 5'-AMP-activated protein kinase subunit gamma-2, 5'-AMP-activated protein kinase subunit gamma-2 isoform X8, Protein kinase AMP-activated non-catalytic subunit gamma 2, Protein kinase, AMP-activated, gamma 2 non-catalytic subunit, Uncharacterized protein | 8.745663 | 0.523873 | 4 | 0.593616 | 0.510193 |
| Macroautophagy | ABCC1 | ABC transporter C family member 1, ATP-binding cassette, sub-family C, Multidrug resistance-associated protein 1 | 8.745663 | 0.523873 | 4 | 0.593616 | 0.610269 |
| Macroautophagy | UBE2V1 | Ubiquitin conjugating enzyme E2 V1, Ubiquitin-conjugating enzyme E2 variant 1 | 8.745663 | 0.523873 | 4 | 0.593616 | 0.593364 |
| Macroautophagy | MTOR | Nucleoprotein TPR, Serine/threonine-protein kinase mTOR, Serine/threonine-protein kinase TOR | 8.745663 | 0.523873 | 4 | 0.593616 | 0.619026 |
| Macroautophagy | HSP90AA1 | Heat shock protein 90 alpha family class A member 1, Heat shock protein HSP 90-alpha | 8.745663 | 0.523873 | 4 | 0.593616 | 0.624325 |
| Macroautophagy | CSNK2A2 | Casein kinase 2 alpha 2, Casein kinase II subunit alpha' | 8.745663 | 0.523873 | 4 | 0.593616 | 0.53622 |
| Macroautophagy | HDAC6 | Histone deacetylase 6, Histone deacetylase 6, isoform G | 8.745663 | 0.523873 | 4 | 0.593616 | 0.531859 |
| Macroautophagy | RPTOR | Regulatory associated protein of MTOR complex 1, Regulatory associated protein of MTOR, complex 1, Regulatory-associated protein of mTOR, Regulatory-associated protein of MTOR, complex 1, WD_REPEATS_REGION domain-containing protein | 8.745663 | 0.523873 | 4 | 0.593616 | 0.543357 |
| Macroautophagy | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 8.745663 | 0.523873 | 4 | 0.593616 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS4X | 40S ribosomal protein S4, 40S ribosomal protein S4, X isoform | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL35 | 60S ribosomal protein L35 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS15 | 40S ribosomal protein S15 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL7A | 60S ribosomal protein L7-A, 60S ribosomal protein L7a | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL19 | 60S ribosomal protein L19, Ribosomal protein L19 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL26 | 60S ribosomal protein L26, GEO07453p1, KOW domain-containing protein, Ribosomal protein L26 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL10 | 60S ribosomal protein L10, MGC89303 protein | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL32 | 60S ribosomal protein L32, Hypothetical LOC496894, Ribosomal protein L32, Uncharacterized protein | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL29 | 60S ribosomal protein L29 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL31 | 60S ribosomal protein L31 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS27 | 40S ribosomal protein S27 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.549907 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL10A | 60S ribosomal protein L10a, Ribosomal protein | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS7 | 40S ribosomal protein S7 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL13A | 60S ribosomal protein L13a | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS12 | 40S ribosomal protein S12 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL35A | 60S ribosomal protein L35-A, 60S ribosomal protein L35a | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS26 | 40S ribosomal protein S26 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS18 | 40S ribosomal protein S18 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL6 | 60S ribosomal protein L6 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS8 | 40S ribosomal protein S8 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL23A | 60S ribosomal protein L23-A, 60S ribosomal protein L23a, FI01658p, MGC89958 protein, Ribosomal protein L23a, Ribosomal_L23eN domain-containing protein | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS29 | 40S ribosomal protein S29 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPLP1 | 60S acidic ribosomal protein P1, Ribosomal protein, large, P1 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL3 | 60S ribosomal protein L3, LOC100145083 protein, Ribosomal protein L3, Uncharacterized protein | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL24 | 60S ribosomal protein L24, Ribosomal protein L24, TRASH domain-containing protein | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL17 | 60S ribosomal protein L17 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL11 | 60S ribosomal protein L11 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS16 | 40S ribosomal protein S16, Ribosomal protein S16, Uncharacterized protein | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL30 | 60S ribosomal protein L30 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL15 | 60S ribosomal protein L15, Ribosomal protein L15 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS23 | 40S ribosomal protein S23, 40S ribosomal protein S23-A | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS15A | 40S ribosomal protein S15a | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL36 | 60S ribosomal protein L36 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL22 | 60S ribosomal protein L22, 60S ribosomal protein L22 1, Ribosomal protein L22, Uncharacterized protein | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS14 | 40S ribosomal protein S14, Ribosomal protein S14, Uncharacterized protein | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL34 | 60S ribosomal protein L34 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS11 | 40S ribosomal protein S11 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS20 | 40S ribosomal protein S20 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL37A | 60S ribosomal protein L37-A, 60S ribosomal protein L37a, MGC89854 protein, Probable 60S ribosomal protein L37-A, Ribosomal protein L37a | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS19 | 40S ribosomal protein S19, Uncharacterized protein | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL13 | 60S ribosomal protein L13 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS13 | 40S ribosomal protein S13 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | FAU | 40S ribosomal protein S30 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS9 | 40S ribosomal protein S9 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS25 | 40S ribosomal protein S25 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL9 | 60S ribosomal protein L9 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | MTREX | Exosome RNA helicase MTR4, Mtr4 exosome RNA helicase | 11.558279 | 0.860727 | 4 | 0.907143 | 0.528009 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL28 | 60S ribosomal protein L28 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL41 | 60S ribosomal protein L41 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS24 | 40S ribosomal protein S24 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL14 | 60S ribosomal protein L14 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | NS1 | Guanine nucleotide-binding protein-like 3 homolog | 11.558279 | 0.860727 | 4 | 0.907143 | 0.507872 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS10 | 40S ribosomal protein S10, Ribosomal protein S10, S10_plectin domain-containing protein | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL8 | 60S ribosomal protein L8 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL23 | 60S ribosomal protein L23 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL39 | 60S ribosomal protein L39, LOC100135132 protein, Ribosomal protein L39 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | JAK2 | Janus kinase 2, Tyrosine-protein kinase, Tyrosine-protein kinase JAK2 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.573235 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS6 | 40S ribosomal protein S6 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.566528 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL27A | 60S ribosomal protein L27-A, 60S ribosomal protein L27a | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL21 | 60S ribosomal protein L21 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS4Y1 | 40S ribosomal protein S4, 40S ribosomal protein S4, Y isoform 1 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS28 | 40S ribosomal protein S28 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL5 | 60S ribosomal protein L5, Ribosomal protein L5 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS5 | 40S ribosomal protein S5, 40S ribosomal protein S5 [Cleaved into: 40S ribosomal protein S5, N-terminally processed], 40S ribosomal protein S5-A, Ribosomal protein S5 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL37 | 60S ribosomal protein L37, Ribosomal protein L37 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPSA | 40S ribosomal protein SA | 11.558279 | 0.860727 | 4 | 0.907143 | 0.521531 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL38 | 60S ribosomal protein L38 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL7 | 60S ribosomal protein L7, MGC79754 protein, Ribosomal protein L7, Uncharacterized protein | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS3 | 40S ribosomal protein S3, DNA-(apurinic or apyrimidinic site) lyase | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL4 | 60S ribosomal protein L4, Ribos_L4_asso_C domain-containing protein, Ribosomal protein L4 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL18 | 60S ribosomal protein L18, Ribosomal protein L18 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS3A | 40S ribosomal protein S3a | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL36A | 60S ribosomal protein L36-A, 60S ribosomal protein L36a, IP17351p, ribosomal protein L36a, Uncharacterized protein | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL18A | 60S ribosomal protein L18-A, 60S ribosomal protein L18a | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPLP0 | 60S acidic ribosomal protein P0 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL12 | 60S ribosomal protein L12 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPL27 | 60S ribosomal protein L27 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS17 | 40S ribosomal protein S17 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS2 | 40S ribosomal protein S2 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| Major pathway of rRNA processing in the nucleolus and cytosol | RPS21 | 40S ribosomal protein S21 | 11.558279 | 0.860727 | 4 | 0.907143 | 0.579609 |
| MAP2K and MAPK activation | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 9.065926 | 0.495804 | 2.839452 | 0.423152 | 0.594807 |
| MAP2K and MAPK activation | FN1 | Fibronectin | 9.065926 | 0.495804 | 2.839452 | 0.423152 | 0.578096 |
| MAP2K and MAPK activation | VWF | von Willebrand factor | 9.065926 | 0.495804 | 2.839452 | 0.423152 | 0.548181 |
| MAP2K and MAPK activation | YWHAB | 14-3-3 protein beta/alpha, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta | 9.065926 | 0.495804 | 2.839452 | 0.423152 | 0.513843 |
| MAP2K and MAPK activation | RAF1 | Non-specific serine/threonine protein kinase, RAF proto-oncogene serine/threonine-protein kinase | 9.065926 | 0.495804 | 2.839452 | 0.423152 | 0.573556 |
| MAP2K and MAPK activation | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 9.065926 | 0.495804 | 2.839452 | 0.423152 | 0.541254 |
| MAP2K and MAPK activation | RAP1A | RAP1A, member of RAS oncogene family, Ras-related protein Rap-1A, Uncharacterized protein | 9.065926 | 0.495804 | 2.839452 | 0.423152 | 0.530823 |
| MAP2K and MAPK activation | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 9.065926 | 0.495804 | 2.839452 | 0.423152 | 0.505988 |
| MAP2K and MAPK activation | BRAF | Non-specific serine/threonine protein kinase, Serine/threonine-protein kinase B-raf | 9.065926 | 0.495804 | 2.839452 | 0.423152 | 0.590074 |
| MAP2K and MAPK activation | MAP2K1 | Dual specificity mitogen-activated protein kinase kinase 1, MAP2K1 protein, Mitogen-activated protein kinase kinase 1, Protein kinase domain-containing protein | 9.065926 | 0.495804 | 2.839452 | 0.423152 | 0.545374 |
| MAPK6/MAPK4 signaling | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.515416 |
| MAPK6/MAPK4 signaling | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.369394 | 0.563141 | 4 | 0.645427 | 0.515416 |
| MAPK6/MAPK4 signaling | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.515416 |
| MAPK6/MAPK4 signaling | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.515416 |
| MAPK6/MAPK4 signaling | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.515416 |
| MAPK6/MAPK4 signaling | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.515416 |
| MAPK6/MAPK4 signaling | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.54735 |
| MAPK6/MAPK4 signaling | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.54735 |
| MAPK6/MAPK4 signaling | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.54735 |
| MAPK6/MAPK4 signaling | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.577481 |
| MAPK6/MAPK4 signaling | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.369394 | 0.563141 | 4 | 0.645427 | 0.515416 |
| MAPK6/MAPK4 signaling | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.515416 |
| MAPK6/MAPK4 signaling | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.515416 |
| MAPK6/MAPK4 signaling | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.573417 |
| MAPK6/MAPK4 signaling | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.515416 |
| MAPK6/MAPK4 signaling | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.515416 |
| MAPK6/MAPK4 signaling | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.54735 |
| MAPK6/MAPK4 signaling | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.54735 |
| MAPK6/MAPK4 signaling | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.546881 |
| MAPK6/MAPK4 signaling | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.54735 |
| MAPK6/MAPK4 signaling | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.572294 |
| MAPK6/MAPK4 signaling | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.565601 |
| MAPK6/MAPK4 signaling | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.577847 |
| MAPK6/MAPK4 signaling | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.54735 |
| MAPK6/MAPK4 signaling | CDK1 | Cell division control protein 2, Cyclin dependent kinase 1, Cyclin-dependent kinase 1 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.514008 |
| MAPK6/MAPK4 signaling | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.547953 |
| MAPK6/MAPK4 signaling | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.5197 |
| MAPK6/MAPK4 signaling | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.369394 | 0.563141 | 4 | 0.645427 | 0.579609 |
| MAPK6/MAPK4 signaling | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.515416 |
| MAPK6/MAPK4 signaling | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.515416 |
| MAPK6/MAPK4 signaling | CCND3 | Cyclin D3, Cyclin N-terminal domain-containing protein, G1/S-specific cyclin-D3 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.562307 |
| MAPK6/MAPK4 signaling | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.515416 |
| MAPK6/MAPK4 signaling | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.57244 |
| MAPK6/MAPK4 signaling | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.54735 |
| MAPK6/MAPK4 signaling | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.54735 |
| MAPK6/MAPK4 signaling | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.58858 |
| MAPK6/MAPK4 signaling | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.369394 | 0.563141 | 4 | 0.645427 | 0.568645 |
| MAPK6/MAPK4 signaling | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.515416 |
| MAPK6/MAPK4 signaling | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.369394 | 0.563141 | 4 | 0.645427 | 0.54735 |
| Maturation of replicase proteins | 1A | Replicase polyprotein 1a | 12.767545 | 1 | 0.701304 | ||
| Maturation of replicase proteins | REP | Rab proteins geranylgeranyltransferase component A, Replicase polyprotein 1ab | 12.767545 | 1 | 0.746505 | ||
| Metabolism of polyamines | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.515416 |
| Metabolism of polyamines | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.515416 |
| Metabolism of polyamines | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.577481 |
| Metabolism of polyamines | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.515416 |
| Metabolism of polyamines | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.546881 |
| Metabolism of polyamines | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.54735 |
| Metabolism of polyamines | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.565601 |
| Metabolism of polyamines | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.515416 |
| Metabolism of polyamines | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.515416 |
| Metabolism of polyamines | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.573417 |
| Metabolism of polyamines | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.515416 |
| Metabolism of polyamines | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.54735 |
| Metabolism of polyamines | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 5.585703 | 0.73257 | 4 | 0.534944 | 0.568645 |
| Metabolism of polyamines | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.515416 |
| Metabolism of polyamines | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 5.585703 | 0.73257 | 4 | 0.534944 | 0.515416 |
| Metabolism of polyamines | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.515416 |
| Metabolism of polyamines | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.58858 |
| Metabolism of polyamines | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.54735 |
| Metabolism of polyamines | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.515416 |
| Metabolism of polyamines | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.54735 |
| Metabolism of polyamines | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.515416 |
| Metabolism of polyamines | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.515416 |
| Metabolism of polyamines | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.5197 |
| Metabolism of polyamines | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.54735 |
| Metabolism of polyamines | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 5.585703 | 0.73257 | 4 | 0.534944 | 0.515416 |
| Metabolism of polyamines | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.572294 |
| Metabolism of polyamines | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.577847 |
| Metabolism of polyamines | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.515416 |
| Metabolism of polyamines | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.54735 |
| Metabolism of polyamines | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.54735 |
| Metabolism of polyamines | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.54735 |
| Metabolism of polyamines | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.547953 |
| Metabolism of polyamines | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.54735 |
| Metabolism of polyamines | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.515416 |
| Metabolism of polyamines | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.57244 |
| Metabolism of polyamines | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 5.585703 | 0.73257 | 4 | 0.534944 | 0.54735 |
| Mitotic Prometaphase | CDK1 | Cell division control protein 2, Cyclin dependent kinase 1, Cyclin-dependent kinase 1 | 8.989683 | 0.498926 | 4 | 0.593731 | 0.514008 |
| Mitotic Prometaphase | AURKB | Aurora kinase, Aurora kinase B | 8.989683 | 0.498926 | 4 | 0.593731 | 0.524412 |
| Mitotic Prometaphase | KIF2C | Kinesin-like protein, Kinesin-like protein KIF2C | 8.989683 | 0.498926 | 4 | 0.593731 | 0.611637 |
| Mitotic Prometaphase | CRK | 14_3_3 domain-containing protein, Adapter molecule crk, Adapter molecule Crk, CRK proto-oncogene, adaptor protein, Uncharacterized protein, V-crk avian sarcoma virus CT10 oncogene homolog | 8.989683 | 0.498926 | 4 | 0.593731 | 0.513369 |
| Mitotic Prometaphase | HSP90AA1 | Heat shock protein 90 alpha family class A member 1, Heat shock protein HSP 90-alpha | 8.989683 | 0.498926 | 4 | 0.593731 | 0.624325 |
| Mitotic Prometaphase | CCNB1 | Cyclin B1, G2/mitotic-specific cyclin-B1, Uncharacterized protein | 8.989683 | 0.498926 | 4 | 0.593731 | 0.513205 |
| Mitotic Prometaphase | PLK1 | Serine/threonine-protein kinase PLK, Serine/threonine-protein kinase PLK1 | 8.989683 | 0.498926 | 4 | 0.593731 | 0.504187 |
| Mitotic Prometaphase | INCENP | Inner centromere protein, Inner centromere protein, isoform A, Uncharacterized protein | 8.989683 | 0.498926 | 4 | 0.593731 | 0.527883 |
| Mitotic Prometaphase | CSNK2A1 | Casein kinase 2, alpha 1 polypeptide, Casein kinase II subunit alpha, Protein kinase domain-containing protein | 8.989683 | 0.498926 | 4 | 0.593731 | 0.538737 |
| Mitotic Prometaphase | PPP1CC | Serine/threonine-protein phosphatase, Serine/threonine-protein phosphatase PP1-gamma catalytic subunit | 8.989683 | 0.498926 | 4 | 0.593731 | 0.503303 |
| Mitotic Prometaphase | CSNK2A2 | Casein kinase 2 alpha 2, Casein kinase II subunit alpha' | 8.989683 | 0.498926 | 4 | 0.593731 | 0.53622 |
| Mitotic Prometaphase | CENPE | Centromere protein E, Centromere-associated protein E, Kinesin motor domain-containing protein | 8.989683 | 0.498926 | 4 | 0.593731 | 0.564477 |
| Mitotic Prometaphase | RPS27 | 40S ribosomal protein S27 | 8.989683 | 0.498926 | 4 | 0.593731 | 0.549907 |
| Mitotic Prometaphase | PPP2R5A | Serine/threonine protein phosphatase 2A regulatory subunit, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform | 8.989683 | 0.498926 | 4 | 0.593731 | 0.559184 |
| Mitotic Prometaphase | NEK7 | NIMA related kinase 7, Protein kinase domain-containing protein, Serine/threonine-protein kinase Nek7 | 8.989683 | 0.498926 | 4 | 0.593731 | 0.513171 |
| Mitotic Prometaphase | HDAC8 | Histone deacetylase, Histone deacetylase 8, WD repeat and SOCS box containing 2 | 8.989683 | 0.498926 | 4 | 0.593731 | 0.535733 |
| Mitotic Prometaphase | CSNK2B | Casein kinase II subunit beta | 8.989683 | 0.498926 | 4 | 0.593731 | 0.537688 |
| MTOR signalling | RPS6 | 40S ribosomal protein S6 | 9.54151 | 0.584349 | 4 | 0.664911 | 0.566528 |
| MTOR signalling | RPTOR | Regulatory associated protein of MTOR complex 1, Regulatory associated protein of MTOR, complex 1, Regulatory-associated protein of mTOR, Regulatory-associated protein of MTOR, complex 1, WD_REPEATS_REGION domain-containing protein | 9.54151 | 0.584349 | 4 | 0.664911 | 0.543357 |
| MTOR signalling | EEF2K | Eukaryotic elongation factor 2 kinase | 9.54151 | 0.584349 | 4 | 0.664911 | 0.532313 |
| MTOR signalling | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 9.54151 | 0.584349 | 4 | 0.664911 | 0.552041 |
| MTOR signalling | PRKAB1 | 5'-AMP-activated protein kinase subunit beta-1, 5'AMP-activated protein kinase beta-1 non-catalytic subunit, Protein kinase AMP-activated non-catalytic subunit beta 1, Protein kinase, AMP-activated, beta 1 non-catalytic subunit | 9.54151 | 0.584349 | 4 | 0.664911 | 0.500481 |
| MTOR signalling | PRKAG1 | 5'-AMP-activated protein kinase subunit gamma-1, Protein kinase, AMP-activated, gamma 1 non-catalytic subunit | 9.54151 | 0.584349 | 4 | 0.664911 | 0.515113 |
| MTOR signalling | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 9.54151 | 0.584349 | 4 | 0.664911 | 0.589663 |
| MTOR signalling | RPS6KB1 | Ribosomal protein S6 kinase, Ribosomal protein S6 kinase beta-1 | 9.54151 | 0.584349 | 4 | 0.664911 | 0.529306 |
| MTOR signalling | PRKAG2 | 5'-AMP-activated protein kinase subunit gamma-2, 5'-AMP-activated protein kinase subunit gamma-2 isoform X8, Protein kinase AMP-activated non-catalytic subunit gamma 2, Protein kinase, AMP-activated, gamma 2 non-catalytic subunit, Uncharacterized protein | 9.54151 | 0.584349 | 4 | 0.664911 | 0.510193 |
| MTOR signalling | FPR1 | FK506-binding protein 1, fMet-Leu-Phe receptor, Formyl peptide receptor 1 | 9.54151 | 0.584349 | 4 | 0.664911 | 0.511829 |
| MTOR signalling | FKBP1A | Peptidyl-prolyl cis-trans isomerase FKBP1A, Peptidylprolyl isomerase | 9.54151 | 0.584349 | 4 | 0.664911 | 0.534375 |
| MTOR signalling | STK11 | Non-specific serine/threonine protein kinase, Serine/threonine-protein kinase STK11 | 9.54151 | 0.584349 | 4 | 0.664911 | 0.504599 |
| MTOR signalling | MTOR | Nucleoprotein TPR, Serine/threonine-protein kinase mTOR, Serine/threonine-protein kinase TOR | 9.54151 | 0.584349 | 4 | 0.664911 | 0.619026 |
| MTOR signalling | YWHAB | 14-3-3 protein beta/alpha, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta | 9.54151 | 0.584349 | 4 | 0.664911 | 0.513843 |
| MyD88:MAL(TIRAP) cascade initiated on plasma membrane | MAPK8 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 8 | 8.663037 | 0.39878 | 3.38468 | 0.445768 | 0.503967 |
| MyD88:MAL(TIRAP) cascade initiated on plasma membrane | MAPK10 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 10 | 8.663037 | 0.39878 | 3.38468 | 0.445768 | 0.506109 |
| MyD88:MAL(TIRAP) cascade initiated on plasma membrane | UBE2V1 | Ubiquitin conjugating enzyme E2 V1, Ubiquitin-conjugating enzyme E2 variant 1 | 8.663037 | 0.39878 | 3.38468 | 0.445768 | 0.593364 |
| MyD88:MAL(TIRAP) cascade initiated on plasma membrane | MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | 8.663037 | 0.39878 | 3.38468 | 0.445768 | 0.519182 |
| MyD88:MAL(TIRAP) cascade initiated on plasma membrane | IRAK4 | Interleukin-1 receptor-associated kinase 4 | 8.663037 | 0.39878 | 3.38468 | 0.445768 | 0.50686 |
| MyD88:MAL(TIRAP) cascade initiated on plasma membrane | MAP2K1 | Dual specificity mitogen-activated protein kinase kinase 1, MAP2K1 protein, Mitogen-activated protein kinase kinase 1, Protein kinase domain-containing protein | 8.663037 | 0.39878 | 3.38468 | 0.445768 | 0.545374 |
| MyD88:MAL(TIRAP) cascade initiated on plasma membrane | RIPK2 | Receptor-interacting serine-threonine kinase 2, Receptor-interacting serine/threonine-protein kinase 2 | 8.663037 | 0.39878 | 3.38468 | 0.445768 | 0.530344 |
| MyD88:MAL(TIRAP) cascade initiated on plasma membrane | BTK | Tyrosine-protein kinase, Tyrosine-protein kinase BTK | 8.663037 | 0.39878 | 3.38468 | 0.445768 | 0.602174 |
| MyD88:MAL(TIRAP) cascade initiated on plasma membrane | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 8.663037 | 0.39878 | 3.38468 | 0.445768 | 0.541254 |
| MyD88:MAL(TIRAP) cascade initiated on plasma membrane | MAPK14 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 14 | 8.663037 | 0.39878 | 3.38468 | 0.445768 | 0.591941 |
| MyD88:MAL(TIRAP) cascade initiated on plasma membrane | MDM2 | E3 ubiquitin-protein ligase Mdm2 | 8.663037 | 0.39878 | 3.38468 | 0.445768 | 0.659771 |
| MyD88:MAL(TIRAP) cascade initiated on plasma membrane | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 8.663037 | 0.39878 | 3.38468 | 0.445768 | 0.579609 |
| MyD88:MAL(TIRAP) cascade initiated on plasma membrane | IKBKB | I-kappa-B kinase, Inhibitor of nuclear factor kappa-B kinase subunit beta | 8.663037 | 0.39878 | 3.38468 | 0.445768 | 0.518113 |
| MyD88:MAL(TIRAP) cascade initiated on plasma membrane | APP | ABPP, Amyloid-beta A4 protein, Amyloid-beta precursor protein | 8.663037 | 0.39878 | 3.38468 | 0.445768 | 0.506343 |
| MyD88:MAL(TIRAP) cascade initiated on plasma membrane | MAPK11 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 11 | 8.663037 | 0.39878 | 3.38468 | 0.445768 | 0.567539 |
| NCAM signaling for neurite out-growth | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 8.091213 | 0.612893 | 4 | 0.604363 | 0.505988 |
| NCAM signaling for neurite out-growth | FGFR1 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 1 | 8.091213 | 0.612893 | 4 | 0.604363 | 0.55821 |
| NCAM signaling for neurite out-growth | PTK2 | Focal adhesion kinase 1, Non-specific protein-tyrosine kinase, Serine/threonine-protein kinase PTK2/STK2 | 8.091213 | 0.612893 | 4 | 0.604363 | 0.503121 |
| NCAM signaling for neurite out-growth | CACNB2 | Brain protein I3, Calcium channel voltage-dependent subunit beta 2, Voltage-dependent L-type calcium channel subunit beta-2 | 8.091213 | 0.612893 | 4 | 0.604363 | 0.596371 |
| NCAM signaling for neurite out-growth | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 8.091213 | 0.612893 | 4 | 0.604363 | 0.541254 |
| NCAM signaling for neurite out-growth | CACNA1C | Voltage-dependent L-type calcium channel subunit alpha, Voltage-dependent L-type calcium channel subunit alpha-1C | 8.091213 | 0.612893 | 4 | 0.604363 | 0.585419 |
| NCAM signaling for neurite out-growth | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 8.091213 | 0.612893 | 4 | 0.604363 | 0.594807 |
| NCAM signaling for neurite out-growth | CACNA1G | Voltage-dependent T-type calcium channel subunit alpha-1G | 8.091213 | 0.612893 | 4 | 0.604363 | 0.579421 |
| NCAM signaling for neurite out-growth | CACNA1D | Voltage-dependent L-type calcium channel subunit alpha, Voltage-dependent L-type calcium channel subunit alpha-1D | 8.091213 | 0.612893 | 4 | 0.604363 | 0.585385 |
| NCAM signaling for neurite out-growth | CACNB1 | Calcium channel voltage-dependent subunit beta 1, GuKc domain-containing protein, Voltage-dependent L-type calcium channel subunit beta-1 | 8.091213 | 0.612893 | 4 | 0.604363 | 0.664934 |
| NCAM signaling for neurite out-growth | CACNA1H | Voltage-dependent T-type calcium channel subunit alpha-1H | 8.091213 | 0.612893 | 4 | 0.604363 | 0.553666 |
| NCAM signaling for neurite out-growth | CACNB4 | Calcium voltage-gated channel auxiliary subunit beta 4, SH3 domain-containing protein, Voltage-dependent L-type calcium channel subunit beta-4 | 8.091213 | 0.612893 | 4 | 0.604363 | 0.598629 |
| NCAM signaling for neurite out-growth | CACNB3 | Calcium channel voltage-dependent subunit beta 3, Voltage-dependent L-type calcium channel subunit beta-3 | 8.091213 | 0.612893 | 4 | 0.604363 | 0.592713 |
| NCAM signaling for neurite out-growth | CACNA1S | Voltage-dependent L-type calcium channel subunit alpha-1S | 8.091213 | 0.612893 | 4 | 0.604363 | 0.596399 |
| NCAM signaling for neurite out-growth | CACNA1I | Voltage-dependent T-type calcium channel subunit alpha-1I | 8.091213 | 0.612893 | 4 | 0.604363 | 0.58751 |
| NCAM1 interactions | CACNB3 | Calcium channel voltage-dependent subunit beta 3, Voltage-dependent L-type calcium channel subunit beta-3 | 9.751164 | 0.853869 | 4 | 0.810487 | 0.592713 |
| NCAM1 interactions | CACNA1H | Voltage-dependent T-type calcium channel subunit alpha-1H | 9.751164 | 0.853869 | 4 | 0.810487 | 0.553666 |
| NCAM1 interactions | CACNA1D | Voltage-dependent L-type calcium channel subunit alpha, Voltage-dependent L-type calcium channel subunit alpha-1D | 9.751164 | 0.853869 | 4 | 0.810487 | 0.585385 |
| NCAM1 interactions | CACNA1G | Voltage-dependent T-type calcium channel subunit alpha-1G | 9.751164 | 0.853869 | 4 | 0.810487 | 0.579421 |
| NCAM1 interactions | CACNA1I | Voltage-dependent T-type calcium channel subunit alpha-1I | 9.751164 | 0.853869 | 4 | 0.810487 | 0.58751 |
| NCAM1 interactions | CACNA1S | Voltage-dependent L-type calcium channel subunit alpha-1S | 9.751164 | 0.853869 | 4 | 0.810487 | 0.596399 |
| NCAM1 interactions | CACNB2 | Brain protein I3, Calcium channel voltage-dependent subunit beta 2, Voltage-dependent L-type calcium channel subunit beta-2 | 9.751164 | 0.853869 | 4 | 0.810487 | 0.596371 |
| NCAM1 interactions | CACNB4 | Calcium voltage-gated channel auxiliary subunit beta 4, SH3 domain-containing protein, Voltage-dependent L-type calcium channel subunit beta-4 | 9.751164 | 0.853869 | 4 | 0.810487 | 0.598629 |
| NCAM1 interactions | CACNB1 | Calcium channel voltage-dependent subunit beta 1, GuKc domain-containing protein, Voltage-dependent L-type calcium channel subunit beta-1 | 9.751164 | 0.853869 | 4 | 0.810487 | 0.664934 |
| NCAM1 interactions | CACNA1C | Voltage-dependent L-type calcium channel subunit alpha, Voltage-dependent L-type calcium channel subunit alpha-1C | 9.751164 | 0.853869 | 4 | 0.810487 | 0.585419 |
| Neddylation | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.54735 |
| Neddylation | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.54735 |
| Neddylation | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.54735 |
| Neddylation | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.57244 |
| Neddylation | DCUN1D1 | DCN1-like protein, DCN1-like protein 1 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.554972 |
| Neddylation | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.515416 |
| Neddylation | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.577847 |
| Neddylation | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.515416 |
| Neddylation | HDAC8 | Histone deacetylase, Histone deacetylase 8, WD repeat and SOCS box containing 2 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.535733 |
| Neddylation | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.515416 |
| Neddylation | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.515416 |
| Neddylation | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.515416 |
| Neddylation | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.515416 |
| Neddylation | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.58858 |
| Neddylation | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.54735 |
| Neddylation | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.52081 | 0.574837 | 4 | 0.659087 | 0.579609 |
| Neddylation | DDB1 | DNA damage-binding protein 1 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.510276 |
| Neddylation | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.547953 |
| Neddylation | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.54735 |
| Neddylation | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.52081 | 0.574837 | 4 | 0.659087 | 0.568645 |
| Neddylation | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.572294 |
| Neddylation | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.52081 | 0.574837 | 4 | 0.659087 | 0.515416 |
| Neddylation | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.54735 |
| Neddylation | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.546881 |
| Neddylation | MAP2K5 | Dual specificity mitogen-activated protein kinase kinase 5, Mitogen-activated protein kinase kinase 5 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.500391 |
| Neddylation | CSN2 | Beta-casein, COP9 signalosome complex subunit 2 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.537553 |
| Neddylation | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.54735 |
| Neddylation | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.5197 |
| Neddylation | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.565601 |
| Neddylation | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.515416 |
| Neddylation | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.577481 |
| Neddylation | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.515416 |
| Neddylation | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.54735 |
| Neddylation | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.515416 |
| Neddylation | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.515416 |
| Neddylation | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.54735 |
| Neddylation | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.515416 |
| Neddylation | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.54735 |
| Neddylation | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.515416 |
| Neddylation | VHL | Protein Vhl, Uncharacterized protein, von Hippel-Lindau disease tumor suppressor, von Hippel-Lindau disease tumor suppressor isoform 1, von Hippel-Lindau tumor suppressor | 9.52081 | 0.574837 | 4 | 0.659087 | 0.588856 |
| Neddylation | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.515416 |
| Neddylation | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.52081 | 0.574837 | 4 | 0.659087 | 0.515416 |
| Neddylation | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.52081 | 0.574837 | 4 | 0.659087 | 0.573417 |
| Negative feedback regulation of MAPK pathway | MAP2K1 | Dual specificity mitogen-activated protein kinase kinase 1, MAP2K1 protein, Mitogen-activated protein kinase kinase 1, Protein kinase domain-containing protein | 8.666523 | 0.737795 | 2.5179 | 0.438425 | 0.545374 |
| Negative feedback regulation of MAPK pathway | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 8.666523 | 0.737795 | 2.5179 | 0.438425 | 0.541254 |
| Negative feedback regulation of MAPK pathway | BRAF | Non-specific serine/threonine protein kinase, Serine/threonine-protein kinase B-raf | 8.666523 | 0.737795 | 2.5179 | 0.438425 | 0.590074 |
| Negative feedback regulation of MAPK pathway | RAF1 | Non-specific serine/threonine protein kinase, RAF proto-oncogene serine/threonine-protein kinase | 8.666523 | 0.737795 | 2.5179 | 0.438425 | 0.573556 |
| Negative regulation of NMDA receptor-mediated neuronal transmission | GRIN2A | Glutamate receptor, Glutamate receptor ionotropic, NMDA 2A | 7.912485 | 0.597503 | 2.74713 | 0.403449 | 0.527756 |
| Negative regulation of NMDA receptor-mediated neuronal transmission | GRIN2B | Glutamate receptor, Glutamate receptor ionotropic, NMDA 2B | 7.912485 | 0.597503 | 2.74713 | 0.403449 | 0.56443 |
| Negative regulation of NMDA receptor-mediated neuronal transmission | GRIN2D | Glutamate receptor, Glutamate receptor ionotropic, NMDA 2D | 7.912485 | 0.597503 | 2.74713 | 0.403449 | 0.556318 |
| Negative regulation of NMDA receptor-mediated neuronal transmission | GRIN1 | Glutamate receptor, Glutamate receptor ionotropic, NMDA 1 | 7.912485 | 0.597503 | 2.74713 | 0.403449 | 0.573676 |
| Negative regulation of NMDA receptor-mediated neuronal transmission | GRIN2C | Glutamate receptor, Glutamate receptor ionotropic, NMDA 2C | 7.912485 | 0.597503 | 2.74713 | 0.403449 | 0.557006 |
| Negative regulation of NOTCH4 signaling | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.54735 |
| Negative regulation of NOTCH4 signaling | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.515416 |
| Negative regulation of NOTCH4 signaling | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.577847 |
| Negative regulation of NOTCH4 signaling | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.54735 |
| Negative regulation of NOTCH4 signaling | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.54735 |
| Negative regulation of NOTCH4 signaling | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.808717 | 0.797455 | 4 | 0.785249 | 0.579609 |
| Negative regulation of NOTCH4 signaling | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.57244 |
| Negative regulation of NOTCH4 signaling | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.547953 |
| Negative regulation of NOTCH4 signaling | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.54735 |
| Negative regulation of NOTCH4 signaling | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.572294 |
| Negative regulation of NOTCH4 signaling | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.5197 |
| Negative regulation of NOTCH4 signaling | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.515416 |
| Negative regulation of NOTCH4 signaling | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.808717 | 0.797455 | 4 | 0.785249 | 0.515416 |
| Negative regulation of NOTCH4 signaling | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.565601 |
| Negative regulation of NOTCH4 signaling | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.515416 |
| Negative regulation of NOTCH4 signaling | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.515416 |
| Negative regulation of NOTCH4 signaling | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.577481 |
| Negative regulation of NOTCH4 signaling | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.515416 |
| Negative regulation of NOTCH4 signaling | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.515416 |
| Negative regulation of NOTCH4 signaling | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.515416 |
| Negative regulation of NOTCH4 signaling | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.515416 |
| Negative regulation of NOTCH4 signaling | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.54735 |
| Negative regulation of NOTCH4 signaling | YWHAZ | 14_3_3 domain-containing protein, 14-3-3 protein zeta, 14-3-3 protein zeta/delta, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta | 9.808717 | 0.797455 | 4 | 0.785249 | 0.602005 |
| Negative regulation of NOTCH4 signaling | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.54735 |
| Negative regulation of NOTCH4 signaling | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.515416 |
| Negative regulation of NOTCH4 signaling | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.808717 | 0.797455 | 4 | 0.785249 | 0.568645 |
| Negative regulation of NOTCH4 signaling | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.58858 |
| Negative regulation of NOTCH4 signaling | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.515416 |
| Negative regulation of NOTCH4 signaling | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.515416 |
| Negative regulation of NOTCH4 signaling | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.54735 |
| Negative regulation of NOTCH4 signaling | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.54735 |
| Negative regulation of NOTCH4 signaling | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.573417 |
| Negative regulation of NOTCH4 signaling | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.808717 | 0.797455 | 4 | 0.785249 | 0.515416 |
| Negative regulation of NOTCH4 signaling | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.515416 |
| Negative regulation of NOTCH4 signaling | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.589663 |
| Negative regulation of NOTCH4 signaling | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.515416 |
| Negative regulation of NOTCH4 signaling | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.808717 | 0.797455 | 4 | 0.785249 | 0.546881 |
| Negative regulation of the PI3K/AKT network | ERBB3 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-3 | 8.068382 | 0.43582 | 4 | 0.514649 | 0.551662 |
| Negative regulation of the PI3K/AKT network | ERBB2 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-2 | 8.068382 | 0.43582 | 4 | 0.514649 | 0.589497 |
| Negative regulation of the PI3K/AKT network | ESR2 | Estrogen receptor 2, Estrogen receptor beta | 8.068382 | 0.43582 | 4 | 0.514649 | 0.53484 |
| Negative regulation of the PI3K/AKT network | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 8.068382 | 0.43582 | 4 | 0.514649 | 0.589663 |
| Negative regulation of the PI3K/AKT network | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 8.068382 | 0.43582 | 4 | 0.514649 | 0.594807 |
| Negative regulation of the PI3K/AKT network | INSR | Insulin receptor, Tyrosine-protein kinase receptor | 8.068382 | 0.43582 | 4 | 0.514649 | 0.563226 |
| Negative regulation of the PI3K/AKT network | FGFR4 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 4 | 8.068382 | 0.43582 | 4 | 0.514649 | 0.555253 |
| Negative regulation of the PI3K/AKT network | MDM2 | E3 ubiquitin-protein ligase Mdm2 | 8.068382 | 0.43582 | 4 | 0.514649 | 0.659771 |
| Negative regulation of the PI3K/AKT network | MET | Hepatocyte growth factor receptor, HGF/SF receptor | 8.068382 | 0.43582 | 4 | 0.514649 | 0.5914 |
| Negative regulation of the PI3K/AKT network | ERBB4 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-4 | 8.068382 | 0.43582 | 4 | 0.514649 | 0.550399 |
| Negative regulation of the PI3K/AKT network | ESR1 | Estrogen receptor | 8.068382 | 0.43582 | 4 | 0.514649 | 0.561602 |
| Negative regulation of the PI3K/AKT network | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 8.068382 | 0.43582 | 4 | 0.514649 | 0.541254 |
| Negative regulation of the PI3K/AKT network | IRAK4 | Interleukin-1 receptor-associated kinase 4 | 8.068382 | 0.43582 | 4 | 0.514649 | 0.50686 |
| Negative regulation of the PI3K/AKT network | PDGFRB | Platelet-derived growth factor receptor beta, Receptor protein-tyrosine kinase | 8.068382 | 0.43582 | 4 | 0.514649 | 0.550695 |
| Negative regulation of the PI3K/AKT network | FLT3 | Receptor protein-tyrosine kinase, Receptor-type tyrosine-protein kinase FLT3 | 8.068382 | 0.43582 | 4 | 0.514649 | 0.529563 |
| Negative regulation of the PI3K/AKT network | EGFR | Epidermal growth factor receptor, Receptor protein-tyrosine kinase | 8.068382 | 0.43582 | 4 | 0.514649 | 0.625817 |
| Negative regulation of the PI3K/AKT network | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 8.068382 | 0.43582 | 4 | 0.514649 | 0.552041 |
| Negative regulation of the PI3K/AKT network | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 8.068382 | 0.43582 | 4 | 0.514649 | 0.646924 |
| Negative regulation of the PI3K/AKT network | FGFR3 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 3 | 8.068382 | 0.43582 | 4 | 0.514649 | 0.560785 |
| Negative regulation of the PI3K/AKT network | LCK | Proto-oncogene tyrosine-protein kinase LCK, Tyrosine-protein kinase, Tyrosine-protein kinase Lck | 8.068382 | 0.43582 | 4 | 0.514649 | 0.543278 |
| Negative regulation of the PI3K/AKT network | PIK3CD | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 8.068382 | 0.43582 | 4 | 0.514649 | 0.632077 |
| Negative regulation of the PI3K/AKT network | FGFR1 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 1 | 8.068382 | 0.43582 | 4 | 0.514649 | 0.55821 |
| Negative regulation of the PI3K/AKT network | PPP2R5A | Serine/threonine protein phosphatase 2A regulatory subunit, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform | 8.068382 | 0.43582 | 4 | 0.514649 | 0.559184 |
| Negative regulation of the PI3K/AKT network | FGFR2 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 2 | 8.068382 | 0.43582 | 4 | 0.514649 | 0.577012 |
| Negative regulation of the PI3K/AKT network | PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 8.068382 | 0.43582 | 4 | 0.514649 | 0.589385 |
| Negative regulation of the PI3K/AKT network | PDGFRA | Platelet-derived growth factor receptor alpha | 8.068382 | 0.43582 | 4 | 0.514649 | 0.532138 |
| Negative regulation of the PI3K/AKT network | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 8.068382 | 0.43582 | 4 | 0.514649 | 0.660919 |
| Negative regulation of the PI3K/AKT network | KIT | Mast/stem cell growth factor receptor, Mast/stem cell growth factor receptor Kit | 8.068382 | 0.43582 | 4 | 0.514649 | 0.562353 |
| Negative regulation of the PI3K/AKT network | FGF1 | Fibroblast growth factor 1, Multifunctional fusion protein [Includes: Fibroblast growth factor 1, Putative fibroblast growth factor 1 | 8.068382 | 0.43582 | 4 | 0.514649 | 0.509844 |
| Neurotransmitter receptors and postsynaptic signal transmission | CHRNA3 | Cholinergic receptor nicotinic alpha 3 subunit, Cholinergic receptor, nicotinic, alpha 3, Neuronal acetylcholine receptor subunit alpha-3 | 7.350272 | 0.409387 | 4 | 0.464385 | 0.52477 |
| Neurotransmitter receptors and postsynaptic signal transmission | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 7.350272 | 0.409387 | 4 | 0.464385 | 0.594807 |
| Neurotransmitter receptors and postsynaptic signal transmission | ERBB4 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-4 | 7.350272 | 0.409387 | 4 | 0.464385 | 0.550399 |
| Neurotransmitter receptors and postsynaptic signal transmission | KCNJ5 | Cardiac inward rectifier, G protein-activated inward rectifier potassium channel 4, Potassium inwardly rectifying channel subfamily J member 5 | 7.350272 | 0.409387 | 4 | 0.464385 | 0.565449 |
| Neurotransmitter receptors and postsynaptic signal transmission | PRKAG2 | 5'-AMP-activated protein kinase subunit gamma-2, 5'-AMP-activated protein kinase subunit gamma-2 isoform X8, Protein kinase AMP-activated non-catalytic subunit gamma 2, Protein kinase, AMP-activated, gamma 2 non-catalytic subunit, Uncharacterized protein | 7.350272 | 0.409387 | 4 | 0.464385 | 0.510193 |
| Neurotransmitter receptors and postsynaptic signal transmission | ADCY7 | Adenylate cyclase type 7 | 7.350272 | 0.409387 | 4 | 0.464385 | 0.550655 |
| Neurotransmitter receptors and postsynaptic signal transmission | HTR3B | 5-hydroxytryptamine, 5-hydroxytryptamine receptor 3B | 7.350272 | 0.409387 | 4 | 0.464385 | 0.501204 |
| Neurotransmitter receptors and postsynaptic signal transmission | CACNG2 | Voltage-dependent calcium channel gamma-2 subunit | 7.350272 | 0.409387 | 4 | 0.464385 | 0.593521 |
| Neurotransmitter receptors and postsynaptic signal transmission | KCNJ3 | G protein-activated inward rectifier potassium channel 1 | 7.350272 | 0.409387 | 4 | 0.464385 | 0.56031 |
| Neurotransmitter receptors and postsynaptic signal transmission | GABRA2 | GABA(A) receptor subunit alpha-2, Gamma-aminobutyric acid receptor subunit alpha-2 | 7.350272 | 0.409387 | 4 | 0.464385 | 0.519767 |
| Neurotransmitter receptors and postsynaptic signal transmission | GRIN1 | Glutamate receptor, Glutamate receptor ionotropic, NMDA 1 | 7.350272 | 0.409387 | 4 | 0.464385 | 0.573676 |
| Neurotransmitter receptors and postsynaptic signal transmission | GRIN3A | Glutamate receptor, Glutamate receptor ionotropic, NMDA 3A | 7.350272 | 0.409387 | 4 | 0.464385 | 0.524922 |
| Neurotransmitter receptors and postsynaptic signal transmission | CHRNB4 | Cholinergic receptor nicotinic beta 4 subunit, Cholinergic receptor, nicotinic, beta 4, Neuronal acetylcholine receptor subunit beta-4 | 7.350272 | 0.409387 | 4 | 0.464385 | 0.507113 |
| Neurotransmitter receptors and postsynaptic signal transmission | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 7.350272 | 0.409387 | 4 | 0.464385 | 0.505988 |
| Neurotransmitter receptors and postsynaptic signal transmission | GRIN2C | Glutamate receptor, Glutamate receptor ionotropic, NMDA 2C | 7.350272 | 0.409387 | 4 | 0.464385 | 0.557006 |
| Neurotransmitter receptors and postsynaptic signal transmission | CACNG3 | Calcium voltage-gated channel auxiliary subunit gamma 3, Voltage-dependent calcium channel gamma-3 subunit | 7.350272 | 0.409387 | 4 | 0.464385 | 0.598629 |
| Neurotransmitter receptors and postsynaptic signal transmission | GRIN3B | Glutamate receptor ionotropic, NMDA 3B | 7.350272 | 0.409387 | 4 | 0.464385 | 0.524714 |
| Neurotransmitter receptors and postsynaptic signal transmission | MAPT | Microtubule-associated protein, Microtubule-associated protein tau | 7.350272 | 0.409387 | 4 | 0.464385 | 0.507322 |
| Neurotransmitter receptors and postsynaptic signal transmission | GNAI2 | G protein subunit alpha i2, Guanine nucleotide binding protein, Guanine nucleotide-binding protein G(i) subunit alpha-2 | 7.350272 | 0.409387 | 4 | 0.464385 | 0.512918 |
| Neurotransmitter receptors and postsynaptic signal transmission | GABBR2 | G_PROTEIN_RECEP_F3_4 domain-containing protein, Gabbr2 protein, Gamma-aminobutyric acid, Gamma-aminobutyric acid type B receptor subunit 2 | 7.350272 | 0.409387 | 4 | 0.464385 | 0.542461 |
| Neurotransmitter receptors and postsynaptic signal transmission | GABRG2 | GABA(A) receptor subunit gamma-2, Gamma-aminobutyric acid, Gamma-aminobutyric acid receptor subunit gamma-2, Gamma-aminobutyric acid type A receptor subunit gamma2 | 7.350272 | 0.409387 | 4 | 0.464385 | 0.522585 |
| Neurotransmitter receptors and postsynaptic signal transmission | CACNG8 | Voltage-dependent calcium channel gamma-8 subunit | 7.350272 | 0.409387 | 4 | 0.464385 | 0.579696 |
| Neurotransmitter receptors and postsynaptic signal transmission | PRKCG | Protein kinase C, Protein kinase C gamma type, Sperm-associated antigen 9 | 7.350272 | 0.409387 | 4 | 0.464385 | 0.500344 |
| Neurotransmitter receptors and postsynaptic signal transmission | GABRA3 | Gamma-aminobutyric acid, Gamma-aminobutyric acid receptor subunit alpha-3, Gamma-aminobutyric acid type A receptor alpha3 subunit, Gamma-aminobutyric acid type A receptor subunit alpha3, Uncharacterized protein | 7.350272 | 0.409387 | 4 | 0.464385 | 0.516173 |
| Neurotransmitter receptors and postsynaptic signal transmission | GABRB3 | Gabrb3 protein, Gamma-aminobutyric acid receptor subunit beta-3, Gamma-aminobutyric acid type A receptor beta3 subunit, Gamma-aminobutyric acid type A receptor subunit beta3 | 7.350272 | 0.409387 | 4 | 0.464385 | 0.53034 |
| Neurotransmitter receptors and postsynaptic signal transmission | GABRA1 | GABA(A) receptor subunit alpha-1, Gamma-aminobutyric acid receptor subunit alpha-1 | 7.350272 | 0.409387 | 4 | 0.464385 | 0.508013 |
| Neurotransmitter receptors and postsynaptic signal transmission | GABRA5 | Gamma-aminobutyric acid, Gamma-aminobutyric acid receptor subunit alpha-5, Gamma-aminobutyric acid type A receptor alpha5 subunit, Gamma-aminobutyric acid type A receptor subunit alpha5, Uncharacterized protein | 7.350272 | 0.409387 | 4 | 0.464385 | 0.519175 |
| Neurotransmitter receptors and postsynaptic signal transmission | PRKAB1 | 5'-AMP-activated protein kinase subunit beta-1, 5'AMP-activated protein kinase beta-1 non-catalytic subunit, Protein kinase AMP-activated non-catalytic subunit beta 1, Protein kinase, AMP-activated, beta 1 non-catalytic subunit | 7.350272 | 0.409387 | 4 | 0.464385 | 0.500481 |
| Neurotransmitter receptors and postsynaptic signal transmission | F9 | Christmas factor, Coagulation factor IX | 7.350272 | 0.409387 | 4 | 0.464385 | 0.632191 |
| Neurotransmitter receptors and postsynaptic signal transmission | GRIN2B | Glutamate receptor, Glutamate receptor ionotropic, NMDA 2B | 7.350272 | 0.409387 | 4 | 0.464385 | 0.56443 |
| Neurotransmitter receptors and postsynaptic signal transmission | PRKCA | Protein kinase C, Protein kinase C alpha type, Protein kinase C, alpha | 7.350272 | 0.409387 | 4 | 0.464385 | 0.52202 |
| Neurotransmitter receptors and postsynaptic signal transmission | GABRA6 | Gamma-aminobutyric acid receptor subunit alpha-6, Gamma-aminobutyric acid type A receptor alpha6 subunit, Gamma-aminobutyric acid type A receptor subunit alpha6 | 7.350272 | 0.409387 | 4 | 0.464385 | 0.50028 |
| Neurotransmitter receptors and postsynaptic signal transmission | CACNG4 | Voltage-dependent calcium channel gamma-4 subunit | 7.350272 | 0.409387 | 4 | 0.464385 | 0.598629 |
| Neurotransmitter receptors and postsynaptic signal transmission | PRKCB | Protein kinase C, Protein kinase C beta type | 7.350272 | 0.409387 | 4 | 0.464385 | 0.520598 |
| Neurotransmitter receptors and postsynaptic signal transmission | MDM2 | E3 ubiquitin-protein ligase Mdm2 | 7.350272 | 0.409387 | 4 | 0.464385 | 0.659771 |
| Neurotransmitter receptors and postsynaptic signal transmission | GRIN2A | Glutamate receptor, Glutamate receptor ionotropic, NMDA 2A | 7.350272 | 0.409387 | 4 | 0.464385 | 0.527756 |
| Neurotransmitter receptors and postsynaptic signal transmission | ADCY9 | Adenylate cyclase 9, Adenylate cyclase type 9 | 7.350272 | 0.409387 | 4 | 0.464385 | 0.550534 |
| Neurotransmitter receptors and postsynaptic signal transmission | GRIN2D | Glutamate receptor, Glutamate receptor ionotropic, NMDA 2D | 7.350272 | 0.409387 | 4 | 0.464385 | 0.556318 |
| Neurotransmitter receptors and postsynaptic signal transmission | GABBR1 | Gamma-aminobutyric acid, Gamma-aminobutyric acid type B receptor subunit 1 | 7.350272 | 0.409387 | 4 | 0.464385 | 0.528076 |
| Neurotransmitter receptors and postsynaptic signal transmission | PRKAR2A | cAMP-dependent protein kinase type II-alpha regulatory subunit, Protein kinase cAMP-dependent type II regulatory subunit alpha, Protein kinase, cAMP-dependent, regulatory subunit type II alpha, Uncharacterized protein | 7.350272 | 0.409387 | 4 | 0.464385 | 0.527065 |
| Neurotransmitter receptors and postsynaptic signal transmission | PRKAG1 | 5'-AMP-activated protein kinase subunit gamma-1, Protein kinase, AMP-activated, gamma 1 non-catalytic subunit | 7.350272 | 0.409387 | 4 | 0.464385 | 0.515113 |
| Neurotransmitter receptors and postsynaptic signal transmission | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 7.350272 | 0.409387 | 4 | 0.464385 | 0.541254 |
| NIK-->noncanonical NF-kB signaling | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.486616 | 0.756711 | 4 | 0.74826 | 0.515416 |
| NIK-->noncanonical NF-kB signaling | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.515416 |
| NIK-->noncanonical NF-kB signaling | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.486616 | 0.756711 | 4 | 0.74826 | 0.515416 |
| NIK-->noncanonical NF-kB signaling | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.547953 |
| NIK-->noncanonical NF-kB signaling | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.54735 |
| NIK-->noncanonical NF-kB signaling | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.565601 |
| NIK-->noncanonical NF-kB signaling | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.515416 |
| NIK-->noncanonical NF-kB signaling | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.515416 |
| NIK-->noncanonical NF-kB signaling | MAP3K14 | Mitogen-activated protein kinase kinase kinase 14 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.556123 |
| NIK-->noncanonical NF-kB signaling | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.54735 |
| NIK-->noncanonical NF-kB signaling | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.486616 | 0.756711 | 4 | 0.74826 | 0.568645 |
| NIK-->noncanonical NF-kB signaling | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.54735 |
| NIK-->noncanonical NF-kB signaling | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.57244 |
| NIK-->noncanonical NF-kB signaling | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.54735 |
| NIK-->noncanonical NF-kB signaling | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.54735 |
| NIK-->noncanonical NF-kB signaling | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.515416 |
| NIK-->noncanonical NF-kB signaling | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.577847 |
| NIK-->noncanonical NF-kB signaling | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.486616 | 0.756711 | 4 | 0.74826 | 0.579609 |
| NIK-->noncanonical NF-kB signaling | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.573417 |
| NIK-->noncanonical NF-kB signaling | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.515416 |
| NIK-->noncanonical NF-kB signaling | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.515416 |
| NIK-->noncanonical NF-kB signaling | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.54735 |
| NIK-->noncanonical NF-kB signaling | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.515416 |
| NIK-->noncanonical NF-kB signaling | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.54735 |
| NIK-->noncanonical NF-kB signaling | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.5197 |
| NIK-->noncanonical NF-kB signaling | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.515416 |
| NIK-->noncanonical NF-kB signaling | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.546881 |
| NIK-->noncanonical NF-kB signaling | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.58858 |
| NIK-->noncanonical NF-kB signaling | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.515416 |
| NIK-->noncanonical NF-kB signaling | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.515416 |
| NIK-->noncanonical NF-kB signaling | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.54735 |
| NIK-->noncanonical NF-kB signaling | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.577481 |
| NIK-->noncanonical NF-kB signaling | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.54735 |
| NIK-->noncanonical NF-kB signaling | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.515416 |
| NIK-->noncanonical NF-kB signaling | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.515416 |
| NIK-->noncanonical NF-kB signaling | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.54735 |
| NIK-->noncanonical NF-kB signaling | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.515416 |
| NIK-->noncanonical NF-kB signaling | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.486616 | 0.756711 | 4 | 0.74826 | 0.572294 |
| Noncanonical activation of NOTCH3 | PSEN2 | Presenilin, Presenilin-2 | 9.082324 | 0.91358 | 4 | 0.805837 | 0.607393 |
| Noncanonical activation of NOTCH3 | APH1B | Aph-1 homolog B, gamma-secretase subunit, APH-1B', Gamma-secretase subunit APH-1B, Uncharacterized protein | 9.082324 | 0.91358 | 4 | 0.805837 | 0.60749 |
| Noncanonical activation of NOTCH3 | NOTCH3 | Neurogenic locus notch homolog protein 3 | 9.082324 | 0.91358 | 4 | 0.805837 | 0.555427 |
| Noncanonical activation of NOTCH3 | APH1A | Anterior pharynx defective 1 homolog A, Anterior pharynx defective 1a homolog, Aph-1 homolog A, gamma-secretase subunit, Gamma-secretase subunit APH-1A, Uncharacterized protein | 9.082324 | 0.91358 | 4 | 0.805837 | 0.60749 |
| Noncanonical activation of NOTCH3 | NCSTN | Nicastrin | 9.082324 | 0.91358 | 4 | 0.805837 | 0.60749 |
| Noncanonical activation of NOTCH3 | PSEN1 | Presenilin, Presenilin-1 | 9.082324 | 0.91358 | 4 | 0.805837 | 0.611222 |
| Noncanonical activation of NOTCH3 | PSENEN | Gamma-secretase subunit PEN-2 | 9.082324 | 0.91358 | 4 | 0.805837 | 0.60725 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS19 | 40S ribosomal protein S19, Uncharacterized protein | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL21 | 60S ribosomal protein L21 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS9 | 40S ribosomal protein S9 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS25 | 40S ribosomal protein S25 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS8 | 40S ribosomal protein S8 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL10A | 60S ribosomal protein L10a, Ribosomal protein | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPLP0 | 60S acidic ribosomal protein P0 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS12 | 40S ribosomal protein S12 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL14 | 60S ribosomal protein L14 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL23 | 60S ribosomal protein L23 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL27A | 60S ribosomal protein L27-A, 60S ribosomal protein L27a | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS17 | 40S ribosomal protein S17 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL37 | 60S ribosomal protein L37, Ribosomal protein L37 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS21 | 40S ribosomal protein S21 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL7 | 60S ribosomal protein L7, MGC79754 protein, Ribosomal protein L7, Uncharacterized protein | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL39 | 60S ribosomal protein L39, LOC100135132 protein, Ribosomal protein L39 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL5 | 60S ribosomal protein L5, Ribosomal protein L5 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS7 | 40S ribosomal protein S7 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS3 | 40S ribosomal protein S3, DNA-(apurinic or apyrimidinic site) lyase | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS20 | 40S ribosomal protein S20 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS29 | 40S ribosomal protein S29 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL37A | 60S ribosomal protein L37-A, 60S ribosomal protein L37a, MGC89854 protein, Probable 60S ribosomal protein L37-A, Ribosomal protein L37a | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS27 | 40S ribosomal protein S27 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.549907 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL13 | 60S ribosomal protein L13 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | FAU | 40S ribosomal protein S30 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL35A | 60S ribosomal protein L35-A, 60S ribosomal protein L35a | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS16 | 40S ribosomal protein S16, Ribosomal protein S16, Uncharacterized protein | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS18 | 40S ribosomal protein S18 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS26 | 40S ribosomal protein S26 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL15 | 60S ribosomal protein L15, Ribosomal protein L15 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS13 | 40S ribosomal protein S13 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL38 | 60S ribosomal protein L38 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL29 | 60S ribosomal protein L29 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL27 | 60S ribosomal protein L27 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS6 | 40S ribosomal protein S6 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.566528 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL19 | 60S ribosomal protein L19, Ribosomal protein L19 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL23A | 60S ribosomal protein L23-A, 60S ribosomal protein L23a, FI01658p, MGC89958 protein, Ribosomal protein L23a, Ribosomal_L23eN domain-containing protein | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS10 | 40S ribosomal protein S10, Ribosomal protein S10, S10_plectin domain-containing protein | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS28 | 40S ribosomal protein S28 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL36A | 60S ribosomal protein L36-A, 60S ribosomal protein L36a, IP17351p, ribosomal protein L36a, Uncharacterized protein | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS23 | 40S ribosomal protein S23, 40S ribosomal protein S23-A | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL4 | 60S ribosomal protein L4, Ribos_L4_asso_C domain-containing protein, Ribosomal protein L4 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL36 | 60S ribosomal protein L36 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPLP1 | 60S acidic ribosomal protein P1, Ribosomal protein, large, P1 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL32 | 60S ribosomal protein L32, Hypothetical LOC496894, Ribosomal protein L32, Uncharacterized protein | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL17 | 60S ribosomal protein L17 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL41 | 60S ribosomal protein L41 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS14 | 40S ribosomal protein S14, Ribosomal protein S14, Uncharacterized protein | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL30 | 60S ribosomal protein L30 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL6 | 60S ribosomal protein L6 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS4X | 40S ribosomal protein S4, 40S ribosomal protein S4, X isoform | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL10 | 60S ribosomal protein L10, MGC89303 protein | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL8 | 60S ribosomal protein L8 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL18A | 60S ribosomal protein L18-A, 60S ribosomal protein L18a | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL3 | 60S ribosomal protein L3, LOC100145083 protein, Ribosomal protein L3, Uncharacterized protein | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL22 | 60S ribosomal protein L22, 60S ribosomal protein L22 1, Ribosomal protein L22, Uncharacterized protein | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL31 | 60S ribosomal protein L31 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS5 | 40S ribosomal protein S5, 40S ribosomal protein S5 [Cleaved into: 40S ribosomal protein S5, N-terminally processed], 40S ribosomal protein S5-A, Ribosomal protein S5 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL12 | 60S ribosomal protein L12 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS24 | 40S ribosomal protein S24 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL26 | 60S ribosomal protein L26, GEO07453p1, KOW domain-containing protein, Ribosomal protein L26 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPSA | 40S ribosomal protein SA | 11.3716 | 0.868827 | 4 | 0.901562 | 0.521531 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS15A | 40S ribosomal protein S15a | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS3A | 40S ribosomal protein S3a | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL18 | 60S ribosomal protein L18, Ribosomal protein L18 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS4Y1 | 40S ribosomal protein S4, 40S ribosomal protein S4, Y isoform 1 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL7A | 60S ribosomal protein L7-A, 60S ribosomal protein L7a | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS11 | 40S ribosomal protein S11 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL11 | 60S ribosomal protein L11 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL28 | 60S ribosomal protein L28 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL34 | 60S ribosomal protein L34 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL35 | 60S ribosomal protein L35 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL24 | 60S ribosomal protein L24, Ribosomal protein L24, TRASH domain-containing protein | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL13A | 60S ribosomal protein L13a | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPL9 | 60S ribosomal protein L9 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS15 | 40S ribosomal protein S15 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | RPS2 | 40S ribosomal protein S2 | 11.3716 | 0.868827 | 4 | 0.901562 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPLP0 | 60S acidic ribosomal protein P0 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPLP1 | 60S acidic ribosomal protein P1, Ribosomal protein, large, P1 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL23 | 60S ribosomal protein L23 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL21 | 60S ribosomal protein L21 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL35A | 60S ribosomal protein L35-A, 60S ribosomal protein L35a | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPSA | 40S ribosomal protein SA | 11.617737 | 0.902944 | 4 | 0.931319 | 0.521531 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL37A | 60S ribosomal protein L37-A, 60S ribosomal protein L37a, MGC89854 protein, Probable 60S ribosomal protein L37-A, Ribosomal protein L37a | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS4X | 40S ribosomal protein S4, 40S ribosomal protein S4, X isoform | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS10 | 40S ribosomal protein S10, Ribosomal protein S10, S10_plectin domain-containing protein | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS25 | 40S ribosomal protein S25 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS27 | 40S ribosomal protein S27 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.549907 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL24 | 60S ribosomal protein L24, Ribosomal protein L24, TRASH domain-containing protein | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS17 | 40S ribosomal protein S17 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS29 | 40S ribosomal protein S29 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL11 | 60S ribosomal protein L11 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS7 | 40S ribosomal protein S7 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL23A | 60S ribosomal protein L23-A, 60S ribosomal protein L23a, FI01658p, MGC89958 protein, Ribosomal protein L23a, Ribosomal_L23eN domain-containing protein | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL10A | 60S ribosomal protein L10a, Ribosomal protein | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS3A | 40S ribosomal protein S3a | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS3 | 40S ribosomal protein S3, DNA-(apurinic or apyrimidinic site) lyase | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL36A | 60S ribosomal protein L36-A, 60S ribosomal protein L36a, IP17351p, ribosomal protein L36a, Uncharacterized protein | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS12 | 40S ribosomal protein S12 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL27 | 60S ribosomal protein L27 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL31 | 60S ribosomal protein L31 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS21 | 40S ribosomal protein S21 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS13 | 40S ribosomal protein S13 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL18 | 60S ribosomal protein L18, Ribosomal protein L18 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL13 | 60S ribosomal protein L13 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL18A | 60S ribosomal protein L18-A, 60S ribosomal protein L18a | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL3 | 60S ribosomal protein L3, LOC100145083 protein, Ribosomal protein L3, Uncharacterized protein | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL22 | 60S ribosomal protein L22, 60S ribosomal protein L22 1, Ribosomal protein L22, Uncharacterized protein | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL30 | 60S ribosomal protein L30 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS16 | 40S ribosomal protein S16, Ribosomal protein S16, Uncharacterized protein | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS18 | 40S ribosomal protein S18 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS28 | 40S ribosomal protein S28 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS8 | 40S ribosomal protein S8 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL19 | 60S ribosomal protein L19, Ribosomal protein L19 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS6 | 40S ribosomal protein S6 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.566528 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS24 | 40S ribosomal protein S24 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL32 | 60S ribosomal protein L32, Hypothetical LOC496894, Ribosomal protein L32, Uncharacterized protein | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL4 | 60S ribosomal protein L4, Ribos_L4_asso_C domain-containing protein, Ribosomal protein L4 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL35 | 60S ribosomal protein L35 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS14 | 40S ribosomal protein S14, Ribosomal protein S14, Uncharacterized protein | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL12 | 60S ribosomal protein L12 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL26 | 60S ribosomal protein L26, GEO07453p1, KOW domain-containing protein, Ribosomal protein L26 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL14 | 60S ribosomal protein L14 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS2 | 40S ribosomal protein S2 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL7 | 60S ribosomal protein L7, MGC79754 protein, Ribosomal protein L7, Uncharacterized protein | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL41 | 60S ribosomal protein L41 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL15 | 60S ribosomal protein L15, Ribosomal protein L15 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL13A | 60S ribosomal protein L13a | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL5 | 60S ribosomal protein L5, Ribosomal protein L5 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL8 | 60S ribosomal protein L8 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL39 | 60S ribosomal protein L39, LOC100135132 protein, Ribosomal protein L39 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS15A | 40S ribosomal protein S15a | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL28 | 60S ribosomal protein L28 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL17 | 60S ribosomal protein L17 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL27A | 60S ribosomal protein L27-A, 60S ribosomal protein L27a | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL9 | 60S ribosomal protein L9 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS15 | 40S ribosomal protein S15 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL37 | 60S ribosomal protein L37, Ribosomal protein L37 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL38 | 60S ribosomal protein L38 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL7A | 60S ribosomal protein L7-A, 60S ribosomal protein L7a | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS5 | 40S ribosomal protein S5, 40S ribosomal protein S5 [Cleaved into: 40S ribosomal protein S5, N-terminally processed], 40S ribosomal protein S5-A, Ribosomal protein S5 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL6 | 60S ribosomal protein L6 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS20 | 40S ribosomal protein S20 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | FAU | 40S ribosomal protein S30 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL10 | 60S ribosomal protein L10, MGC89303 protein | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL34 | 60S ribosomal protein L34 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS9 | 40S ribosomal protein S9 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS26 | 40S ribosomal protein S26 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL36 | 60S ribosomal protein L36 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS23 | 40S ribosomal protein S23, 40S ribosomal protein S23-A | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPL29 | 60S ribosomal protein L29 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS4Y1 | 40S ribosomal protein S4, 40S ribosomal protein S4, Y isoform 1 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS11 | 40S ribosomal protein S11 | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | RPS19 | 40S ribosomal protein S19, Uncharacterized protein | 11.617737 | 0.902944 | 4 | 0.931319 | 0.579609 |
| Notch-HLH transcription pathway | NOTCH3 | Neurogenic locus notch homolog protein 3 | 8.849423 | 0.633944 | 4 | 0.654004 | 0.555427 |
| Notch-HLH transcription pathway | HDAC4 | Histone deacetylase, Histone deacetylase 4 | 8.849423 | 0.633944 | 4 | 0.654004 | 0.528897 |
| Notch-HLH transcription pathway | HDAC6 | Histone deacetylase 6, Histone deacetylase 6, isoform G | 8.849423 | 0.633944 | 4 | 0.654004 | 0.531859 |
| Notch-HLH transcription pathway | HDAC9 | Histone deacetylase, Histone deacetylase 9 | 8.849423 | 0.633944 | 4 | 0.654004 | 0.525623 |
| Notch-HLH transcription pathway | RBPJ | Recombination signal binding protein for immunoglobulin kappa J region, Recombination signal-binding protein for immunoglobulin kappa J region, Recombining binding protein suppressor of hairless, Uncharacterized protein | 8.849423 | 0.633944 | 4 | 0.654004 | 0.647854 |
| Notch-HLH transcription pathway | HDAC7 | Hist_deacetyl domain-containing protein, Histone deacetylase, Histone deacetylase 7 | 8.849423 | 0.633944 | 4 | 0.654004 | 0.522826 |
| Notch-HLH transcription pathway | HDAC10 | Histone deacetylase 10, Polyamine deacetylase HDAC10 | 8.849423 | 0.633944 | 4 | 0.654004 | 0.525881 |
| Notch-HLH transcription pathway | HDAC8 | Histone deacetylase, Histone deacetylase 8, WD repeat and SOCS box containing 2 | 8.849423 | 0.633944 | 4 | 0.654004 | 0.535733 |
| Notch-HLH transcription pathway | HDAC5 | Histone deacetylase, Histone deacetylase 5 | 8.849423 | 0.633944 | 4 | 0.654004 | 0.523224 |
| Notch-HLH transcription pathway | NOTCH1 | Neurogenic locus notch homolog protein 1 | 8.849423 | 0.633944 | 4 | 0.654004 | 0.591875 |
| Notch-HLH transcription pathway | HDAC1 | Histone deacetylase 1, Histone deacetylase HDAC1 | 8.849423 | 0.633944 | 4 | 0.654004 | 0.540105 |
| Notch-HLH transcription pathway | HDAC3 | Histone deacetylase, Histone deacetylase 3 | 8.849423 | 0.633944 | 4 | 0.654004 | 0.528637 |
| Notch-HLH transcription pathway | HDAC2 | Histone deacetylase 2 | 8.849423 | 0.633944 | 4 | 0.654004 | 0.52963 |
| Notch-HLH transcription pathway | HDAC11 | Hdac11 protein, Hist_deacetyl domain-containing protein, Histone deacetylase 11, Histone deacetylase 11, isoform A | 8.849423 | 0.633944 | 4 | 0.654004 | 0.524904 |
| NOTCH1 Intracellular Domain Regulates Transcription | HDAC8 | Histone deacetylase, Histone deacetylase 8, WD repeat and SOCS box containing 2 | 8.563785 | 0.5573 | 4 | 0.600946 | 0.535733 |
| NOTCH1 Intracellular Domain Regulates Transcription | CCNC | Cyclin-C | 8.563785 | 0.5573 | 4 | 0.600946 | 0.503295 |
| NOTCH1 Intracellular Domain Regulates Transcription | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 8.563785 | 0.5573 | 4 | 0.600946 | 0.579609 |
| NOTCH1 Intracellular Domain Regulates Transcription | HDAC10 | Histone deacetylase 10, Polyamine deacetylase HDAC10 | 8.563785 | 0.5573 | 4 | 0.600946 | 0.525881 |
| NOTCH1 Intracellular Domain Regulates Transcription | HDAC11 | Hdac11 protein, Hist_deacetyl domain-containing protein, Histone deacetylase 11, Histone deacetylase 11, isoform A | 8.563785 | 0.5573 | 4 | 0.600946 | 0.524904 |
| NOTCH1 Intracellular Domain Regulates Transcription | HDAC5 | Histone deacetylase, Histone deacetylase 5 | 8.563785 | 0.5573 | 4 | 0.600946 | 0.523224 |
| NOTCH1 Intracellular Domain Regulates Transcription | NOTCH1 | Neurogenic locus notch homolog protein 1 | 8.563785 | 0.5573 | 4 | 0.600946 | 0.591875 |
| NOTCH1 Intracellular Domain Regulates Transcription | HDAC4 | Histone deacetylase, Histone deacetylase 4 | 8.563785 | 0.5573 | 4 | 0.600946 | 0.528897 |
| NOTCH1 Intracellular Domain Regulates Transcription | HDAC1 | Histone deacetylase 1, Histone deacetylase HDAC1 | 8.563785 | 0.5573 | 4 | 0.600946 | 0.540105 |
| NOTCH1 Intracellular Domain Regulates Transcription | HDAC7 | Hist_deacetyl domain-containing protein, Histone deacetylase, Histone deacetylase 7 | 8.563785 | 0.5573 | 4 | 0.600946 | 0.522826 |
| NOTCH1 Intracellular Domain Regulates Transcription | HDAC2 | Histone deacetylase 2 | 8.563785 | 0.5573 | 4 | 0.600946 | 0.52963 |
| NOTCH1 Intracellular Domain Regulates Transcription | RBPJ | Recombination signal binding protein for immunoglobulin kappa J region, Recombination signal-binding protein for immunoglobulin kappa J region, Recombining binding protein suppressor of hairless, Uncharacterized protein | 8.563785 | 0.5573 | 4 | 0.600946 | 0.647854 |
| NOTCH1 Intracellular Domain Regulates Transcription | HDAC3 | Histone deacetylase, Histone deacetylase 3 | 8.563785 | 0.5573 | 4 | 0.600946 | 0.528637 |
| NOTCH1 Intracellular Domain Regulates Transcription | HDAC6 | Histone deacetylase 6, Histone deacetylase 6, isoform G | 8.563785 | 0.5573 | 4 | 0.600946 | 0.531859 |
| NOTCH1 Intracellular Domain Regulates Transcription | HDAC9 | Histone deacetylase, Histone deacetylase 9 | 8.563785 | 0.5573 | 4 | 0.600946 | 0.525623 |
| NOTCH2 Activation and Transmission of Signal to the Nucleus | APH1B | Aph-1 homolog B, gamma-secretase subunit, APH-1B', Gamma-secretase subunit APH-1B, Uncharacterized protein | 9.20789 | 0.736457 | 4 | 0.723753 | 0.60749 |
| NOTCH2 Activation and Transmission of Signal to the Nucleus | NOTCH1 | Neurogenic locus notch homolog protein 1 | 9.20789 | 0.736457 | 4 | 0.723753 | 0.591875 |
| NOTCH2 Activation and Transmission of Signal to the Nucleus | PSEN2 | Presenilin, Presenilin-2 | 9.20789 | 0.736457 | 4 | 0.723753 | 0.607393 |
| NOTCH2 Activation and Transmission of Signal to the Nucleus | APH1A | Anterior pharynx defective 1 homolog A, Anterior pharynx defective 1a homolog, Aph-1 homolog A, gamma-secretase subunit, Gamma-secretase subunit APH-1A, Uncharacterized protein | 9.20789 | 0.736457 | 4 | 0.723753 | 0.60749 |
| NOTCH2 Activation and Transmission of Signal to the Nucleus | NCSTN | Nicastrin | 9.20789 | 0.736457 | 4 | 0.723753 | 0.60749 |
| NOTCH2 Activation and Transmission of Signal to the Nucleus | PSEN1 | Presenilin, Presenilin-1 | 9.20789 | 0.736457 | 4 | 0.723753 | 0.611222 |
| NOTCH2 Activation and Transmission of Signal to the Nucleus | PSENEN | Gamma-secretase subunit PEN-2 | 9.20789 | 0.736457 | 4 | 0.723753 | 0.60725 |
| NOTCH2 Activation and Transmission of Signal to the Nucleus | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.20789 | 0.736457 | 4 | 0.723753 | 0.579609 |
| NOTCH3 Activation and Transmission of Signal to the Nucleus | APH1A | Anterior pharynx defective 1 homolog A, Anterior pharynx defective 1a homolog, Aph-1 homolog A, gamma-secretase subunit, Gamma-secretase subunit APH-1A, Uncharacterized protein | 9.645309 | 0.803563 | 4 | 0.779873 | 0.60749 |
| NOTCH3 Activation and Transmission of Signal to the Nucleus | NOTCH3 | Neurogenic locus notch homolog protein 3 | 9.645309 | 0.803563 | 4 | 0.779873 | 0.555427 |
| NOTCH3 Activation and Transmission of Signal to the Nucleus | APH1B | Aph-1 homolog B, gamma-secretase subunit, APH-1B', Gamma-secretase subunit APH-1B, Uncharacterized protein | 9.645309 | 0.803563 | 4 | 0.779873 | 0.60749 |
| NOTCH3 Activation and Transmission of Signal to the Nucleus | EGFR | Epidermal growth factor receptor, Receptor protein-tyrosine kinase | 9.645309 | 0.803563 | 4 | 0.779873 | 0.625817 |
| NOTCH3 Activation and Transmission of Signal to the Nucleus | NCSTN | Nicastrin | 9.645309 | 0.803563 | 4 | 0.779873 | 0.60749 |
| NOTCH3 Activation and Transmission of Signal to the Nucleus | PSEN1 | Presenilin, Presenilin-1 | 9.645309 | 0.803563 | 4 | 0.779873 | 0.611222 |
| NOTCH3 Activation and Transmission of Signal to the Nucleus | PSENEN | Gamma-secretase subunit PEN-2 | 9.645309 | 0.803563 | 4 | 0.779873 | 0.60725 |
| NOTCH3 Activation and Transmission of Signal to the Nucleus | PSEN2 | Presenilin, Presenilin-2 | 9.645309 | 0.803563 | 4 | 0.779873 | 0.607393 |
| NOTCH3 Activation and Transmission of Signal to the Nucleus | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.645309 | 0.803563 | 4 | 0.779873 | 0.579609 |
| NOTCH4 Activation and Transmission of Signal to the Nucleus | NCSTN | Nicastrin | 9.220305 | 0.846778 | 4 | 0.779554 | 0.60749 |
| NOTCH4 Activation and Transmission of Signal to the Nucleus | YWHAZ | 14_3_3 domain-containing protein, 14-3-3 protein zeta, 14-3-3 protein zeta/delta, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta | 9.220305 | 0.846778 | 4 | 0.779554 | 0.602005 |
| NOTCH4 Activation and Transmission of Signal to the Nucleus | PSENEN | Gamma-secretase subunit PEN-2 | 9.220305 | 0.846778 | 4 | 0.779554 | 0.60725 |
| NOTCH4 Activation and Transmission of Signal to the Nucleus | PSEN1 | Presenilin, Presenilin-1 | 9.220305 | 0.846778 | 4 | 0.779554 | 0.611222 |
| NOTCH4 Activation and Transmission of Signal to the Nucleus | PSEN2 | Presenilin, Presenilin-2 | 9.220305 | 0.846778 | 4 | 0.779554 | 0.607393 |
| NOTCH4 Activation and Transmission of Signal to the Nucleus | APH1B | Aph-1 homolog B, gamma-secretase subunit, APH-1B', Gamma-secretase subunit APH-1B, Uncharacterized protein | 9.220305 | 0.846778 | 4 | 0.779554 | 0.60749 |
| NOTCH4 Activation and Transmission of Signal to the Nucleus | APH1A | Anterior pharynx defective 1 homolog A, Anterior pharynx defective 1a homolog, Aph-1 homolog A, gamma-secretase subunit, Gamma-secretase subunit APH-1A, Uncharacterized protein | 9.220305 | 0.846778 | 4 | 0.779554 | 0.60749 |
| NRIF signals cell death from the nucleus | MAPK8 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 8 | 7.913846 | 0.702206 | 4 | 0.639869 | 0.503967 |
| NRIF signals cell death from the nucleus | PSEN2 | Presenilin, Presenilin-2 | 7.913846 | 0.702206 | 4 | 0.639869 | 0.607393 |
| NRIF signals cell death from the nucleus | APH1A | Anterior pharynx defective 1 homolog A, Anterior pharynx defective 1a homolog, Aph-1 homolog A, gamma-secretase subunit, Gamma-secretase subunit APH-1A, Uncharacterized protein | 7.913846 | 0.702206 | 4 | 0.639869 | 0.60749 |
| NRIF signals cell death from the nucleus | NCSTN | Nicastrin | 7.913846 | 0.702206 | 4 | 0.639869 | 0.60749 |
| NRIF signals cell death from the nucleus | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 7.913846 | 0.702206 | 4 | 0.639869 | 0.579609 |
| NRIF signals cell death from the nucleus | APH1B | Aph-1 homolog B, gamma-secretase subunit, APH-1B', Gamma-secretase subunit APH-1B, Uncharacterized protein | 7.913846 | 0.702206 | 4 | 0.639869 | 0.60749 |
| NRIF signals cell death from the nucleus | PSEN1 | Presenilin, Presenilin-1 | 7.913846 | 0.702206 | 4 | 0.639869 | 0.611222 |
| NRIF signals cell death from the nucleus | PSENEN | Gamma-secretase subunit PEN-2 | 7.913846 | 0.702206 | 4 | 0.639869 | 0.60725 |
| Nuclear signaling by ERBB4 | APH1B | Aph-1 homolog B, gamma-secretase subunit, APH-1B', Gamma-secretase subunit APH-1B, Uncharacterized protein | 9.172161 | 0.695745 | 4 | 0.701554 | 0.60749 |
| Nuclear signaling by ERBB4 | EGFR | Epidermal growth factor receptor, Receptor protein-tyrosine kinase | 9.172161 | 0.695745 | 4 | 0.701554 | 0.625817 |
| Nuclear signaling by ERBB4 | PSEN1 | Presenilin, Presenilin-1 | 9.172161 | 0.695745 | 4 | 0.701554 | 0.611222 |
| Nuclear signaling by ERBB4 | YAP1 | Transcriptional coactivator YAP1, Yes1 associated transcriptional regulator | 9.172161 | 0.695745 | 4 | 0.701554 | 0.628833 |
| Nuclear signaling by ERBB4 | PSENEN | Gamma-secretase subunit PEN-2 | 9.172161 | 0.695745 | 4 | 0.701554 | 0.60725 |
| Nuclear signaling by ERBB4 | CSN2 | Beta-casein, COP9 signalosome complex subunit 2 | 9.172161 | 0.695745 | 4 | 0.701554 | 0.537553 |
| Nuclear signaling by ERBB4 | APH1A | Anterior pharynx defective 1 homolog A, Anterior pharynx defective 1a homolog, Aph-1 homolog A, gamma-secretase subunit, Gamma-secretase subunit APH-1A, Uncharacterized protein | 9.172161 | 0.695745 | 4 | 0.701554 | 0.60749 |
| Nuclear signaling by ERBB4 | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 9.172161 | 0.695745 | 4 | 0.701554 | 0.594807 |
| Nuclear signaling by ERBB4 | NCSTN | Nicastrin | 9.172161 | 0.695745 | 4 | 0.701554 | 0.60749 |
| Nuclear signaling by ERBB4 | PGR | Nuclear receptor subfamily 3 group C member 3, Progesterone receptor | 9.172161 | 0.695745 | 4 | 0.701554 | 0.5833 |
| Nuclear signaling by ERBB4 | PSEN2 | Presenilin, Presenilin-2 | 9.172161 | 0.695745 | 4 | 0.701554 | 0.607393 |
| Nuclear signaling by ERBB4 | ERBB4 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-4 | 9.172161 | 0.695745 | 4 | 0.701554 | 0.550399 |
| Nuclear signaling by ERBB4 | ESR1 | Estrogen receptor | 9.172161 | 0.695745 | 4 | 0.701554 | 0.561602 |
| Orc1 removal from chromatin | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.58858 |
| Orc1 removal from chromatin | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.54735 |
| Orc1 removal from chromatin | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.515416 |
| Orc1 removal from chromatin | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.54735 |
| Orc1 removal from chromatin | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.515416 |
| Orc1 removal from chromatin | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.577481 |
| Orc1 removal from chromatin | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.515416 |
| Orc1 removal from chromatin | CDK2 | Cyclin dependent kinase 2, Cyclin-dependent kinase 2, Protein kinase domain-containing protein, Uncharacterized protein | 9.632843 | 0.76502 | 4 | 0.759958 | 0.532011 |
| Orc1 removal from chromatin | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.515416 |
| Orc1 removal from chromatin | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.54735 |
| Orc1 removal from chromatin | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.5197 |
| Orc1 removal from chromatin | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.515416 |
| Orc1 removal from chromatin | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.515416 |
| Orc1 removal from chromatin | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.632843 | 0.76502 | 4 | 0.759958 | 0.568645 |
| Orc1 removal from chromatin | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.515416 |
| Orc1 removal from chromatin | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.515416 |
| Orc1 removal from chromatin | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.632843 | 0.76502 | 4 | 0.759958 | 0.579609 |
| Orc1 removal from chromatin | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.573417 |
| Orc1 removal from chromatin | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.54735 |
| Orc1 removal from chromatin | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.515416 |
| Orc1 removal from chromatin | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.515416 |
| Orc1 removal from chromatin | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.54735 |
| Orc1 removal from chromatin | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.54735 |
| Orc1 removal from chromatin | CCNA1 | Cyclin A1, Cyclin-A1, Cyclin-A2 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.506777 |
| Orc1 removal from chromatin | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.515416 |
| Orc1 removal from chromatin | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.572294 |
| Orc1 removal from chromatin | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.577847 |
| Orc1 removal from chromatin | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.546881 |
| Orc1 removal from chromatin | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.54735 |
| Orc1 removal from chromatin | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.57244 |
| Orc1 removal from chromatin | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.54735 |
| Orc1 removal from chromatin | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.547953 |
| Orc1 removal from chromatin | CCNA2 | Cyclin-A2 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.527977 |
| Orc1 removal from chromatin | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.565601 |
| Orc1 removal from chromatin | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.632843 | 0.76502 | 4 | 0.759958 | 0.515416 |
| Orc1 removal from chromatin | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.54735 |
| Orc1 removal from chromatin | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.632843 | 0.76502 | 4 | 0.759958 | 0.515416 |
| Orc1 removal from chromatin | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.515416 |
| Orc1 removal from chromatin | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.54735 |
| Orc1 removal from chromatin | MCM4 | DNA helicase, DNA replication licensing factor mcm4, DNA replication licensing factor MCM4 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.515269 |
| Orc1 removal from chromatin | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.632843 | 0.76502 | 4 | 0.759958 | 0.515416 |
| Oxidative Stress Induced Senescence | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 8.797875 | 0.454664 | 3.66101 | 0.514102 | 0.579609 |
| Oxidative Stress Induced Senescence | CDK6 | Cyclin dependent kinase 6, Cyclin-dependent kinase, Cyclin-dependent kinase 6 | 8.797875 | 0.454664 | 3.66101 | 0.514102 | 0.509686 |
| Oxidative Stress Induced Senescence | MDM4 | Double minute 4 protein, Protein Mdm4 | 8.797875 | 0.454664 | 3.66101 | 0.514102 | 0.510741 |
| Oxidative Stress Induced Senescence | TP53 | Cellular tumor antigen p53 | 8.797875 | 0.454664 | 3.66101 | 0.514102 | 0.525937 |
| Oxidative Stress Induced Senescence | CBX6 | Chromo domain-containing protein, Chromobox 6, Chromobox protein homolog 6 | 8.797875 | 0.454664 | 3.66101 | 0.514102 | 0.575593 |
| Oxidative Stress Induced Senescence | TNIK | TRAF2 and NCK interacting kinase, TRAF2 and NCK-interacting kinase, Traf2 and NCK-interacting protein kinase, TRAF2 and NCK-interacting protein kinase | 8.797875 | 0.454664 | 3.66101 | 0.514102 | 0.502833 |
| Oxidative Stress Induced Senescence | MAPK14 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 14 | 8.797875 | 0.454664 | 3.66101 | 0.514102 | 0.591941 |
| Oxidative Stress Induced Senescence | MDM2 | E3 ubiquitin-protein ligase Mdm2 | 8.797875 | 0.454664 | 3.66101 | 0.514102 | 0.659771 |
| Oxidative Stress Induced Senescence | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 8.797875 | 0.454664 | 3.66101 | 0.514102 | 0.541254 |
| Oxidative Stress Induced Senescence | CDK4 | Cyclin dependent kinase 4, Cyclin-dependent kinase 4, Cyclin-dependent kinase 4, isoform A | 8.797875 | 0.454664 | 3.66101 | 0.514102 | 0.511296 |
| Oxidative Stress Induced Senescence | MAPK11 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 11 | 8.797875 | 0.454664 | 3.66101 | 0.514102 | 0.567539 |
| Oxidative Stress Induced Senescence | MAPK10 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 10 | 8.797875 | 0.454664 | 3.66101 | 0.514102 | 0.506109 |
| Oxidative Stress Induced Senescence | MAPK8 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 8 | 8.797875 | 0.454664 | 3.66101 | 0.514102 | 0.503967 |
| Oxidative Stress Induced Senescence | EZH2 | [Histone H3]-lysine(27) N-trimethyltransferase, Histone-lysine N-methyltransferase EZH2 | 8.797875 | 0.454664 | 3.66101 | 0.514102 | 0.53644 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.54735 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.54735 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.515416 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.572294 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.523819 | 0.703969 | 4 | 0.723808 | 0.515416 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.54735 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.515416 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.546881 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.523819 | 0.703969 | 4 | 0.723808 | 0.515416 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.515416 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.547953 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.515416 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.577847 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.515416 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.577481 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.54735 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.573417 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.57244 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | VHL | Protein Vhl, Uncharacterized protein, von Hippel-Lindau disease tumor suppressor, von Hippel-Lindau disease tumor suppressor isoform 1, von Hippel-Lindau tumor suppressor | 9.523819 | 0.703969 | 4 | 0.723808 | 0.588856 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.54735 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.515416 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.58858 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.54735 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.515416 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.54735 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.54735 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.515416 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.515416 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.54735 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.515416 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.515416 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.565601 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.523819 | 0.703969 | 4 | 0.723808 | 0.579609 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.515416 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.5197 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.515416 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.523819 | 0.703969 | 4 | 0.723808 | 0.568645 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.523819 | 0.703969 | 4 | 0.723808 | 0.54735 |
| p38MAPK events | MAPK14 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 14 | 8.90043 | 0.730576 | 4 | 0.704952 | 0.591941 |
| p38MAPK events | MAPK13 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 13 | 8.90043 | 0.730576 | 4 | 0.704952 | 0.573201 |
| p38MAPK events | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 8.90043 | 0.730576 | 4 | 0.704952 | 0.594807 |
| p38MAPK events | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 8.90043 | 0.730576 | 4 | 0.704952 | 0.505988 |
| p38MAPK events | MAPK11 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 11 | 8.90043 | 0.730576 | 4 | 0.704952 | 0.567539 |
| p38MAPK events | MAPK12 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 12 | 8.90043 | 0.730576 | 4 | 0.704952 | 0.573393 |
| p53-Dependent G1 DNA Damage Response | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.577481 |
| p53-Dependent G1 DNA Damage Response | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.515416 |
| p53-Dependent G1 DNA Damage Response | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.515416 |
| p53-Dependent G1 DNA Damage Response | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.515416 |
| p53-Dependent G1 DNA Damage Response | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 7.274686 | 0.758427 | 4 | 0.635006 | 0.568645 |
| p53-Dependent G1 DNA Damage Response | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.54735 |
| p53-Dependent G1 DNA Damage Response | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.546881 |
| p53-Dependent G1 DNA Damage Response | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.573417 |
| p53-Dependent G1 DNA Damage Response | MDM4 | Double minute 4 protein, Protein Mdm4 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.510741 |
| p53-Dependent G1 DNA Damage Response | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.54735 |
| p53-Dependent G1 DNA Damage Response | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.58858 |
| p53-Dependent G1 DNA Damage Response | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.54735 |
| p53-Dependent G1 DNA Damage Response | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.54735 |
| p53-Dependent G1 DNA Damage Response | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.515416 |
| p53-Dependent G1 DNA Damage Response | TP53 | Cellular tumor antigen p53 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.525937 |
| p53-Dependent G1 DNA Damage Response | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.54735 |
| p53-Dependent G1 DNA Damage Response | MDM2 | E3 ubiquitin-protein ligase Mdm2 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.659771 |
| p53-Dependent G1 DNA Damage Response | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.515416 |
| p53-Dependent G1 DNA Damage Response | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.54735 |
| p53-Dependent G1 DNA Damage Response | CCNE1 | Cyclin E1, Cyclin N-terminal domain-containing protein, G1/S-specific cyclin-E1 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.526741 |
| p53-Dependent G1 DNA Damage Response | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.547953 |
| p53-Dependent G1 DNA Damage Response | CCNA1 | Cyclin A1, Cyclin-A1, Cyclin-A2 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.506777 |
| p53-Dependent G1 DNA Damage Response | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 7.274686 | 0.758427 | 4 | 0.635006 | 0.515416 |
| p53-Dependent G1 DNA Damage Response | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.515416 |
| p53-Dependent G1 DNA Damage Response | CDK2 | Cyclin dependent kinase 2, Cyclin-dependent kinase 2, Protein kinase domain-containing protein, Uncharacterized protein | 7.274686 | 0.758427 | 4 | 0.635006 | 0.532011 |
| p53-Dependent G1 DNA Damage Response | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.515416 |
| p53-Dependent G1 DNA Damage Response | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.54735 |
| p53-Dependent G1 DNA Damage Response | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.54735 |
| p53-Dependent G1 DNA Damage Response | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 7.274686 | 0.758427 | 4 | 0.635006 | 0.515416 |
| p53-Dependent G1 DNA Damage Response | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 7.274686 | 0.758427 | 4 | 0.635006 | 0.579609 |
| p53-Dependent G1 DNA Damage Response | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.572294 |
| p53-Dependent G1 DNA Damage Response | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.577847 |
| p53-Dependent G1 DNA Damage Response | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.565601 |
| p53-Dependent G1 DNA Damage Response | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.54735 |
| p53-Dependent G1 DNA Damage Response | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.515416 |
| p53-Dependent G1 DNA Damage Response | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.515416 |
| p53-Dependent G1 DNA Damage Response | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.515416 |
| p53-Dependent G1 DNA Damage Response | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.5197 |
| p53-Dependent G1 DNA Damage Response | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.515416 |
| p53-Dependent G1 DNA Damage Response | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.57244 |
| p53-Dependent G1 DNA Damage Response | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.54735 |
| p53-Dependent G1 DNA Damage Response | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.515416 |
| p53-Dependent G1 DNA Damage Response | CCNA2 | Cyclin-A2 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.527977 |
| p53-Dependent G1 DNA Damage Response | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.515416 |
| p53-Dependent G1 DNA Damage Response | CCNE2 | Cyclin E2, Cyclin N-terminal domain-containing protein, G1/S-specific cyclin-E2 | 7.274686 | 0.758427 | 4 | 0.635006 | 0.503983 |
| Packaging of Eight RNA Segments | PB1 | Protein PB1-F2, RNA-directed RNA polymerase catalytic subunit | 7.405237 | 0.528279 | 0.508942 | ||
| Paradoxical activation of RAF signaling by kinase inactive BRAF | RAP1A | RAP1A, member of RAS oncogene family, Ras-related protein Rap-1A, Uncharacterized protein | 8.880042 | 0.512627 | 4 | 0.594925 | 0.530823 |
| Paradoxical activation of RAF signaling by kinase inactive BRAF | FN1 | Fibronectin | 8.880042 | 0.512627 | 4 | 0.594925 | 0.578096 |
| Paradoxical activation of RAF signaling by kinase inactive BRAF | MAP2K1 | Dual specificity mitogen-activated protein kinase kinase 1, MAP2K1 protein, Mitogen-activated protein kinase kinase 1, Protein kinase domain-containing protein | 8.880042 | 0.512627 | 4 | 0.594925 | 0.545374 |
| Paradoxical activation of RAF signaling by kinase inactive BRAF | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 8.880042 | 0.512627 | 4 | 0.594925 | 0.541254 |
| Paradoxical activation of RAF signaling by kinase inactive BRAF | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 8.880042 | 0.512627 | 4 | 0.594925 | 0.505988 |
| Paradoxical activation of RAF signaling by kinase inactive BRAF | JAK2 | Janus kinase 2, Tyrosine-protein kinase, Tyrosine-protein kinase JAK2 | 8.880042 | 0.512627 | 4 | 0.594925 | 0.573235 |
| Paradoxical activation of RAF signaling by kinase inactive BRAF | RAF1 | Non-specific serine/threonine protein kinase, RAF proto-oncogene serine/threonine-protein kinase | 8.880042 | 0.512627 | 4 | 0.594925 | 0.573556 |
| Paradoxical activation of RAF signaling by kinase inactive BRAF | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 8.880042 | 0.512627 | 4 | 0.594925 | 0.594807 |
| Paradoxical activation of RAF signaling by kinase inactive BRAF | BRAF | Non-specific serine/threonine protein kinase, Serine/threonine-protein kinase B-raf | 8.880042 | 0.512627 | 4 | 0.594925 | 0.590074 |
| Paradoxical activation of RAF signaling by kinase inactive BRAF | YWHAB | 14-3-3 protein beta/alpha, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta | 8.880042 | 0.512627 | 4 | 0.594925 | 0.513843 |
| Paradoxical activation of RAF signaling by kinase inactive BRAF | VWF | von Willebrand factor | 8.880042 | 0.512627 | 4 | 0.594925 | 0.548181 |
| PCP/CE pathway | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.5197 |
| PCP/CE pathway | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.54735 |
| PCP/CE pathway | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.515416 |
| PCP/CE pathway | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.54735 |
| PCP/CE pathway | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.515416 |
| PCP/CE pathway | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.57244 |
| PCP/CE pathway | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.515416 |
| PCP/CE pathway | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.573417 |
| PCP/CE pathway | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.515416 |
| PCP/CE pathway | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.54735 |
| PCP/CE pathway | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.54735 |
| PCP/CE pathway | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 8.131456 | 0.659158 | 4 | 0.629572 | 0.515416 |
| PCP/CE pathway | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.565601 |
| PCP/CE pathway | PRKCB | Protein kinase C, Protein kinase C beta type | 8.131456 | 0.659158 | 4 | 0.629572 | 0.520598 |
| PCP/CE pathway | PRKCG | Protein kinase C, Protein kinase C gamma type, Sperm-associated antigen 9 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.500344 |
| PCP/CE pathway | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.515416 |
| PCP/CE pathway | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.515416 |
| PCP/CE pathway | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 8.131456 | 0.659158 | 4 | 0.629572 | 0.579609 |
| PCP/CE pathway | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.54735 |
| PCP/CE pathway | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.54735 |
| PCP/CE pathway | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.515416 |
| PCP/CE pathway | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.54735 |
| PCP/CE pathway | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.515416 |
| PCP/CE pathway | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.54735 |
| PCP/CE pathway | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.515416 |
| PCP/CE pathway | DVL2 | Dishevelled segment polarity protein 2, Segment polarity protein dishevelled homolog DVL-2, Uncharacterized protein | 8.131456 | 0.659158 | 4 | 0.629572 | 0.529242 |
| PCP/CE pathway | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.515416 |
| PCP/CE pathway | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.572294 |
| PCP/CE pathway | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.54735 |
| PCP/CE pathway | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.577481 |
| PCP/CE pathway | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.54735 |
| PCP/CE pathway | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.515416 |
| PCP/CE pathway | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.515416 |
| PCP/CE pathway | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.547953 |
| PCP/CE pathway | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.515416 |
| PCP/CE pathway | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.546881 |
| PCP/CE pathway | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 8.131456 | 0.659158 | 4 | 0.629572 | 0.568645 |
| PCP/CE pathway | PRKCA | Protein kinase C, Protein kinase C alpha type, Protein kinase C, alpha | 8.131456 | 0.659158 | 4 | 0.629572 | 0.52202 |
| PCP/CE pathway | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.577847 |
| PCP/CE pathway | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 8.131456 | 0.659158 | 4 | 0.629572 | 0.515416 |
| PCP/CE pathway | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 8.131456 | 0.659158 | 4 | 0.629572 | 0.58858 |
| Peptide chain elongation | RPL36A | 60S ribosomal protein L36-A, 60S ribosomal protein L36a, IP17351p, ribosomal protein L36a, Uncharacterized protein | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL41 | 60S ribosomal protein L41 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS28 | 40S ribosomal protein S28 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS8 | 40S ribosomal protein S8 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL35 | 60S ribosomal protein L35 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL27A | 60S ribosomal protein L27-A, 60S ribosomal protein L27a | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL18A | 60S ribosomal protein L18-A, 60S ribosomal protein L18a | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS23 | 40S ribosomal protein S23, 40S ribosomal protein S23-A | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS21 | 40S ribosomal protein S21 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL24 | 60S ribosomal protein L24, Ribosomal protein L24, TRASH domain-containing protein | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS4Y1 | 40S ribosomal protein S4, 40S ribosomal protein S4, Y isoform 1 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS9 | 40S ribosomal protein S9 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL26 | 60S ribosomal protein L26, GEO07453p1, KOW domain-containing protein, Ribosomal protein L26 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS16 | 40S ribosomal protein S16, Ribosomal protein S16, Uncharacterized protein | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS10 | 40S ribosomal protein S10, Ribosomal protein S10, S10_plectin domain-containing protein | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL18 | 60S ribosomal protein L18, Ribosomal protein L18 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS3A | 40S ribosomal protein S3a | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL13A | 60S ribosomal protein L13a | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS5 | 40S ribosomal protein S5, 40S ribosomal protein S5 [Cleaved into: 40S ribosomal protein S5, N-terminally processed], 40S ribosomal protein S5-A, Ribosomal protein S5 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS27 | 40S ribosomal protein S27 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.549907 |
| Peptide chain elongation | RPL7 | 60S ribosomal protein L7, MGC79754 protein, Ribosomal protein L7, Uncharacterized protein | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL37 | 60S ribosomal protein L37, Ribosomal protein L37 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL17 | 60S ribosomal protein L17 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL14 | 60S ribosomal protein L14 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPSA | 40S ribosomal protein SA | 11.710621 | 0.907231 | 4 | 0.938254 | 0.521531 |
| Peptide chain elongation | RPL19 | 60S ribosomal protein L19, Ribosomal protein L19 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL28 | 60S ribosomal protein L28 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS19 | 40S ribosomal protein S19, Uncharacterized protein | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL6 | 60S ribosomal protein L6 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL11 | 60S ribosomal protein L11 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS14 | 40S ribosomal protein S14, Ribosomal protein S14, Uncharacterized protein | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS18 | 40S ribosomal protein S18 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS29 | 40S ribosomal protein S29 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL21 | 60S ribosomal protein L21 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL15 | 60S ribosomal protein L15, Ribosomal protein L15 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS11 | 40S ribosomal protein S11 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS15 | 40S ribosomal protein S15 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL12 | 60S ribosomal protein L12 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL29 | 60S ribosomal protein L29 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL36 | 60S ribosomal protein L36 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL10A | 60S ribosomal protein L10a, Ribosomal protein | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL37A | 60S ribosomal protein L37-A, 60S ribosomal protein L37a, MGC89854 protein, Probable 60S ribosomal protein L37-A, Ribosomal protein L37a | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | FAU | 40S ribosomal protein S30 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS13 | 40S ribosomal protein S13 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS12 | 40S ribosomal protein S12 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS25 | 40S ribosomal protein S25 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS24 | 40S ribosomal protein S24 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL30 | 60S ribosomal protein L30 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL38 | 60S ribosomal protein L38 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPLP1 | 60S acidic ribosomal protein P1, Ribosomal protein, large, P1 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL8 | 60S ribosomal protein L8 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL5 | 60S ribosomal protein L5, Ribosomal protein L5 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL34 | 60S ribosomal protein L34 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS26 | 40S ribosomal protein S26 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL31 | 60S ribosomal protein L31 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL23 | 60S ribosomal protein L23 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL10 | 60S ribosomal protein L10, MGC89303 protein | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPLP0 | 60S acidic ribosomal protein P0 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL35A | 60S ribosomal protein L35-A, 60S ribosomal protein L35a | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL32 | 60S ribosomal protein L32, Hypothetical LOC496894, Ribosomal protein L32, Uncharacterized protein | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS2 | 40S ribosomal protein S2 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS6 | 40S ribosomal protein S6 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.566528 |
| Peptide chain elongation | RPS7 | 40S ribosomal protein S7 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL22 | 60S ribosomal protein L22, 60S ribosomal protein L22 1, Ribosomal protein L22, Uncharacterized protein | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL3 | 60S ribosomal protein L3, LOC100145083 protein, Ribosomal protein L3, Uncharacterized protein | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS15A | 40S ribosomal protein S15a | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL39 | 60S ribosomal protein L39, LOC100135132 protein, Ribosomal protein L39 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL4 | 60S ribosomal protein L4, Ribos_L4_asso_C domain-containing protein, Ribosomal protein L4 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL7A | 60S ribosomal protein L7-A, 60S ribosomal protein L7a | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS20 | 40S ribosomal protein S20 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL23A | 60S ribosomal protein L23-A, 60S ribosomal protein L23a, FI01658p, MGC89958 protein, Ribosomal protein L23a, Ribosomal_L23eN domain-containing protein | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS3 | 40S ribosomal protein S3, DNA-(apurinic or apyrimidinic site) lyase | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS17 | 40S ribosomal protein S17 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL13 | 60S ribosomal protein L13 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPS4X | 40S ribosomal protein S4, 40S ribosomal protein S4, X isoform | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide chain elongation | RPL27 | 60S ribosomal protein L27 | 11.710621 | 0.907231 | 4 | 0.938254 | 0.579609 |
| Peptide ligand-binding receptors | UTS2R | G_PROTEIN_RECEP_F1_2 domain-containing protein, Urotensin II receptor, Urotensin-2 receptor | 9.901344 | 0.420855 | 4 | 0.601727 | 0.594282 |
| Peptide ligand-binding receptors | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.901344 | 0.420855 | 4 | 0.601727 | 0.515416 |
| Peptide ligand-binding receptors | KNG1 | Bradykinin, Kininogen 1, Kininogen-1, Kininogen-1 [Cleaved into: Kininogen-1 heavy chain; Bradykinin; Kininogen-1 light chain], Uncharacterized protein | 9.901344 | 0.420855 | 4 | 0.601727 | 0.566192 |
| Peptide ligand-binding receptors | SSTR2 | Somatostatin receptor type 2 | 9.901344 | 0.420855 | 4 | 0.601727 | 0.595919 |
| Peptide ligand-binding receptors | ACKR3 | Atypical chemokine receptor 3 | 9.901344 | 0.420855 | 4 | 0.601727 | 0.521317 |
| Peptide ligand-binding receptors | AGTR1B | Angiotensin II receptor, type 1b | 9.901344 | 0.420855 | 4 | 0.601727 | 0.614033 |
| Peptide ligand-binding receptors | APLN | Apelin, APJ endogenous ligand | 9.901344 | 0.420855 | 4 | 0.601727 | 0.502238 |
| Peptide ligand-binding receptors | CXCR4 | C-X-C chemokine receptor type 4, G_PROTEIN_RECEP_F1_2 domain-containing protein | 9.901344 | 0.420855 | 4 | 0.601727 | 0.512583 |
| Peptide ligand-binding receptors | MCHR2 | Melanin concentrating hormone receptor 2, Melanin concentrating hormone receptor 2a, Melanin-concentrating hormone receptor 2 | 9.901344 | 0.420855 | 4 | 0.601727 | 0.520996 |
| Peptide ligand-binding receptors | SSTR5 | G_PROTEIN_RECEP_F1_2 domain-containing protein, Somatostatin receptor 5, Somatostatin receptor type 5 | 9.901344 | 0.420855 | 4 | 0.601727 | 0.563799 |
| Peptide ligand-binding receptors | C3 | C3-beta-c, Complement C3, Complement C3 [Cleaved into: Complement C3 beta chain; C3-beta-c, Uncharacterized protein | 9.901344 | 0.420855 | 4 | 0.601727 | 0.586567 |
| Peptide ligand-binding receptors | CXCR3 | C-X-C chemokine receptor type 3 | 9.901344 | 0.420855 | 4 | 0.601727 | 0.537539 |
| Peptide ligand-binding receptors | F2 | Activation peptide fragment 1, Prothrombin | 9.901344 | 0.420855 | 4 | 0.601727 | 0.5402 |
| Peptide ligand-binding receptors | MC1R | Melanocyte-stimulating hormone receptor | 9.901344 | 0.420855 | 4 | 0.601727 | 0.526117 |
| Peptide ligand-binding receptors | BDKRB1 | B1 bradykinin receptor, Bradykinin receptor B1 | 9.901344 | 0.420855 | 4 | 0.601727 | 0.547577 |
| Peptide ligand-binding receptors | OPRK1 | Kappa-type opioid receptor, Opioid receptor kappa 1, Opioid receptor, kappa 1 | 9.901344 | 0.420855 | 4 | 0.601727 | 0.590275 |
| Peptide ligand-binding receptors | KISS1R | G protein-coupled receptor 54, KiSS-1 receptor, KISS1 receptor | 9.901344 | 0.420855 | 4 | 0.601727 | 0.63445 |
| Peptide ligand-binding receptors | NPY2R | Neuropeptide Y receptor type 2, NPY-Y2 receptor | 9.901344 | 0.420855 | 4 | 0.601727 | 0.534429 |
| Peptide ligand-binding receptors | AGTR2 | Angiotensin II receptor, type 2, Angiotensin II type-2 receptor, Type-2 angiotensin II receptor | 9.901344 | 0.420855 | 4 | 0.601727 | 0.611168 |
| Peptide ligand-binding receptors | HCRTR2 | Hypocretin receptor type 2, Orexin receptor type 2 | 9.901344 | 0.420855 | 4 | 0.601727 | 0.558726 |
| Peptide ligand-binding receptors | CCR5 | C-C chemokine receptor type 5, Chemokine receptor | 9.901344 | 0.420855 | 4 | 0.601727 | 0.581451 |
| Peptide ligand-binding receptors | EDNRB | Endothelin receptor non-selective type, Endothelin receptor type B | 9.901344 | 0.420855 | 4 | 0.601727 | 0.518386 |
| Peptide ligand-binding receptors | NPY4R | Neuropeptide Y receptor 4, Neuropeptide Y receptor type 1, Neuropeptide Y receptor type 4 | 9.901344 | 0.420855 | 4 | 0.601727 | 0.586341 |
| Peptide ligand-binding receptors | FPR1 | FK506-binding protein 1, fMet-Leu-Phe receptor, Formyl peptide receptor 1 | 9.901344 | 0.420855 | 4 | 0.601727 | 0.511829 |
| Peptide ligand-binding receptors | F2R | CWC22 homolog, spliceosome-associated protein, Proteinase-activated receptor 1 | 9.901344 | 0.420855 | 4 | 0.601727 | 0.58059 |
| Peptide ligand-binding receptors | TACR2 | NK-2 receptor, Substance-K receptor | 9.901344 | 0.420855 | 4 | 0.601727 | 0.601846 |
| Peptide ligand-binding receptors | CCKBR | Cholecystokinin-2 receptor, Gastrin/cholecystokinin type B receptor | 9.901344 | 0.420855 | 4 | 0.601727 | 0.593267 |
| Peptide ligand-binding receptors | AVPR2 | AVPR V2, Vasopressin V2 receptor | 9.901344 | 0.420855 | 4 | 0.601727 | 0.569629 |
| Peptide ligand-binding receptors | OPRM1 | Mu-type opioid receptor | 9.901344 | 0.420855 | 4 | 0.601727 | 0.614279 |
| Peptide ligand-binding receptors | NPY5R | Neuropeptide Y receptor type 5, NPY-Y5 receptor | 9.901344 | 0.420855 | 4 | 0.601727 | 0.567119 |
| Peptide ligand-binding receptors | F2RL3 | F2R like thrombin or trypsin receptor 3, F2R-like thrombin or trypsin receptor 3, G_PROTEIN_RECEP_F1_2 domain-containing protein, Proteinase-activated receptor 4 | 9.901344 | 0.420855 | 4 | 0.601727 | 0.537847 |
| Peptide ligand-binding receptors | AGTR1 | Agtr1.2 protein, Angiotensin II receptor type 1, Type-1 angiotensin II receptor, Type-1A angiotensin II receptor | 9.901344 | 0.420855 | 4 | 0.601727 | 0.648107 |
| Peptide ligand-binding receptors | SSTR4 | G_PROTEIN_RECEP_F1_2 domain-containing protein, Somatostatin receptor type 4 | 9.901344 | 0.420855 | 4 | 0.601727 | 0.53719 |
| Peptide ligand-binding receptors | GHSR | GH-releasing peptide receptor, Growth hormone secretagogue receptor type 1 | 9.901344 | 0.420855 | 4 | 0.601727 | 0.542668 |
| Peptide ligand-binding receptors | HCRTR1 | Hypocretin receptor 1, Orexin receptor type 1 | 9.901344 | 0.420855 | 4 | 0.601727 | 0.541349 |
| Peptide ligand-binding receptors | OXTR | Isotocin receptor-like 2, Oxytocin receptor, Oxytocin-like receptor | 9.901344 | 0.420855 | 4 | 0.601727 | 0.526619 |
| Peptide ligand-binding receptors | NMUR1 | Neuromedin U receptor 1, Neuromedin U receptor type 1, Neuromedin-U receptor 1 | 9.901344 | 0.420855 | 4 | 0.601727 | 0.62859 |
| Peptide ligand-binding receptors | SSTR1 | Somatostatin receptor type 1 | 9.901344 | 0.420855 | 4 | 0.601727 | 0.557663 |
| Peptide ligand-binding receptors | AVPR1A | Arginine vasopressin receptor 1A, G_PROTEIN_RECEP_F1_2 domain-containing protein, Vasopressin V1a receptor | 9.901344 | 0.420855 | 4 | 0.601727 | 0.54747 |
| Peptide ligand-binding receptors | APLNR | Apelin receptor, Apelin receptor 1, APLNR protein | 9.901344 | 0.420855 | 4 | 0.601727 | 0.600344 |
| Peptide ligand-binding receptors | CCKAR | Cholecystokinin receptor type A, Cholecystokinin-2 receptor | 9.901344 | 0.420855 | 4 | 0.601727 | 0.582813 |
| Peptide ligand-binding receptors | SSTR3 | Somatostatin receptor 3, Somatostatin receptor type 3 | 9.901344 | 0.420855 | 4 | 0.601727 | 0.543809 |
| Peptide ligand-binding receptors | MC4R | Melanocortin receptor 4 | 9.901344 | 0.420855 | 4 | 0.601727 | 0.598533 |
| Peptide ligand-binding receptors | C5AR1 | C5a anaphylatoxin chemotactic receptor, C5a anaphylatoxin chemotactic receptor 1 | 9.901344 | 0.420855 | 4 | 0.601727 | 0.500177 |
| Peptide ligand-binding receptors | NPY1R | Neuropeptide Y receptor type 1 | 9.901344 | 0.420855 | 4 | 0.601727 | 0.527768 |
| Peptide ligand-binding receptors | OPRL1 | Kappa-type 3 opioid receptor, Nociceptin receptor | 9.901344 | 0.420855 | 4 | 0.601727 | 0.654503 |
| Peptide ligand-binding receptors | OPRD1 | Delta-type opioid receptor, G_PROTEIN_RECEP_F1_2 domain-containing protein, Opioid receptor delta 1 | 9.901344 | 0.420855 | 4 | 0.601727 | 0.598587 |
| Peptide ligand-binding receptors | BDKRB2 | B1 bradykinin receptor, B2 bradykinin receptor | 9.901344 | 0.420855 | 4 | 0.601727 | 0.518514 |
| Peptide ligand-binding receptors | TACR1 | NK-1 receptor, Substance-P receptor | 9.901344 | 0.420855 | 4 | 0.601727 | 0.738091 |
| Peptide ligand-binding receptors | MCHR1 | G-protein coupled receptor 24, Melanin-concentrating hormone receptor 1 | 9.901344 | 0.420855 | 4 | 0.601727 | 0.678762 |
| Peptide ligand-binding receptors | APP | ABPP, Amyloid-beta A4 protein, Amyloid-beta precursor protein | 9.901344 | 0.420855 | 4 | 0.601727 | 0.506343 |
| Peptide ligand-binding receptors | AVPR1B | G_PROTEIN_RECEP_F1_2 domain-containing protein, Vasopressin V1b receptor | 9.901344 | 0.420855 | 4 | 0.601727 | 0.514485 |
| Peptide ligand-binding receptors | MC5R | Melanocortin receptor 5, mRNA cap guanine-N7 methyltransferase | 9.901344 | 0.420855 | 4 | 0.601727 | 0.501364 |
| Peptide ligand-binding receptors | CCR3 | C-C chemokine receptor type 3, C-C motif chemokine receptor 3, Probable C-C chemokine receptor type 3 | 9.901344 | 0.420855 | 4 | 0.601727 | 0.617373 |
| Peptide ligand-binding receptors | CCR2 | C-C chemokine receptor type 2, C-C chemokine receptor type 2 isoform B, C-C motif chemokine receptor 2, Chemokine, G_PROTEIN_RECEP_F1_2 domain-containing protein | 9.901344 | 0.420855 | 4 | 0.601727 | 0.562183 |
| Peptide ligand-binding receptors | NMUR2 | Neuromedin U receptor 2, Neuromedin U receptor type 2, Neuromedin-U receptor 2 | 9.901344 | 0.420855 | 4 | 0.601727 | 0.612605 |
| Peptide ligand-binding receptors | TACR3 | Neuromedin-K receptor, NK-3 receptor | 9.901344 | 0.420855 | 4 | 0.601727 | 0.602621 |
| Phase 0 - rapid depolarisation | CACNB2 | Brain protein I3, Calcium channel voltage-dependent subunit beta 2, Voltage-dependent L-type calcium channel subunit beta-2 | 9.999487 | 0.587858 | 4 | 0.690292 | 0.596371 |
| Phase 0 - rapid depolarisation | CACNG7 | Voltage-dependent calcium channel gamma-7 subunit | 9.999487 | 0.587858 | 4 | 0.690292 | 0.598629 |
| Phase 0 - rapid depolarisation | SCN2A | Sodium channel protein, Sodium channel protein type 2 subunit alpha | 9.999487 | 0.587858 | 4 | 0.690292 | 0.538824 |
| Phase 0 - rapid depolarisation | SCN4A | Sodium channel protein, Sodium channel protein type 4 subunit alpha | 9.999487 | 0.587858 | 4 | 0.690292 | 0.695739 |
| Phase 0 - rapid depolarisation | CACNA1C | Voltage-dependent L-type calcium channel subunit alpha, Voltage-dependent L-type calcium channel subunit alpha-1C | 9.999487 | 0.587858 | 4 | 0.690292 | 0.585419 |
| Phase 0 - rapid depolarisation | CACNB1 | Calcium channel voltage-dependent subunit beta 1, GuKc domain-containing protein, Voltage-dependent L-type calcium channel subunit beta-1 | 9.999487 | 0.587858 | 4 | 0.690292 | 0.664934 |
| Phase 0 - rapid depolarisation | CACNA2D2 | Calcium voltage-gated channel auxiliary subunit alpha2delta 2, Voltage-dependent calcium channel subunit alpha-2/delta-2, VWFA domain-containing protein | 9.999487 | 0.587858 | 4 | 0.690292 | 0.58576 |
| Phase 0 - rapid depolarisation | CACNG6 | Voltage-dependent calcium channel gamma-6 subunit | 9.999487 | 0.587858 | 4 | 0.690292 | 0.598629 |
| Phase 0 - rapid depolarisation | CACNG8 | Voltage-dependent calcium channel gamma-8 subunit | 9.999487 | 0.587858 | 4 | 0.690292 | 0.579696 |
| Phase 0 - rapid depolarisation | SCN9A | Sodium channel protein, Sodium channel protein type 9 subunit alpha | 9.999487 | 0.587858 | 4 | 0.690292 | 0.589531 |
| Phase 0 - rapid depolarisation | CACNG4 | Voltage-dependent calcium channel gamma-4 subunit | 9.999487 | 0.587858 | 4 | 0.690292 | 0.598629 |
| Phase 2 - plateau phase | CACNB2 | Brain protein I3, Calcium channel voltage-dependent subunit beta 2, Voltage-dependent L-type calcium channel subunit beta-2 | 10.512441 | 0.812879 | 4 | 0.829265 | 0.596371 |
| Phase 2 - plateau phase | CACNA2D2 | Calcium voltage-gated channel auxiliary subunit alpha2delta 2, Voltage-dependent calcium channel subunit alpha-2/delta-2, VWFA domain-containing protein | 10.512441 | 0.812879 | 4 | 0.829265 | 0.58576 |
| Phase 2 - plateau phase | CACNG8 | Voltage-dependent calcium channel gamma-8 subunit | 10.512441 | 0.812879 | 4 | 0.829265 | 0.579696 |
| Phase 2 - plateau phase | CACNA1C | Voltage-dependent L-type calcium channel subunit alpha, Voltage-dependent L-type calcium channel subunit alpha-1C | 10.512441 | 0.812879 | 4 | 0.829265 | 0.585419 |
| Phase 2 - plateau phase | CACNG4 | Voltage-dependent calcium channel gamma-4 subunit | 10.512441 | 0.812879 | 4 | 0.829265 | 0.598629 |
| Phase 2 - plateau phase | CACNG6 | Voltage-dependent calcium channel gamma-6 subunit | 10.512441 | 0.812879 | 4 | 0.829265 | 0.598629 |
| Phase 2 - plateau phase | CACNB1 | Calcium channel voltage-dependent subunit beta 1, GuKc domain-containing protein, Voltage-dependent L-type calcium channel subunit beta-1 | 10.512441 | 0.812879 | 4 | 0.829265 | 0.664934 |
| Phase 2 - plateau phase | CACNG7 | Voltage-dependent calcium channel gamma-7 subunit | 10.512441 | 0.812879 | 4 | 0.829265 | 0.598629 |
| PI3K events in ERBB2 signaling | ERBB4 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-4 | 9.847765 | 0.726888 | 3.573565 | 0.671812 | 0.550399 |
| PI3K events in ERBB2 signaling | ERBB3 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-3 | 9.847765 | 0.726888 | 3.573565 | 0.671812 | 0.551662 |
| PI3K events in ERBB2 signaling | EGFR | Epidermal growth factor receptor, Receptor protein-tyrosine kinase | 9.847765 | 0.726888 | 3.573565 | 0.671812 | 0.625817 |
| PI3K events in ERBB2 signaling | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.847765 | 0.726888 | 3.573565 | 0.671812 | 0.660919 |
| PI3K events in ERBB2 signaling | ERBB2 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-2 | 9.847765 | 0.726888 | 3.573565 | 0.671812 | 0.589497 |
| PI3K events in ERBB2 signaling | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 9.847765 | 0.726888 | 3.573565 | 0.671812 | 0.646924 |
| PI3K events in ERBB4 signaling | ERBB4 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-4 | 10.222502 | 0.829438 | 2.714429 | 0.558214 | 0.550399 |
| PI3K events in ERBB4 signaling | EGFR | Epidermal growth factor receptor, Receptor protein-tyrosine kinase | 10.222502 | 0.829438 | 2.714429 | 0.558214 | 0.625817 |
| PI3K events in ERBB4 signaling | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 10.222502 | 0.829438 | 2.714429 | 0.558214 | 0.660919 |
| PI3K events in ERBB4 signaling | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 10.222502 | 0.829438 | 2.714429 | 0.558214 | 0.646924 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 9.696631 | 0.451014 | 4 | 0.606246 | 0.552041 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.696631 | 0.451014 | 4 | 0.606246 | 0.660919 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 9.696631 | 0.451014 | 4 | 0.606246 | 0.646924 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | PDGFRA | Platelet-derived growth factor receptor alpha | 9.696631 | 0.451014 | 4 | 0.606246 | 0.532138 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | PPP2R5A | Serine/threonine protein phosphatase 2A regulatory subunit, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform | 9.696631 | 0.451014 | 4 | 0.606246 | 0.559184 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | EGFR | Epidermal growth factor receptor, Receptor protein-tyrosine kinase | 9.696631 | 0.451014 | 4 | 0.606246 | 0.625817 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.696631 | 0.451014 | 4 | 0.606246 | 0.589385 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 9.696631 | 0.451014 | 4 | 0.606246 | 0.589663 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | ESR1 | Estrogen receptor | 9.696631 | 0.451014 | 4 | 0.606246 | 0.561602 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | INSR | Insulin receptor, Tyrosine-protein kinase receptor | 9.696631 | 0.451014 | 4 | 0.606246 | 0.563226 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | FGFR2 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 2 | 9.696631 | 0.451014 | 4 | 0.606246 | 0.577012 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | PDGFRB | Platelet-derived growth factor receptor beta, Receptor protein-tyrosine kinase | 9.696631 | 0.451014 | 4 | 0.606246 | 0.550695 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | MDM2 | E3 ubiquitin-protein ligase Mdm2 | 9.696631 | 0.451014 | 4 | 0.606246 | 0.659771 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | LCK | Proto-oncogene tyrosine-protein kinase LCK, Tyrosine-protein kinase, Tyrosine-protein kinase Lck | 9.696631 | 0.451014 | 4 | 0.606246 | 0.543278 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | FGF1 | Fibroblast growth factor 1, Multifunctional fusion protein [Includes: Fibroblast growth factor 1, Putative fibroblast growth factor 1 | 9.696631 | 0.451014 | 4 | 0.606246 | 0.509844 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | FGFR1 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 1 | 9.696631 | 0.451014 | 4 | 0.606246 | 0.55821 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 9.696631 | 0.451014 | 4 | 0.606246 | 0.594807 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | FGFR3 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 3 | 9.696631 | 0.451014 | 4 | 0.606246 | 0.560785 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | ERBB3 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-3 | 9.696631 | 0.451014 | 4 | 0.606246 | 0.551662 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | ERBB2 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-2 | 9.696631 | 0.451014 | 4 | 0.606246 | 0.589497 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | MET | Hepatocyte growth factor receptor, HGF/SF receptor | 9.696631 | 0.451014 | 4 | 0.606246 | 0.5914 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 9.696631 | 0.451014 | 4 | 0.606246 | 0.541254 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | IRAK4 | Interleukin-1 receptor-associated kinase 4 | 9.696631 | 0.451014 | 4 | 0.606246 | 0.50686 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | PIK3CD | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.696631 | 0.451014 | 4 | 0.606246 | 0.632077 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | FLT3 | Receptor protein-tyrosine kinase, Receptor-type tyrosine-protein kinase FLT3 | 9.696631 | 0.451014 | 4 | 0.606246 | 0.529563 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | KIT | Mast/stem cell growth factor receptor, Mast/stem cell growth factor receptor Kit | 9.696631 | 0.451014 | 4 | 0.606246 | 0.562353 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | ESR2 | Estrogen receptor 2, Estrogen receptor beta | 9.696631 | 0.451014 | 4 | 0.606246 | 0.53484 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | ERBB4 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-4 | 9.696631 | 0.451014 | 4 | 0.606246 | 0.550399 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | FGFR4 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 4 | 9.696631 | 0.451014 | 4 | 0.606246 | 0.555253 |
| PIP3 activates AKT signaling | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.54735 |
| PIP3 activates AKT signaling | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.577481 |
| PIP3 activates AKT signaling | MET | Hepatocyte growth factor receptor, HGF/SF receptor | 9.788402 | 0.522192 | 4 | 0.646569 | 0.5914 |
| PIP3 activates AKT signaling | FLT3 | Receptor protein-tyrosine kinase, Receptor-type tyrosine-protein kinase FLT3 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.529563 |
| PIP3 activates AKT signaling | PDGFRA | Platelet-derived growth factor receptor alpha | 9.788402 | 0.522192 | 4 | 0.646569 | 0.532138 |
| PIP3 activates AKT signaling | PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.788402 | 0.522192 | 4 | 0.646569 | 0.589385 |
| PIP3 activates AKT signaling | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.788402 | 0.522192 | 4 | 0.646569 | 0.579609 |
| PIP3 activates AKT signaling | ERBB4 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-4 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.550399 |
| PIP3 activates AKT signaling | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.54735 |
| PIP3 activates AKT signaling | IRAK4 | Interleukin-1 receptor-associated kinase 4 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.50686 |
| PIP3 activates AKT signaling | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.546881 |
| PIP3 activates AKT signaling | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.515416 |
| PIP3 activates AKT signaling | LCK | Proto-oncogene tyrosine-protein kinase LCK, Tyrosine-protein kinase, Tyrosine-protein kinase Lck | 9.788402 | 0.522192 | 4 | 0.646569 | 0.543278 |
| PIP3 activates AKT signaling | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.589663 |
| PIP3 activates AKT signaling | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.788402 | 0.522192 | 4 | 0.646569 | 0.568645 |
| PIP3 activates AKT signaling | HDAC2 | Histone deacetylase 2 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.52963 |
| PIP3 activates AKT signaling | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.58858 |
| PIP3 activates AKT signaling | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.54735 |
| PIP3 activates AKT signaling | ERBB3 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-3 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.551662 |
| PIP3 activates AKT signaling | BAD | BCL2 associated agonist of cell death, Bcl2-associated agonist of cell death | 9.788402 | 0.522192 | 4 | 0.646569 | 0.596467 |
| PIP3 activates AKT signaling | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.515416 |
| PIP3 activates AKT signaling | HDAC1 | Histone deacetylase 1, Histone deacetylase HDAC1 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.540105 |
| PIP3 activates AKT signaling | HDAC5 | Histone deacetylase, Histone deacetylase 5 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.523224 |
| PIP3 activates AKT signaling | PPP2R5A | Serine/threonine protein phosphatase 2A regulatory subunit, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform | 9.788402 | 0.522192 | 4 | 0.646569 | 0.559184 |
| PIP3 activates AKT signaling | CSNK2B | Casein kinase II subunit beta | 9.788402 | 0.522192 | 4 | 0.646569 | 0.537688 |
| PIP3 activates AKT signaling | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.547953 |
| PIP3 activates AKT signaling | PIK3CD | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.788402 | 0.522192 | 4 | 0.646569 | 0.632077 |
| PIP3 activates AKT signaling | FGFR4 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 4 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.555253 |
| PIP3 activates AKT signaling | CHD4 | Chromodomain-helicase-DNA-binding protein 4, DNA helicase | 9.788402 | 0.522192 | 4 | 0.646569 | 0.566824 |
| PIP3 activates AKT signaling | INSR | Insulin receptor, Tyrosine-protein kinase receptor | 9.788402 | 0.522192 | 4 | 0.646569 | 0.563226 |
| PIP3 activates AKT signaling | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.515416 |
| PIP3 activates AKT signaling | RCOR1 | REST corepressor 1, Uncharacterized protein | 9.788402 | 0.522192 | 4 | 0.646569 | 0.529174 |
| PIP3 activates AKT signaling | ESR1 | Estrogen receptor | 9.788402 | 0.522192 | 4 | 0.646569 | 0.561602 |
| PIP3 activates AKT signaling | USP13 | Ubiquitin carboxyl-terminal hydrolase, Ubiquitin carboxyl-terminal hydrolase 13 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.538123 |
| PIP3 activates AKT signaling | EGFR | Epidermal growth factor receptor, Receptor protein-tyrosine kinase | 9.788402 | 0.522192 | 4 | 0.646569 | 0.625817 |
| PIP3 activates AKT signaling | TNKS | Poly [ADP-ribose] polymerase, Poly [ADP-ribose] polymerase tankyrase, Poly [ADP-ribose] polymerase tankyrase-1 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.506213 |
| PIP3 activates AKT signaling | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.788402 | 0.522192 | 4 | 0.646569 | 0.515416 |
| PIP3 activates AKT signaling | TP53 | Cellular tumor antigen p53 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.525937 |
| PIP3 activates AKT signaling | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.572294 |
| PIP3 activates AKT signaling | FGFR3 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 3 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.560785 |
| PIP3 activates AKT signaling | FGF1 | Fibroblast growth factor 1, Multifunctional fusion protein [Includes: Fibroblast growth factor 1, Putative fibroblast growth factor 1 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.509844 |
| PIP3 activates AKT signaling | HDAC3 | Histone deacetylase, Histone deacetylase 3 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.528637 |
| PIP3 activates AKT signaling | CBX6 | Chromo domain-containing protein, Chromobox 6, Chromobox protein homolog 6 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.575593 |
| PIP3 activates AKT signaling | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.565601 |
| PIP3 activates AKT signaling | MDM2 | E3 ubiquitin-protein ligase Mdm2 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.659771 |
| PIP3 activates AKT signaling | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.541254 |
| PIP3 activates AKT signaling | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 9.788402 | 0.522192 | 4 | 0.646569 | 0.552041 |
| PIP3 activates AKT signaling | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 9.788402 | 0.522192 | 4 | 0.646569 | 0.646924 |
| PIP3 activates AKT signaling | XIAP | E3 ubiquitin-protein ligase XIAP, RING-type domain-containing protein, X-linked inhibitor of apoptosis | 9.788402 | 0.522192 | 4 | 0.646569 | 0.589894 |
| PIP3 activates AKT signaling | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.54735 |
| PIP3 activates AKT signaling | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.515416 |
| PIP3 activates AKT signaling | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.5197 |
| PIP3 activates AKT signaling | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.577847 |
| PIP3 activates AKT signaling | HDAC7 | Hist_deacetyl domain-containing protein, Histone deacetylase, Histone deacetylase 7 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.522826 |
| PIP3 activates AKT signaling | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.788402 | 0.522192 | 4 | 0.646569 | 0.660919 |
| PIP3 activates AKT signaling | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.57244 |
| PIP3 activates AKT signaling | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.515416 |
| PIP3 activates AKT signaling | ERBB2 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-2 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.589497 |
| PIP3 activates AKT signaling | FGFR2 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 2 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.577012 |
| PIP3 activates AKT signaling | PDGFRB | Platelet-derived growth factor receptor beta, Receptor protein-tyrosine kinase | 9.788402 | 0.522192 | 4 | 0.646569 | 0.550695 |
| PIP3 activates AKT signaling | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 9.788402 | 0.522192 | 4 | 0.646569 | 0.594807 |
| PIP3 activates AKT signaling | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.515416 |
| PIP3 activates AKT signaling | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.54735 |
| PIP3 activates AKT signaling | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.515416 |
| PIP3 activates AKT signaling | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.788402 | 0.522192 | 4 | 0.646569 | 0.515416 |
| PIP3 activates AKT signaling | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.54735 |
| PIP3 activates AKT signaling | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.515416 |
| PIP3 activates AKT signaling | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.515416 |
| PIP3 activates AKT signaling | KIT | Mast/stem cell growth factor receptor, Mast/stem cell growth factor receptor Kit | 9.788402 | 0.522192 | 4 | 0.646569 | 0.562353 |
| PIP3 activates AKT signaling | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.54735 |
| PIP3 activates AKT signaling | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.515416 |
| PIP3 activates AKT signaling | EZH2 | [Histone H3]-lysine(27) N-trimethyltransferase, Histone-lysine N-methyltransferase EZH2 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.53644 |
| PIP3 activates AKT signaling | CSNK2A2 | Casein kinase 2 alpha 2, Casein kinase II subunit alpha' | 9.788402 | 0.522192 | 4 | 0.646569 | 0.53622 |
| PIP3 activates AKT signaling | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.515416 |
| PIP3 activates AKT signaling | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.54735 |
| PIP3 activates AKT signaling | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.573417 |
| PIP3 activates AKT signaling | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.515416 |
| PIP3 activates AKT signaling | MTOR | Nucleoprotein TPR, Serine/threonine-protein kinase mTOR, Serine/threonine-protein kinase TOR | 9.788402 | 0.522192 | 4 | 0.646569 | 0.619026 |
| PIP3 activates AKT signaling | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.54735 |
| PIP3 activates AKT signaling | ESR2 | Estrogen receptor 2, Estrogen receptor beta | 9.788402 | 0.522192 | 4 | 0.646569 | 0.53484 |
| PIP3 activates AKT signaling | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.515416 |
| PIP3 activates AKT signaling | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.54735 |
| PIP3 activates AKT signaling | FGFR1 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 1 | 9.788402 | 0.522192 | 4 | 0.646569 | 0.55821 |
| PIP3 activates AKT signaling | CSNK2A1 | Casein kinase 2, alpha 1 polypeptide, Casein kinase II subunit alpha, Protein kinase domain-containing protein | 9.788402 | 0.522192 | 4 | 0.646569 | 0.538737 |
| PIP3 activates AKT signaling | RPTOR | Regulatory associated protein of MTOR complex 1, Regulatory associated protein of MTOR, complex 1, Regulatory-associated protein of mTOR, Regulatory-associated protein of MTOR, complex 1, WD_REPEATS_REGION domain-containing protein | 9.788402 | 0.522192 | 4 | 0.646569 | 0.543357 |
| PKA-mediated phosphorylation of CREB | ADCY7 | Adenylate cyclase type 7 | 6.588257 | 0.576838 | 3.332167 | 0.423851 | 0.550655 |
| PKA-mediated phosphorylation of CREB | PRKAR2A | cAMP-dependent protein kinase type II-alpha regulatory subunit, Protein kinase cAMP-dependent type II regulatory subunit alpha, Protein kinase, cAMP-dependent, regulatory subunit type II alpha, Uncharacterized protein | 6.588257 | 0.576838 | 3.332167 | 0.423851 | 0.527065 |
| PKA-mediated phosphorylation of CREB | ADCY9 | Adenylate cyclase 9, Adenylate cyclase type 9 | 6.588257 | 0.576838 | 3.332167 | 0.423851 | 0.550534 |
| PKA activation | ADCY9 | Adenylate cyclase 9, Adenylate cyclase type 9 | 9.4896 | 0.607068 | 3.19968 | 0.53882 | 0.550534 |
| PKA activation | PRKAR2A | cAMP-dependent protein kinase type II-alpha regulatory subunit, Protein kinase cAMP-dependent type II regulatory subunit alpha, Protein kinase, cAMP-dependent, regulatory subunit type II alpha, Uncharacterized protein | 9.4896 | 0.607068 | 3.19968 | 0.53882 | 0.527065 |
| PKA activation | ADCY7 | Adenylate cyclase type 7 | 9.4896 | 0.607068 | 3.19968 | 0.53882 | 0.550655 |
| PKA activation in glucagon signalling | PRKAR2A | cAMP-dependent protein kinase type II-alpha regulatory subunit, Protein kinase cAMP-dependent type II regulatory subunit alpha, Protein kinase, cAMP-dependent, regulatory subunit type II alpha, Uncharacterized protein | 9.180021 | 0.63764 | 3.697636 | 0.622041 | 0.527065 |
| PKA activation in glucagon signalling | ADCY7 | Adenylate cyclase type 7 | 9.180021 | 0.63764 | 3.697636 | 0.622041 | 0.550655 |
| PKA activation in glucagon signalling | ADCY9 | Adenylate cyclase 9, Adenylate cyclase type 9 | 9.180021 | 0.63764 | 3.697636 | 0.622041 | 0.550534 |
| Platelet Aggregation (Plug Formation) | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 8.652709 | 0.538753 | 4 | 0.59626 | 0.594807 |
| Platelet Aggregation (Plug Formation) | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 8.652709 | 0.538753 | 4 | 0.59626 | 0.552041 |
| Platelet Aggregation (Plug Formation) | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 8.652709 | 0.538753 | 4 | 0.59626 | 0.589663 |
| Platelet Aggregation (Plug Formation) | PTK2 | Focal adhesion kinase 1, Non-specific protein-tyrosine kinase, Serine/threonine-protein kinase PTK2/STK2 | 8.652709 | 0.538753 | 4 | 0.59626 | 0.503121 |
| Platelet Aggregation (Plug Formation) | RAP1A | RAP1A, member of RAS oncogene family, Ras-related protein Rap-1A, Uncharacterized protein | 8.652709 | 0.538753 | 4 | 0.59626 | 0.530823 |
| Platelet Aggregation (Plug Formation) | SHC | DShc protein | 8.652709 | 0.538753 | 4 | 0.59626 | 0.605 |
| Platelet Aggregation (Plug Formation) | SYK | Tyrosine-protein kinase, Tyrosine-protein kinase SYK | 8.652709 | 0.538753 | 4 | 0.59626 | 0.557587 |
| Platelet Aggregation (Plug Formation) | CRK | 14_3_3 domain-containing protein, Adapter molecule crk, Adapter molecule Crk, CRK proto-oncogene, adaptor protein, Uncharacterized protein, V-crk avian sarcoma virus CT10 oncogene homolog | 8.652709 | 0.538753 | 4 | 0.59626 | 0.513369 |
| Platelet Aggregation (Plug Formation) | ADRA2B | Adrenoceptor alpha 2B, Alpha-2B adrenergic receptor, G_PROTEIN_RECEP_F1_2 domain-containing protein | 8.652709 | 0.538753 | 4 | 0.59626 | 0.524078 |
| Platelet Aggregation (Plug Formation) | ADRA2A | Adrenoceptor alpha 2A, Alpha-2A adrenergic receptor | 8.652709 | 0.538753 | 4 | 0.59626 | 0.530746 |
| Platelet Aggregation (Plug Formation) | F2 | Activation peptide fragment 1, Prothrombin | 8.652709 | 0.538753 | 4 | 0.59626 | 0.5402 |
| Platelet Aggregation (Plug Formation) | MPL | Thrombopoietin receptor | 8.652709 | 0.538753 | 4 | 0.59626 | 0.544974 |
| Platelet Aggregation (Plug Formation) | SHC1 | SHC-transforming protein 1 | 8.652709 | 0.538753 | 4 | 0.59626 | 0.521858 |
| Platelet Aggregation (Plug Formation) | FN1 | Fibronectin | 8.652709 | 0.538753 | 4 | 0.59626 | 0.578096 |
| Platelet Aggregation (Plug Formation) | ADRA2C | Adrenoceptor alpha 2C, Alpha-2C adrenergic receptor | 8.652709 | 0.538753 | 4 | 0.59626 | 0.528655 |
| Platelet Aggregation (Plug Formation) | VWF | von Willebrand factor | 8.652709 | 0.538753 | 4 | 0.59626 | 0.548181 |
| Platelet Aggregation (Plug Formation) | PTPN1 | Protein-tyrosine-phosphatase, Tyrosine-protein phosphatase non-receptor type, Tyrosine-protein phosphatase non-receptor type 1 | 8.652709 | 0.538753 | 4 | 0.59626 | 0.510671 |
| Potential therapeutics for SARS | CHD4 | Chromodomain-helicase-DNA-binding protein 4, DNA helicase | 8.632104 | 0.44271 | 3.890271 | 0.532165 | 0.566824 |
| Potential therapeutics for SARS | NR3C1 | Glucocorticoid receptor | 8.632104 | 0.44271 | 3.890271 | 0.532165 | 0.59851 |
| Potential therapeutics for SARS | HSP90AB1 | Heat shock protein 90 alpha family class B member 1, Heat shock protein 90kDa alpha, Heat shock protein HSP 90-beta, Heat shock protein HSP 90-beta isoform a | 8.632104 | 0.44271 | 3.890271 | 0.532165 | 0.569354 |
| Potential therapeutics for SARS | HDAC1 | Histone deacetylase 1, Histone deacetylase HDAC1 | 8.632104 | 0.44271 | 3.890271 | 0.532165 | 0.540105 |
| Potential therapeutics for SARS | RCOR1 | REST corepressor 1, Uncharacterized protein | 8.632104 | 0.44271 | 3.890271 | 0.532165 | 0.529174 |
| Potential therapeutics for SARS | CYSLTR1 | Cysteinyl leukotriene receptor 1 | 8.632104 | 0.44271 | 3.890271 | 0.532165 | 0.552802 |
| Potential therapeutics for SARS | ROCK2 | Rho-associated protein kinase, Rho-associated protein kinase 2 | 8.632104 | 0.44271 | 3.890271 | 0.532165 | 0.516942 |
| Potential therapeutics for SARS | FURIN | Furin, Furin, paired basic amino acid cleaving enzyme, Trans Golgi network protease furin | 8.632104 | 0.44271 | 3.890271 | 0.532165 | 0.715659 |
| Potential therapeutics for SARS | JAK3 | Tyrosine-protein kinase, Tyrosine-protein kinase JAK3 | 8.632104 | 0.44271 | 3.890271 | 0.532165 | 0.537649 |
| Potential therapeutics for SARS | SIGMAR1 | Sigma non-opioid intracellular receptor 1 | 8.632104 | 0.44271 | 3.890271 | 0.532165 | 0.654919 |
| Potential therapeutics for SARS | BRD4 | Bromodomain-containing protein 4 | 8.632104 | 0.44271 | 3.890271 | 0.532165 | 0.570264 |
| Potential therapeutics for SARS | JAK2 | Janus kinase 2, Tyrosine-protein kinase, Tyrosine-protein kinase JAK2 | 8.632104 | 0.44271 | 3.890271 | 0.532165 | 0.573235 |
| Potential therapeutics for SARS | TBK1 | Protein kinase domain-containing protein, Serine/threonine-protein kinase TBK1, Similar to TANK-binding kinase 1, TANK binding kinase 1, TANK-binding kinase 1 | 8.632104 | 0.44271 | 3.890271 | 0.532165 | 0.510594 |
| Potential therapeutics for SARS | FKBP1A | Peptidyl-prolyl cis-trans isomerase FKBP1A, Peptidylprolyl isomerase | 8.632104 | 0.44271 | 3.890271 | 0.532165 | 0.534375 |
| Potential therapeutics for SARS | HSP90AA1 | Heat shock protein 90 alpha family class A member 1, Heat shock protein HSP 90-alpha | 8.632104 | 0.44271 | 3.890271 | 0.532165 | 0.624325 |
| Potential therapeutics for SARS | HDAC2 | Histone deacetylase 2 | 8.632104 | 0.44271 | 3.890271 | 0.532165 | 0.52963 |
| Potential therapeutics for SARS | CRBN | Protein cereblon | 8.632104 | 0.44271 | 3.890271 | 0.532165 | 0.508911 |
| Potential therapeutics for SARS | S1PR1 | Sphingosine 1-phosphate receptor 1 | 8.632104 | 0.44271 | 3.890271 | 0.532165 | 0.519942 |
| Potential therapeutics for SARS | JAK1 | Solute carrier family 39, Tyrosine-protein kinase, Tyrosine-protein kinase JAK1 | 8.632104 | 0.44271 | 3.890271 | 0.532165 | 0.551488 |
| Presynaptic depolarization and calcium channel opening | CACNA2D1 | Dihydropyridine receptor alpha 2 subunit | 10.508413 | 0.960989 | 4 | 0.903112 | 0.605387 |
| Presynaptic depolarization and calcium channel opening | CACNA1B | Ca_chan_IQ domain-containing protein, Voltage-dependent N-type calcium channel subunit alpha, Voltage-dependent N-type calcium channel subunit alpha-1B | 10.508413 | 0.960989 | 4 | 0.903112 | 0.592016 |
| Presynaptic depolarization and calcium channel opening | CACNB1 | Calcium channel voltage-dependent subunit beta 1, GuKc domain-containing protein, Voltage-dependent L-type calcium channel subunit beta-1 | 10.508413 | 0.960989 | 4 | 0.903112 | 0.664934 |
| Presynaptic depolarization and calcium channel opening | CACNA2D2 | Calcium voltage-gated channel auxiliary subunit alpha2delta 2, Voltage-dependent calcium channel subunit alpha-2/delta-2, VWFA domain-containing protein | 10.508413 | 0.960989 | 4 | 0.903112 | 0.58576 |
| Presynaptic depolarization and calcium channel opening | CACNG2 | Voltage-dependent calcium channel gamma-2 subunit | 10.508413 | 0.960989 | 4 | 0.903112 | 0.593521 |
| Presynaptic depolarization and calcium channel opening | CACNA2D3 | Calcium voltage-gated channel auxiliary subunit alpha2delta 3, Voltage-dependent calcium channel subunit alpha-2/delta-3, VWFA domain-containing protein | 10.508413 | 0.960989 | 4 | 0.903112 | 0.598629 |
| Presynaptic depolarization and calcium channel opening | CACNB3 | Calcium channel voltage-dependent subunit beta 3, Voltage-dependent L-type calcium channel subunit beta-3 | 10.508413 | 0.960989 | 4 | 0.903112 | 0.592713 |
| Presynaptic depolarization and calcium channel opening | CACNB2 | Brain protein I3, Calcium channel voltage-dependent subunit beta 2, Voltage-dependent L-type calcium channel subunit beta-2 | 10.508413 | 0.960989 | 4 | 0.903112 | 0.596371 |
| Presynaptic depolarization and calcium channel opening | CACNA1A | Voltage-dependent P/Q-type calcium channel subunit alpha, Voltage-dependent P/Q-type calcium channel subunit alpha-1A | 10.508413 | 0.960989 | 4 | 0.903112 | 0.600805 |
| Presynaptic depolarization and calcium channel opening | CACNB4 | Calcium voltage-gated channel auxiliary subunit beta 4, SH3 domain-containing protein, Voltage-dependent L-type calcium channel subunit beta-4 | 10.508413 | 0.960989 | 4 | 0.903112 | 0.598629 |
| Presynaptic depolarization and calcium channel opening | CACNA1E | Voltage-dependent R-type calcium channel subunit alpha, Voltage-dependent R-type calcium channel subunit alpha-1E | 10.508413 | 0.960989 | 4 | 0.903112 | 0.598263 |
| Presynaptic depolarization and calcium channel opening | CACNG4 | Voltage-dependent calcium channel gamma-4 subunit | 10.508413 | 0.960989 | 4 | 0.903112 | 0.598629 |
| RAF activation | BRAF | Non-specific serine/threonine protein kinase, Serine/threonine-protein kinase B-raf | 8.926679 | 0.530594 | 3.610625 | 0.547294 | 0.590074 |
| RAF activation | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 8.926679 | 0.530594 | 3.610625 | 0.547294 | 0.505988 |
| RAF activation | YWHAB | 14-3-3 protein beta/alpha, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta | 8.926679 | 0.530594 | 3.610625 | 0.547294 | 0.513843 |
| RAF activation | RAF1 | Non-specific serine/threonine protein kinase, RAF proto-oncogene serine/threonine-protein kinase | 8.926679 | 0.530594 | 3.610625 | 0.547294 | 0.573556 |
| RAF activation | JAK2 | Janus kinase 2, Tyrosine-protein kinase, Tyrosine-protein kinase JAK2 | 8.926679 | 0.530594 | 3.610625 | 0.547294 | 0.573235 |
| RAF activation | PPP2R5A | Serine/threonine protein phosphatase 2A regulatory subunit, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform | 8.926679 | 0.530594 | 3.610625 | 0.547294 | 0.559184 |
| RAF activation | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 8.926679 | 0.530594 | 3.610625 | 0.547294 | 0.594807 |
| RAF activation | PPP1CC | Serine/threonine-protein phosphatase, Serine/threonine-protein phosphatase PP1-gamma catalytic subunit | 8.926679 | 0.530594 | 3.610625 | 0.547294 | 0.503303 |
| RAF activation | MAP2K1 | Dual specificity mitogen-activated protein kinase kinase 1, MAP2K1 protein, Mitogen-activated protein kinase kinase 1, Protein kinase domain-containing protein | 8.926679 | 0.530594 | 3.610625 | 0.547294 | 0.545374 |
| RAF/MAP kinase cascade | PTK2 | Focal adhesion kinase 1, Non-specific protein-tyrosine kinase, Serine/threonine-protein kinase PTK2/STK2 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.503121 |
| RAF/MAP kinase cascade | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.364338 | 0.496957 | 4 | 0.612074 | 0.515416 |
| RAF/MAP kinase cascade | FGFR1 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 1 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.55821 |
| RAF/MAP kinase cascade | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.515416 |
| RAF/MAP kinase cascade | MAP2K1 | Dual specificity mitogen-activated protein kinase kinase 1, MAP2K1 protein, Mitogen-activated protein kinase kinase 1, Protein kinase domain-containing protein | 9.364338 | 0.496957 | 4 | 0.612074 | 0.545374 |
| RAF/MAP kinase cascade | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 9.364338 | 0.496957 | 4 | 0.612074 | 0.646924 |
| RAF/MAP kinase cascade | BCL2L1 | Apoptosis regulator Bcl-X, Bcl-2-like protein 1, BCL2-like 1 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.865729 |
| RAF/MAP kinase cascade | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.515416 |
| RAF/MAP kinase cascade | FGF1 | Fibroblast growth factor 1, Multifunctional fusion protein [Includes: Fibroblast growth factor 1, Putative fibroblast growth factor 1 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.509844 |
| RAF/MAP kinase cascade | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.515416 |
| RAF/MAP kinase cascade | ERBB3 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-3 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.551662 |
| RAF/MAP kinase cascade | GRIN1 | Glutamate receptor, Glutamate receptor ionotropic, NMDA 1 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.573676 |
| RAF/MAP kinase cascade | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.515416 |
| RAF/MAP kinase cascade | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.364338 | 0.496957 | 4 | 0.612074 | 0.568645 |
| RAF/MAP kinase cascade | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 9.364338 | 0.496957 | 4 | 0.612074 | 0.505988 |
| RAF/MAP kinase cascade | ERBB4 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-4 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.550399 |
| RAF/MAP kinase cascade | JAK1 | Solute carrier family 39, Tyrosine-protein kinase, Tyrosine-protein kinase JAK1 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.551488 |
| RAF/MAP kinase cascade | MET | Hepatocyte growth factor receptor, HGF/SF receptor | 9.364338 | 0.496957 | 4 | 0.612074 | 0.5914 |
| RAF/MAP kinase cascade | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.515416 |
| RAF/MAP kinase cascade | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.364338 | 0.496957 | 4 | 0.612074 | 0.579609 |
| RAF/MAP kinase cascade | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.54735 |
| RAF/MAP kinase cascade | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.515416 |
| RAF/MAP kinase cascade | PRKCQ | Protein kinase C, Protein kinase C theta type | 9.364338 | 0.496957 | 4 | 0.612074 | 0.523352 |
| RAF/MAP kinase cascade | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.541254 |
| RAF/MAP kinase cascade | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.515416 |
| RAF/MAP kinase cascade | FGFR2 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 2 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.577012 |
| RAF/MAP kinase cascade | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.54735 |
| RAF/MAP kinase cascade | PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.364338 | 0.496957 | 4 | 0.612074 | 0.589385 |
| RAF/MAP kinase cascade | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.515416 |
| RAF/MAP kinase cascade | MAPK12 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 12 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.573393 |
| RAF/MAP kinase cascade | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.515416 |
| RAF/MAP kinase cascade | GRIN2D | Glutamate receptor, Glutamate receptor ionotropic, NMDA 2D | 9.364338 | 0.496957 | 4 | 0.612074 | 0.556318 |
| RAF/MAP kinase cascade | FN1 | Fibronectin | 9.364338 | 0.496957 | 4 | 0.612074 | 0.578096 |
| RAF/MAP kinase cascade | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.54735 |
| RAF/MAP kinase cascade | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.547953 |
| RAF/MAP kinase cascade | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.5197 |
| RAF/MAP kinase cascade | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.577481 |
| RAF/MAP kinase cascade | BRAF | Non-specific serine/threonine protein kinase, Serine/threonine-protein kinase B-raf | 9.364338 | 0.496957 | 4 | 0.612074 | 0.590074 |
| RAF/MAP kinase cascade | PPP2R5A | Serine/threonine protein phosphatase 2A regulatory subunit, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform | 9.364338 | 0.496957 | 4 | 0.612074 | 0.559184 |
| RAF/MAP kinase cascade | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.577847 |
| RAF/MAP kinase cascade | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.515416 |
| RAF/MAP kinase cascade | PDGFRA | Platelet-derived growth factor receptor alpha | 9.364338 | 0.496957 | 4 | 0.612074 | 0.532138 |
| RAF/MAP kinase cascade | PPP1CC | Serine/threonine-protein phosphatase, Serine/threonine-protein phosphatase PP1-gamma catalytic subunit | 9.364338 | 0.496957 | 4 | 0.612074 | 0.503303 |
| RAF/MAP kinase cascade | JAK3 | Tyrosine-protein kinase, Tyrosine-protein kinase JAK3 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.537649 |
| RAF/MAP kinase cascade | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.54735 |
| RAF/MAP kinase cascade | VWF | von Willebrand factor | 9.364338 | 0.496957 | 4 | 0.612074 | 0.548181 |
| RAF/MAP kinase cascade | PDGFRB | Platelet-derived growth factor receptor beta, Receptor protein-tyrosine kinase | 9.364338 | 0.496957 | 4 | 0.612074 | 0.550695 |
| RAF/MAP kinase cascade | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.515416 |
| RAF/MAP kinase cascade | GRIN2A | Glutamate receptor, Glutamate receptor ionotropic, NMDA 2A | 9.364338 | 0.496957 | 4 | 0.612074 | 0.527756 |
| RAF/MAP kinase cascade | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.54735 |
| RAF/MAP kinase cascade | EGFR | Epidermal growth factor receptor, Receptor protein-tyrosine kinase | 9.364338 | 0.496957 | 4 | 0.612074 | 0.625817 |
| RAF/MAP kinase cascade | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.565601 |
| RAF/MAP kinase cascade | FGFR4 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 4 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.555253 |
| RAF/MAP kinase cascade | RET | Proto-oncogene tyrosine-protein kinase receptor Ret | 9.364338 | 0.496957 | 4 | 0.612074 | 0.515319 |
| RAF/MAP kinase cascade | FLT3 | Receptor protein-tyrosine kinase, Receptor-type tyrosine-protein kinase FLT3 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.529563 |
| RAF/MAP kinase cascade | FGFR3 | Fibroblast growth factor receptor, Fibroblast growth factor receptor 3 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.560785 |
| RAF/MAP kinase cascade | SHC | DShc protein | 9.364338 | 0.496957 | 4 | 0.612074 | 0.605 |
| RAF/MAP kinase cascade | FNTB | Protein farnesyltransferase subunit beta | 9.364338 | 0.496957 | 4 | 0.612074 | 0.672613 |
| RAF/MAP kinase cascade | YWHAB | 14-3-3 protein beta/alpha, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta | 9.364338 | 0.496957 | 4 | 0.612074 | 0.513843 |
| RAF/MAP kinase cascade | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.515416 |
| RAF/MAP kinase cascade | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.573417 |
| RAF/MAP kinase cascade | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.54735 |
| RAF/MAP kinase cascade | RAF1 | Non-specific serine/threonine protein kinase, RAF proto-oncogene serine/threonine-protein kinase | 9.364338 | 0.496957 | 4 | 0.612074 | 0.573556 |
| RAF/MAP kinase cascade | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.54735 |
| RAF/MAP kinase cascade | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.364338 | 0.496957 | 4 | 0.612074 | 0.515416 |
| RAF/MAP kinase cascade | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.54735 |
| RAF/MAP kinase cascade | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.572294 |
| RAF/MAP kinase cascade | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.54735 |
| RAF/MAP kinase cascade | KIT | Mast/stem cell growth factor receptor, Mast/stem cell growth factor receptor Kit | 9.364338 | 0.496957 | 4 | 0.612074 | 0.562353 |
| RAF/MAP kinase cascade | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.58858 |
| RAF/MAP kinase cascade | GRIN2B | Glutamate receptor, Glutamate receptor ionotropic, NMDA 2B | 9.364338 | 0.496957 | 4 | 0.612074 | 0.56443 |
| RAF/MAP kinase cascade | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.57244 |
| RAF/MAP kinase cascade | ERBB2 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-2 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.589497 |
| RAF/MAP kinase cascade | TEK | Angiopoietin-1 receptor, Receptor protein-tyrosine kinase | 9.364338 | 0.496957 | 4 | 0.612074 | 0.631911 |
| RAF/MAP kinase cascade | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.54735 |
| RAF/MAP kinase cascade | FNTA | Farnesyl transferase alpha, isoform A, Farnesyltransferase, CAAX box, alpha, Protein farnesyltransferase/geranylgeranyltransferase type-1 subunit alpha, Uncharacterized protein | 9.364338 | 0.496957 | 4 | 0.612074 | 0.662978 |
| RAF/MAP kinase cascade | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 9.364338 | 0.496957 | 4 | 0.612074 | 0.594807 |
| RAF/MAP kinase cascade | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.515416 |
| RAF/MAP kinase cascade | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.546881 |
| RAF/MAP kinase cascade | RAP1A | RAP1A, member of RAS oncogene family, Ras-related protein Rap-1A, Uncharacterized protein | 9.364338 | 0.496957 | 4 | 0.612074 | 0.530823 |
| RAF/MAP kinase cascade | JAK2 | Janus kinase 2, Tyrosine-protein kinase, Tyrosine-protein kinase JAK2 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.573235 |
| RAF/MAP kinase cascade | SHC1 | SHC-transforming protein 1 | 9.364338 | 0.496957 | 4 | 0.612074 | 0.521858 |
| RAF/MAP kinase cascade | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.364338 | 0.496957 | 4 | 0.612074 | 0.660919 |
| Receptor Mediated Mitophagy | CSNK2A2 | Casein kinase 2 alpha 2, Casein kinase II subunit alpha' | 7.952133 | 0.782606 | 3.099592 | 0.528515 | 0.53622 |
| Receptor Mediated Mitophagy | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 7.952133 | 0.782606 | 3.099592 | 0.528515 | 0.594807 |
| Receptor Mediated Mitophagy | CSNK2B | Casein kinase II subunit beta | 7.952133 | 0.782606 | 3.099592 | 0.528515 | 0.537688 |
| Receptor Mediated Mitophagy | ULK1 | Non-specific serine/threonine protein kinase, Serine/threonine-protein kinase, Serine/threonine-protein kinase ULK1 | 7.952133 | 0.782606 | 3.099592 | 0.528515 | 0.511522 |
| Receptor Mediated Mitophagy | CSNK2A1 | Casein kinase 2, alpha 1 polypeptide, Casein kinase II subunit alpha, Protein kinase domain-containing protein | 7.952133 | 0.782606 | 3.099592 | 0.528515 | 0.538737 |
| Recruitment of NuMA to mitotic centrosomes | CDK1 | Cell division control protein 2, Cyclin dependent kinase 1, Cyclin-dependent kinase 1 | 8.654307 | 0.514565 | 3.993284 | 0.583268 | 0.514008 |
| Recruitment of NuMA to mitotic centrosomes | PLK1 | Serine/threonine-protein kinase PLK, Serine/threonine-protein kinase PLK1 | 8.654307 | 0.514565 | 3.993284 | 0.583268 | 0.504187 |
| Recruitment of NuMA to mitotic centrosomes | CRK | 14_3_3 domain-containing protein, Adapter molecule crk, Adapter molecule Crk, CRK proto-oncogene, adaptor protein, Uncharacterized protein, V-crk avian sarcoma virus CT10 oncogene homolog | 8.654307 | 0.514565 | 3.993284 | 0.583268 | 0.513369 |
| Recruitment of NuMA to mitotic centrosomes | HSP90AA1 | Heat shock protein 90 alpha family class A member 1, Heat shock protein HSP 90-alpha | 8.654307 | 0.514565 | 3.993284 | 0.583268 | 0.624325 |
| Recycling pathway of L1 | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 8.795657 | 0.499824 | 3.899775 | 0.569533 | 0.541254 |
| Recycling pathway of L1 | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 8.795657 | 0.499824 | 3.899775 | 0.569533 | 0.594807 |
| Regulated proteolysis of p75NTR | PSENEN | Gamma-secretase subunit PEN-2 | 8.233995 | 0.669199 | 3.444041 | 0.550945 | 0.60725 |
| Regulated proteolysis of p75NTR | NCSTN | Nicastrin | 8.233995 | 0.669199 | 3.444041 | 0.550945 | 0.60749 |
| Regulated proteolysis of p75NTR | PSEN2 | Presenilin, Presenilin-2 | 8.233995 | 0.669199 | 3.444041 | 0.550945 | 0.607393 |
| Regulated proteolysis of p75NTR | PSEN1 | Presenilin, Presenilin-1 | 8.233995 | 0.669199 | 3.444041 | 0.550945 | 0.611222 |
| Regulated proteolysis of p75NTR | APH1B | Aph-1 homolog B, gamma-secretase subunit, APH-1B', Gamma-secretase subunit APH-1B, Uncharacterized protein | 8.233995 | 0.669199 | 3.444041 | 0.550945 | 0.60749 |
| Regulated proteolysis of p75NTR | APH1A | Anterior pharynx defective 1 homolog A, Anterior pharynx defective 1a homolog, Aph-1 homolog A, gamma-secretase subunit, Gamma-secretase subunit APH-1A, Uncharacterized protein | 8.233995 | 0.669199 | 3.444041 | 0.550945 | 0.60749 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.577847 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.515416 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.57244 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.515416 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.515416 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.515416 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.515416 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.515416 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.54735 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.54735 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.54735 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.515416 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.515416 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.572294 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.547953 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.5197 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.54735 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.515416 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.515416 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.515905 | 0.798815 | 4 | 0.770823 | 0.568645 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.515905 | 0.798815 | 4 | 0.770823 | 0.515416 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.515416 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.54735 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.54735 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.58858 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.515905 | 0.798815 | 4 | 0.770823 | 0.579609 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.54735 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.565601 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.573417 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.577481 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.515905 | 0.798815 | 4 | 0.770823 | 0.515416 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.546881 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.515416 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.54735 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.515416 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.54735 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.515905 | 0.798815 | 4 | 0.770823 | 0.54735 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.515416 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.577847 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 7.755508 | 0.770159 | 4 | 0.665677 | 0.568645 |
| Regulation of APC/C activators between G1/S and early anaphase | CDK1 | Cell division control protein 2, Cyclin dependent kinase 1, Cyclin-dependent kinase 1 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.514008 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.5197 |
| Regulation of APC/C activators between G1/S and early anaphase | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 7.755508 | 0.770159 | 4 | 0.665677 | 0.579609 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 7.755508 | 0.770159 | 4 | 0.665677 | 0.515416 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.515416 |
| Regulation of APC/C activators between G1/S and early anaphase | CCNB1 | Cyclin B1, G2/mitotic-specific cyclin-B1, Uncharacterized protein | 7.755508 | 0.770159 | 4 | 0.665677 | 0.513205 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.515416 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.54735 |
| Regulation of APC/C activators between G1/S and early anaphase | CCNA1 | Cyclin A1, Cyclin-A1, Cyclin-A2 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.506777 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.54735 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.515416 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.54735 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.515416 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.515416 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.573417 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.515416 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.547953 |
| Regulation of APC/C activators between G1/S and early anaphase | PLK1 | Serine/threonine-protein kinase PLK, Serine/threonine-protein kinase PLK1 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.504187 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.54735 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 7.755508 | 0.770159 | 4 | 0.665677 | 0.515416 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.546881 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.54735 |
| Regulation of APC/C activators between G1/S and early anaphase | CDK2 | Cyclin dependent kinase 2, Cyclin-dependent kinase 2, Protein kinase domain-containing protein, Uncharacterized protein | 7.755508 | 0.770159 | 4 | 0.665677 | 0.532011 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.515416 |
| Regulation of APC/C activators between G1/S and early anaphase | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.515416 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.54735 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.54735 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.54735 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.577481 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.54735 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.54735 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.515416 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.515416 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.572294 |
| Regulation of APC/C activators between G1/S and early anaphase | CCNA2 | Cyclin-A2 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.527977 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.515416 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.58858 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.57244 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.565601 |
| Regulation of APC/C activators between G1/S and early anaphase | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 7.755508 | 0.770159 | 4 | 0.665677 | 0.515416 |
| Regulation of Apoptosis | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.54735 |
| Regulation of Apoptosis | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.54735 |
| Regulation of Apoptosis | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.5197 |
| Regulation of Apoptosis | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.515416 |
| Regulation of Apoptosis | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.54735 |
| Regulation of Apoptosis | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.577847 |
| Regulation of Apoptosis | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.515416 |
| Regulation of Apoptosis | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.54735 |
| Regulation of Apoptosis | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.54735 |
| Regulation of Apoptosis | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.547953 |
| Regulation of Apoptosis | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.54735 |
| Regulation of Apoptosis | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.515416 |
| Regulation of Apoptosis | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.515416 |
| Regulation of Apoptosis | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.54735 |
| Regulation of Apoptosis | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 4.723092 | 0.779411 | 4 | 0.513864 | 0.515416 |
| Regulation of Apoptosis | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.54735 |
| Regulation of Apoptosis | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.515416 |
| Regulation of Apoptosis | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 4.723092 | 0.779411 | 4 | 0.513864 | 0.515416 |
| Regulation of Apoptosis | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 4.723092 | 0.779411 | 4 | 0.513864 | 0.579609 |
| Regulation of Apoptosis | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.577481 |
| Regulation of Apoptosis | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.58858 |
| Regulation of Apoptosis | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.515416 |
| Regulation of Apoptosis | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.546881 |
| Regulation of Apoptosis | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.515416 |
| Regulation of Apoptosis | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.515416 |
| Regulation of Apoptosis | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.515416 |
| Regulation of Apoptosis | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.573417 |
| Regulation of Apoptosis | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.515416 |
| Regulation of Apoptosis | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.572294 |
| Regulation of Apoptosis | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.54735 |
| Regulation of Apoptosis | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 4.723092 | 0.779411 | 4 | 0.513864 | 0.568645 |
| Regulation of Apoptosis | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.515416 |
| Regulation of Apoptosis | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.515416 |
| Regulation of Apoptosis | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.565601 |
| Regulation of Apoptosis | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.57244 |
| Regulation of Apoptosis | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.515416 |
| Regulation of Apoptosis | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 4.723092 | 0.779411 | 4 | 0.513864 | 0.54735 |
| Regulation of expression of SLITs and ROBOs | RPL6 | 60S ribosomal protein L6 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 11.300142 | 0.825543 | 4 | 0.876234 | 0.515416 |
| Regulation of expression of SLITs and ROBOs | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.577481 |
| Regulation of expression of SLITs and ROBOs | RPS20 | 40S ribosomal protein S20 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL15 | 60S ribosomal protein L15, Ribosomal protein L15 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPS24 | 40S ribosomal protein S24 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 11.300142 | 0.825543 | 4 | 0.876234 | 0.515416 |
| Regulation of expression of SLITs and ROBOs | RPS3A | 40S ribosomal protein S3a | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL27 | 60S ribosomal protein L27 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL14 | 60S ribosomal protein L14 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL29 | 60S ribosomal protein L29 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL37 | 60S ribosomal protein L37, Ribosomal protein L37 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.54735 |
| Regulation of expression of SLITs and ROBOs | RPL28 | 60S ribosomal protein L28 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL22 | 60S ribosomal protein L22, 60S ribosomal protein L22 1, Ribosomal protein L22, Uncharacterized protein | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPS12 | 40S ribosomal protein S12 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.54735 |
| Regulation of expression of SLITs and ROBOs | RPL8 | 60S ribosomal protein L8 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.546881 |
| Regulation of expression of SLITs and ROBOs | RPL11 | 60S ribosomal protein L11 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPS10 | 40S ribosomal protein S10, Ribosomal protein S10, S10_plectin domain-containing protein | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.515416 |
| Regulation of expression of SLITs and ROBOs | RPL37A | 60S ribosomal protein L37-A, 60S ribosomal protein L37a, MGC89854 protein, Probable 60S ribosomal protein L37-A, Ribosomal protein L37a | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPSA | 40S ribosomal protein SA | 11.300142 | 0.825543 | 4 | 0.876234 | 0.521531 |
| Regulation of expression of SLITs and ROBOs | RPL17 | 60S ribosomal protein L17 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | FAU | 40S ribosomal protein S30 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL36 | 60S ribosomal protein L36 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL30 | 60S ribosomal protein L30 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPS14 | 40S ribosomal protein S14, Ribosomal protein S14, Uncharacterized protein | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.515416 |
| Regulation of expression of SLITs and ROBOs | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.565601 |
| Regulation of expression of SLITs and ROBOs | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.5197 |
| Regulation of expression of SLITs and ROBOs | RPL7A | 60S ribosomal protein L7-A, 60S ribosomal protein L7a | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL39 | 60S ribosomal protein L39, LOC100135132 protein, Ribosomal protein L39 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPS23 | 40S ribosomal protein S23, 40S ribosomal protein S23-A | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL13 | 60S ribosomal protein L13 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.515416 |
| Regulation of expression of SLITs and ROBOs | RPL38 | 60S ribosomal protein L38 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.54735 |
| Regulation of expression of SLITs and ROBOs | RPS15A | 40S ribosomal protein S15a | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPS15 | 40S ribosomal protein S15 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.572294 |
| Regulation of expression of SLITs and ROBOs | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.54735 |
| Regulation of expression of SLITs and ROBOs | RPL10 | 60S ribosomal protein L10, MGC89303 protein | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.54735 |
| Regulation of expression of SLITs and ROBOs | RPL5 | 60S ribosomal protein L5, Ribosomal protein L5 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.54735 |
| Regulation of expression of SLITs and ROBOs | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.515416 |
| Regulation of expression of SLITs and ROBOs | RPS11 | 40S ribosomal protein S11 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL21 | 60S ribosomal protein L21 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL23 | 60S ribosomal protein L23 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL18 | 60S ribosomal protein L18, Ribosomal protein L18 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.515416 |
| Regulation of expression of SLITs and ROBOs | RPL13A | 60S ribosomal protein L13a | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL36A | 60S ribosomal protein L36-A, 60S ribosomal protein L36a, IP17351p, ribosomal protein L36a, Uncharacterized protein | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPS26 | 40S ribosomal protein S26 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL35 | 60S ribosomal protein L35 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL35A | 60S ribosomal protein L35-A, 60S ribosomal protein L35a | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPS13 | 40S ribosomal protein S13 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPS7 | 40S ribosomal protein S7 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL12 | 60S ribosomal protein L12 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.515416 |
| Regulation of expression of SLITs and ROBOs | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.515416 |
| Regulation of expression of SLITs and ROBOs | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.515416 |
| Regulation of expression of SLITs and ROBOs | RPL19 | 60S ribosomal protein L19, Ribosomal protein L19 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL18A | 60S ribosomal protein L18-A, 60S ribosomal protein L18a | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.54735 |
| Regulation of expression of SLITs and ROBOs | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.547953 |
| Regulation of expression of SLITs and ROBOs | RPL10A | 60S ribosomal protein L10a, Ribosomal protein | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.515416 |
| Regulation of expression of SLITs and ROBOs | RPS5 | 40S ribosomal protein S5, 40S ribosomal protein S5 [Cleaved into: 40S ribosomal protein S5, N-terminally processed], 40S ribosomal protein S5-A, Ribosomal protein S5 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL34 | 60S ribosomal protein L34 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL3 | 60S ribosomal protein L3, LOC100145083 protein, Ribosomal protein L3, Uncharacterized protein | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL27A | 60S ribosomal protein L27-A, 60S ribosomal protein L27a | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPS21 | 40S ribosomal protein S21 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPS18 | 40S ribosomal protein S18 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPS27 | 40S ribosomal protein S27 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.549907 |
| Regulation of expression of SLITs and ROBOs | RPL7 | 60S ribosomal protein L7, MGC79754 protein, Ribosomal protein L7, Uncharacterized protein | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL32 | 60S ribosomal protein L32, Hypothetical LOC496894, Ribosomal protein L32, Uncharacterized protein | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL26 | 60S ribosomal protein L26, GEO07453p1, KOW domain-containing protein, Ribosomal protein L26 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.58858 |
| Regulation of expression of SLITs and ROBOs | RPS4Y1 | 40S ribosomal protein S4, 40S ribosomal protein S4, Y isoform 1 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.515416 |
| Regulation of expression of SLITs and ROBOs | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.577847 |
| Regulation of expression of SLITs and ROBOs | RPS9 | 40S ribosomal protein S9 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL41 | 60S ribosomal protein L41 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPS28 | 40S ribosomal protein S28 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPLP1 | 60S acidic ribosomal protein P1, Ribosomal protein, large, P1 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.573417 |
| Regulation of expression of SLITs and ROBOs | RPL4 | 60S ribosomal protein L4, Ribos_L4_asso_C domain-containing protein, Ribosomal protein L4 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.54735 |
| Regulation of expression of SLITs and ROBOs | RPS17 | 40S ribosomal protein S17 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.515416 |
| Regulation of expression of SLITs and ROBOs | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.515416 |
| Regulation of expression of SLITs and ROBOs | RPS8 | 40S ribosomal protein S8 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.515416 |
| Regulation of expression of SLITs and ROBOs | RPS16 | 40S ribosomal protein S16, Ribosomal protein S16, Uncharacterized protein | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPS19 | 40S ribosomal protein S19, Uncharacterized protein | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPS4X | 40S ribosomal protein S4, 40S ribosomal protein S4, X isoform | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPS3 | 40S ribosomal protein S3, DNA-(apurinic or apyrimidinic site) lyase | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPS6 | 40S ribosomal protein S6 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.566528 |
| Regulation of expression of SLITs and ROBOs | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 11.300142 | 0.825543 | 4 | 0.876234 | 0.568645 |
| Regulation of expression of SLITs and ROBOs | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.57244 |
| Regulation of expression of SLITs and ROBOs | RPS25 | 40S ribosomal protein S25 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL23A | 60S ribosomal protein L23-A, 60S ribosomal protein L23a, FI01658p, MGC89958 protein, Ribosomal protein L23a, Ribosomal_L23eN domain-containing protein | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.54735 |
| Regulation of expression of SLITs and ROBOs | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.54735 |
| Regulation of expression of SLITs and ROBOs | RPS29 | 40S ribosomal protein S29 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPS2 | 40S ribosomal protein S2 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPLP0 | 60S acidic ribosomal protein P0 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL24 | 60S ribosomal protein L24, Ribosomal protein L24, TRASH domain-containing protein | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of expression of SLITs and ROBOs | RPL31 | 60S ribosomal protein L31 | 11.300142 | 0.825543 | 4 | 0.876234 | 0.579609 |
| Regulation of MECP2 expression and activity | HDAC3 | Histone deacetylase, Histone deacetylase 3 | 7.991802 | 0.561731 | 3.082144 | 0.442021 | 0.528637 |
| Regulation of MECP2 expression and activity | HDAC1 | Histone deacetylase 1, Histone deacetylase HDAC1 | 7.991802 | 0.561731 | 3.082144 | 0.442021 | 0.540105 |
| Regulation of MECP2 expression and activity | AURKB | Aurora kinase, Aurora kinase B | 7.991802 | 0.561731 | 3.082144 | 0.442021 | 0.524412 |
| Regulation of MECP2 expression and activity | HDAC2 | Histone deacetylase 2 | 7.991802 | 0.561731 | 3.082144 | 0.442021 | 0.52963 |
| Regulation of ornithine decarboxylase (ODC) | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.515416 |
| Regulation of ornithine decarboxylase (ODC) | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.515416 |
| Regulation of ornithine decarboxylase (ODC) | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.57244 |
| Regulation of ornithine decarboxylase (ODC) | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.54735 |
| Regulation of ornithine decarboxylase (ODC) | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.262359 | 0.782192 | 4 | 0.749431 | 0.568645 |
| Regulation of ornithine decarboxylase (ODC) | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.54735 |
| Regulation of ornithine decarboxylase (ODC) | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.515416 |
| Regulation of ornithine decarboxylase (ODC) | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.58858 |
| Regulation of ornithine decarboxylase (ODC) | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.262359 | 0.782192 | 4 | 0.749431 | 0.515416 |
| Regulation of ornithine decarboxylase (ODC) | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.54735 |
| Regulation of ornithine decarboxylase (ODC) | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.515416 |
| Regulation of ornithine decarboxylase (ODC) | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.546881 |
| Regulation of ornithine decarboxylase (ODC) | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.515416 |
| Regulation of ornithine decarboxylase (ODC) | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.5197 |
| Regulation of ornithine decarboxylase (ODC) | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.54735 |
| Regulation of ornithine decarboxylase (ODC) | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.515416 |
| Regulation of ornithine decarboxylase (ODC) | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.262359 | 0.782192 | 4 | 0.749431 | 0.515416 |
| Regulation of ornithine decarboxylase (ODC) | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.54735 |
| Regulation of ornithine decarboxylase (ODC) | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.515416 |
| Regulation of ornithine decarboxylase (ODC) | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.54735 |
| Regulation of ornithine decarboxylase (ODC) | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.54735 |
| Regulation of ornithine decarboxylase (ODC) | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.515416 |
| Regulation of ornithine decarboxylase (ODC) | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.565601 |
| Regulation of ornithine decarboxylase (ODC) | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.515416 |
| Regulation of ornithine decarboxylase (ODC) | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.577481 |
| Regulation of ornithine decarboxylase (ODC) | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.54735 |
| Regulation of ornithine decarboxylase (ODC) | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.515416 |
| Regulation of ornithine decarboxylase (ODC) | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.573417 |
| Regulation of ornithine decarboxylase (ODC) | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.515416 |
| Regulation of ornithine decarboxylase (ODC) | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.54735 |
| Regulation of ornithine decarboxylase (ODC) | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.515416 |
| Regulation of ornithine decarboxylase (ODC) | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.572294 |
| Regulation of ornithine decarboxylase (ODC) | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.515416 |
| Regulation of ornithine decarboxylase (ODC) | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.547953 |
| Regulation of ornithine decarboxylase (ODC) | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.577847 |
| Regulation of ornithine decarboxylase (ODC) | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.262359 | 0.782192 | 4 | 0.749431 | 0.54735 |
| Regulation of PTEN stability and activity | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.515416 |
| Regulation of PTEN stability and activity | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.589663 |
| Regulation of PTEN stability and activity | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.515416 |
| Regulation of PTEN stability and activity | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.515416 |
| Regulation of PTEN stability and activity | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.515416 |
| Regulation of PTEN stability and activity | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.54735 |
| Regulation of PTEN stability and activity | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.515416 |
| Regulation of PTEN stability and activity | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.614901 | 0.743819 | 4 | 0.748432 | 0.515416 |
| Regulation of PTEN stability and activity | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.515416 |
| Regulation of PTEN stability and activity | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 9.614901 | 0.743819 | 4 | 0.748432 | 0.552041 |
| Regulation of PTEN stability and activity | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.54735 |
| Regulation of PTEN stability and activity | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.54735 |
| Regulation of PTEN stability and activity | XIAP | E3 ubiquitin-protein ligase XIAP, RING-type domain-containing protein, X-linked inhibitor of apoptosis | 9.614901 | 0.743819 | 4 | 0.748432 | 0.589894 |
| Regulation of PTEN stability and activity | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.577847 |
| Regulation of PTEN stability and activity | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.572294 |
| Regulation of PTEN stability and activity | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.614901 | 0.743819 | 4 | 0.748432 | 0.568645 |
| Regulation of PTEN stability and activity | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.5197 |
| Regulation of PTEN stability and activity | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.614901 | 0.743819 | 4 | 0.748432 | 0.579609 |
| Regulation of PTEN stability and activity | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.515416 |
| Regulation of PTEN stability and activity | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.58858 |
| Regulation of PTEN stability and activity | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.515416 |
| Regulation of PTEN stability and activity | USP13 | Ubiquitin carboxyl-terminal hydrolase, Ubiquitin carboxyl-terminal hydrolase 13 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.538123 |
| Regulation of PTEN stability and activity | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.57244 |
| Regulation of PTEN stability and activity | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.54735 |
| Regulation of PTEN stability and activity | CSNK2A2 | Casein kinase 2 alpha 2, Casein kinase II subunit alpha' | 9.614901 | 0.743819 | 4 | 0.748432 | 0.53622 |
| Regulation of PTEN stability and activity | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.614901 | 0.743819 | 4 | 0.748432 | 0.515416 |
| Regulation of PTEN stability and activity | TNKS | Poly [ADP-ribose] polymerase, Poly [ADP-ribose] polymerase tankyrase, Poly [ADP-ribose] polymerase tankyrase-1 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.506213 |
| Regulation of PTEN stability and activity | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.54735 |
| Regulation of PTEN stability and activity | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.547953 |
| Regulation of PTEN stability and activity | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.54735 |
| Regulation of PTEN stability and activity | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.515416 |
| Regulation of PTEN stability and activity | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.573417 |
| Regulation of PTEN stability and activity | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.577481 |
| Regulation of PTEN stability and activity | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.515416 |
| Regulation of PTEN stability and activity | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.515416 |
| Regulation of PTEN stability and activity | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.54735 |
| Regulation of PTEN stability and activity | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.54735 |
| Regulation of PTEN stability and activity | CSNK2A1 | Casein kinase 2, alpha 1 polypeptide, Casein kinase II subunit alpha, Protein kinase domain-containing protein | 9.614901 | 0.743819 | 4 | 0.748432 | 0.538737 |
| Regulation of PTEN stability and activity | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.515416 |
| Regulation of PTEN stability and activity | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.515416 |
| Regulation of PTEN stability and activity | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.54735 |
| Regulation of PTEN stability and activity | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.565601 |
| Regulation of PTEN stability and activity | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.54735 |
| Regulation of PTEN stability and activity | CSNK2B | Casein kinase II subunit beta | 9.614901 | 0.743819 | 4 | 0.748432 | 0.537688 |
| Regulation of PTEN stability and activity | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.614901 | 0.743819 | 4 | 0.748432 | 0.546881 |
| Regulation of RAS by GAPs | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.515416 |
| Regulation of RAS by GAPs | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.572294 |
| Regulation of RAS by GAPs | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.546881 |
| Regulation of RAS by GAPs | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 9.824358 | 0.731662 | 4 | 0.753159 | 0.505988 |
| Regulation of RAS by GAPs | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.58858 |
| Regulation of RAS by GAPs | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.515416 |
| Regulation of RAS by GAPs | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.54735 |
| Regulation of RAS by GAPs | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.54735 |
| Regulation of RAS by GAPs | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.515416 |
| Regulation of RAS by GAPs | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.515416 |
| Regulation of RAS by GAPs | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.515416 |
| Regulation of RAS by GAPs | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.54735 |
| Regulation of RAS by GAPs | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.515416 |
| Regulation of RAS by GAPs | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.577847 |
| Regulation of RAS by GAPs | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.54735 |
| Regulation of RAS by GAPs | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.565601 |
| Regulation of RAS by GAPs | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.824358 | 0.731662 | 4 | 0.753159 | 0.515416 |
| Regulation of RAS by GAPs | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.54735 |
| Regulation of RAS by GAPs | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.515416 |
| Regulation of RAS by GAPs | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.547953 |
| Regulation of RAS by GAPs | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.824358 | 0.731662 | 4 | 0.753159 | 0.579609 |
| Regulation of RAS by GAPs | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.515416 |
| Regulation of RAS by GAPs | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.573417 |
| Regulation of RAS by GAPs | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.515416 |
| Regulation of RAS by GAPs | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.515416 |
| Regulation of RAS by GAPs | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.515416 |
| Regulation of RAS by GAPs | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.5197 |
| Regulation of RAS by GAPs | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.54735 |
| Regulation of RAS by GAPs | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.54735 |
| Regulation of RAS by GAPs | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.577481 |
| Regulation of RAS by GAPs | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.824358 | 0.731662 | 4 | 0.753159 | 0.568645 |
| Regulation of RAS by GAPs | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.54735 |
| Regulation of RAS by GAPs | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.54735 |
| Regulation of RAS by GAPs | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.824358 | 0.731662 | 4 | 0.753159 | 0.515416 |
| Regulation of RAS by GAPs | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.515416 |
| Regulation of RAS by GAPs | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.515416 |
| Regulation of RAS by GAPs | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.57244 |
| Regulation of RAS by GAPs | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.824358 | 0.731662 | 4 | 0.753159 | 0.54735 |
| Regulation of RUNX2 expression and activity | ESR1 | Estrogen receptor | 9.955204 | 0.712092 | 4 | 0.750124 | 0.561602 |
| Regulation of RUNX2 expression and activity | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.54735 |
| Regulation of RUNX2 expression and activity | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.515416 |
| Regulation of RUNX2 expression and activity | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.955204 | 0.712092 | 4 | 0.750124 | 0.579609 |
| Regulation of RUNX2 expression and activity | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.58858 |
| Regulation of RUNX2 expression and activity | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.547953 |
| Regulation of RUNX2 expression and activity | NR3C1 | Glucocorticoid receptor | 9.955204 | 0.712092 | 4 | 0.750124 | 0.59851 |
| Regulation of RUNX2 expression and activity | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.955204 | 0.712092 | 4 | 0.750124 | 0.515416 |
| Regulation of RUNX2 expression and activity | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.515416 |
| Regulation of RUNX2 expression and activity | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.515416 |
| Regulation of RUNX2 expression and activity | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.572294 |
| Regulation of RUNX2 expression and activity | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.577847 |
| Regulation of RUNX2 expression and activity | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.54735 |
| Regulation of RUNX2 expression and activity | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.515416 |
| Regulation of RUNX2 expression and activity | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.565601 |
| Regulation of RUNX2 expression and activity | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.515416 |
| Regulation of RUNX2 expression and activity | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.955204 | 0.712092 | 4 | 0.750124 | 0.515416 |
| Regulation of RUNX2 expression and activity | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.54735 |
| Regulation of RUNX2 expression and activity | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.54735 |
| Regulation of RUNX2 expression and activity | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.546881 |
| Regulation of RUNX2 expression and activity | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.515416 |
| Regulation of RUNX2 expression and activity | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.577481 |
| Regulation of RUNX2 expression and activity | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.5197 |
| Regulation of RUNX2 expression and activity | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.54735 |
| Regulation of RUNX2 expression and activity | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.515416 |
| Regulation of RUNX2 expression and activity | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.955204 | 0.712092 | 4 | 0.750124 | 0.568645 |
| Regulation of RUNX2 expression and activity | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.515416 |
| Regulation of RUNX2 expression and activity | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.54735 |
| Regulation of RUNX2 expression and activity | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.54735 |
| Regulation of RUNX2 expression and activity | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.54735 |
| Regulation of RUNX2 expression and activity | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.573417 |
| Regulation of RUNX2 expression and activity | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.515416 |
| Regulation of RUNX2 expression and activity | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.515416 |
| Regulation of RUNX2 expression and activity | PPARGC1A | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | 9.955204 | 0.712092 | 4 | 0.750124 | 0.539093 |
| Regulation of RUNX2 expression and activity | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.54735 |
| Regulation of RUNX2 expression and activity | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.515416 |
| Regulation of RUNX2 expression and activity | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.57244 |
| Regulation of RUNX2 expression and activity | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.515416 |
| Regulation of RUNX2 expression and activity | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.54735 |
| Regulation of RUNX2 expression and activity | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.955204 | 0.712092 | 4 | 0.750124 | 0.515416 |
| Regulation of RUNX3 expression and activity | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.515416 |
| Regulation of RUNX3 expression and activity | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.54735 |
| Regulation of RUNX3 expression and activity | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.946907 | 0.767289 | 4 | 0.777295 | 0.568645 |
| Regulation of RUNX3 expression and activity | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.946907 | 0.767289 | 4 | 0.777295 | 0.515416 |
| Regulation of RUNX3 expression and activity | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.54735 |
| Regulation of RUNX3 expression and activity | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.54735 |
| Regulation of RUNX3 expression and activity | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.565601 |
| Regulation of RUNX3 expression and activity | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.54735 |
| Regulation of RUNX3 expression and activity | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.54735 |
| Regulation of RUNX3 expression and activity | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.515416 |
| Regulation of RUNX3 expression and activity | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.54735 |
| Regulation of RUNX3 expression and activity | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.572294 |
| Regulation of RUNX3 expression and activity | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.515416 |
| Regulation of RUNX3 expression and activity | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.58858 |
| Regulation of RUNX3 expression and activity | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.515416 |
| Regulation of RUNX3 expression and activity | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.515416 |
| Regulation of RUNX3 expression and activity | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 9.946907 | 0.767289 | 4 | 0.777295 | 0.594807 |
| Regulation of RUNX3 expression and activity | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.946907 | 0.767289 | 4 | 0.777295 | 0.579609 |
| Regulation of RUNX3 expression and activity | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.515416 |
| Regulation of RUNX3 expression and activity | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.54735 |
| Regulation of RUNX3 expression and activity | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.57244 |
| Regulation of RUNX3 expression and activity | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.547953 |
| Regulation of RUNX3 expression and activity | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.577481 |
| Regulation of RUNX3 expression and activity | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.515416 |
| Regulation of RUNX3 expression and activity | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.546881 |
| Regulation of RUNX3 expression and activity | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.515416 |
| Regulation of RUNX3 expression and activity | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.515416 |
| Regulation of RUNX3 expression and activity | MDM2 | E3 ubiquitin-protein ligase Mdm2 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.659771 |
| Regulation of RUNX3 expression and activity | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.946907 | 0.767289 | 4 | 0.777295 | 0.515416 |
| Regulation of RUNX3 expression and activity | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.515416 |
| Regulation of RUNX3 expression and activity | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.5197 |
| Regulation of RUNX3 expression and activity | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.573417 |
| Regulation of RUNX3 expression and activity | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.54735 |
| Regulation of RUNX3 expression and activity | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.515416 |
| Regulation of RUNX3 expression and activity | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.515416 |
| Regulation of RUNX3 expression and activity | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.515416 |
| Regulation of RUNX3 expression and activity | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.946907 | 0.767289 | 4 | 0.777295 | 0.577847 |
| Regulation of signaling by CBL | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 9.703907 | 0.643672 | 3.812912 | 0.670071 | 0.646924 |
| Regulation of signaling by CBL | YES1 | Tyrosine-protein kinase, Tyrosine-protein kinase yes, Tyrosine-protein kinase Yes | 9.703907 | 0.643672 | 3.812912 | 0.670071 | 0.506641 |
| Regulation of signaling by CBL | PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.703907 | 0.643672 | 3.812912 | 0.670071 | 0.589385 |
| Regulation of signaling by CBL | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.703907 | 0.643672 | 3.812912 | 0.670071 | 0.579609 |
| Regulation of signaling by CBL | PIK3CD | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.703907 | 0.643672 | 3.812912 | 0.670071 | 0.632077 |
| Regulation of signaling by CBL | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.703907 | 0.643672 | 3.812912 | 0.670071 | 0.660919 |
| Regulation of signaling by CBL | SYK | Tyrosine-protein kinase, Tyrosine-protein kinase SYK | 9.703907 | 0.643672 | 3.812912 | 0.670071 | 0.557587 |
| Regulation of signaling by CBL | HCK | Tyrosine-protein kinase, Tyrosine-protein kinase HCK | 9.703907 | 0.643672 | 3.812912 | 0.670071 | 0.515497 |
| Regulation of signaling by CBL | CRK | 14_3_3 domain-containing protein, Adapter molecule crk, Adapter molecule Crk, CRK proto-oncogene, adaptor protein, Uncharacterized protein, V-crk avian sarcoma virus CT10 oncogene homolog | 9.703907 | 0.643672 | 3.812912 | 0.670071 | 0.513369 |
| Regulation of TP53 Activity through Phosphorylation | ATR | Non-specific serine/threonine protein kinase, Serine/threonine-protein kinase ATR | 8.731508 | 0.51859 | 4 | 0.590244 | 0.518847 |
| Regulation of TP53 Activity through Phosphorylation | AURKA | Aurora kinase, Aurora kinase A | 8.731508 | 0.51859 | 4 | 0.590244 | 0.54457 |
| Regulation of TP53 Activity through Phosphorylation | MAPK11 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 11 | 8.731508 | 0.51859 | 4 | 0.590244 | 0.567539 |
| Regulation of TP53 Activity through Phosphorylation | CSNK2A1 | Casein kinase 2, alpha 1 polypeptide, Casein kinase II subunit alpha, Protein kinase domain-containing protein | 8.731508 | 0.51859 | 4 | 0.590244 | 0.538737 |
| Regulation of TP53 Activity through Phosphorylation | BLM | Bloom syndrome protein, Bloom syndrome protein homolog, DNA helicase | 8.731508 | 0.51859 | 4 | 0.590244 | 0.509426 |
| Regulation of TP53 Activity through Phosphorylation | PRKAG1 | 5'-AMP-activated protein kinase subunit gamma-1, Protein kinase, AMP-activated, gamma 1 non-catalytic subunit | 8.731508 | 0.51859 | 4 | 0.590244 | 0.515113 |
| Regulation of TP53 Activity through Phosphorylation | CDK2 | Cyclin dependent kinase 2, Cyclin-dependent kinase 2, Protein kinase domain-containing protein, Uncharacterized protein | 8.731508 | 0.51859 | 4 | 0.590244 | 0.532011 |
| Regulation of TP53 Activity through Phosphorylation | CCNA1 | Cyclin A1, Cyclin-A1, Cyclin-A2 | 8.731508 | 0.51859 | 4 | 0.590244 | 0.506777 |
| Regulation of TP53 Activity through Phosphorylation | HIPK1 | Homeodomain interacting protein kinase 1, Homeodomain-interacting protein kinase 1 | 8.731508 | 0.51859 | 4 | 0.590244 | 0.512719 |
| Regulation of TP53 Activity through Phosphorylation | MDM2 | E3 ubiquitin-protein ligase Mdm2 | 8.731508 | 0.51859 | 4 | 0.590244 | 0.659771 |
| Regulation of TP53 Activity through Phosphorylation | CSNK2A2 | Casein kinase 2 alpha 2, Casein kinase II subunit alpha' | 8.731508 | 0.51859 | 4 | 0.590244 | 0.53622 |
| Regulation of TP53 Activity through Phosphorylation | ATRIP | ATR interacting protein, ATR-interacting protein, Uncharacterized protein | 8.731508 | 0.51859 | 4 | 0.590244 | 0.54377 |
| Regulation of TP53 Activity through Phosphorylation | PRKAG2 | 5'-AMP-activated protein kinase subunit gamma-2, 5'-AMP-activated protein kinase subunit gamma-2 isoform X8, Protein kinase AMP-activated non-catalytic subunit gamma 2, Protein kinase, AMP-activated, gamma 2 non-catalytic subunit, Uncharacterized protein | 8.731508 | 0.51859 | 4 | 0.590244 | 0.510193 |
| Regulation of TP53 Activity through Phosphorylation | MAPK14 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 14 | 8.731508 | 0.51859 | 4 | 0.590244 | 0.591941 |
| Regulation of TP53 Activity through Phosphorylation | AURKB | Aurora kinase, Aurora kinase B | 8.731508 | 0.51859 | 4 | 0.590244 | 0.524412 |
| Regulation of TP53 Activity through Phosphorylation | CSNK2B | Casein kinase II subunit beta | 8.731508 | 0.51859 | 4 | 0.590244 | 0.537688 |
| Regulation of TP53 Activity through Phosphorylation | PRKAB1 | 5'-AMP-activated protein kinase subunit beta-1, 5'AMP-activated protein kinase beta-1 non-catalytic subunit, Protein kinase AMP-activated non-catalytic subunit beta 1, Protein kinase, AMP-activated, beta 1 non-catalytic subunit | 8.731508 | 0.51859 | 4 | 0.590244 | 0.500481 |
| Regulation of TP53 Activity through Phosphorylation | TP53 | Cellular tumor antigen p53 | 8.731508 | 0.51859 | 4 | 0.590244 | 0.525937 |
| Regulation of TP53 Activity through Phosphorylation | TP53RK | EKC/KEOPS complex subunit Tp53rk, EKC/KEOPS complex subunit TP53RK, Non-specific serine/threonine protein kinase | 8.731508 | 0.51859 | 4 | 0.590244 | 0.504969 |
| Regulation of TP53 Activity through Phosphorylation | PDE10A | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A, Histone acetyltransferase, Phosphodiesterase | 8.731508 | 0.51859 | 4 | 0.590244 | 0.660672 |
| Regulation of TP53 Activity through Phosphorylation | CCNA2 | Cyclin-A2 | 8.731508 | 0.51859 | 4 | 0.590244 | 0.527977 |
| Regulation of TP53 Activity through Phosphorylation | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 8.731508 | 0.51859 | 4 | 0.590244 | 0.579609 |
| Regulation of TP53 Activity through Phosphorylation | STK11 | Non-specific serine/threonine protein kinase, Serine/threonine-protein kinase STK11 | 8.731508 | 0.51859 | 4 | 0.590244 | 0.504599 |
| Regulation of TP53 Activity through Phosphorylation | MDM4 | Double minute 4 protein, Protein Mdm4 | 8.731508 | 0.51859 | 4 | 0.590244 | 0.510741 |
| Regulation of TP53 Degradation | MTOR | Nucleoprotein TPR, Serine/threonine-protein kinase mTOR, Serine/threonine-protein kinase TOR | 8.814101 | 0.557317 | 3.508277 | 0.538405 | 0.619026 |
| Regulation of TP53 Degradation | MDM4 | Double minute 4 protein, Protein Mdm4 | 8.814101 | 0.557317 | 3.508277 | 0.538405 | 0.510741 |
| Regulation of TP53 Degradation | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 8.814101 | 0.557317 | 3.508277 | 0.538405 | 0.552041 |
| Regulation of TP53 Degradation | CDK2 | Cyclin dependent kinase 2, Cyclin-dependent kinase 2, Protein kinase domain-containing protein, Uncharacterized protein | 8.814101 | 0.557317 | 3.508277 | 0.538405 | 0.532011 |
| Regulation of TP53 Degradation | CCNA1 | Cyclin A1, Cyclin-A1, Cyclin-A2 | 8.814101 | 0.557317 | 3.508277 | 0.538405 | 0.506777 |
| Regulation of TP53 Degradation | CDK1 | Cell division control protein 2, Cyclin dependent kinase 1, Cyclin-dependent kinase 1 | 8.814101 | 0.557317 | 3.508277 | 0.538405 | 0.514008 |
| Regulation of TP53 Degradation | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 8.814101 | 0.557317 | 3.508277 | 0.538405 | 0.589663 |
| Regulation of TP53 Degradation | TP53 | Cellular tumor antigen p53 | 8.814101 | 0.557317 | 3.508277 | 0.538405 | 0.525937 |
| Regulation of TP53 Degradation | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 8.814101 | 0.557317 | 3.508277 | 0.538405 | 0.579609 |
| Regulation of TP53 Degradation | CCNA2 | Cyclin-A2 | 8.814101 | 0.557317 | 3.508277 | 0.538405 | 0.527977 |
| Regulation of TP53 Degradation | MDM2 | E3 ubiquitin-protein ligase Mdm2 | 8.814101 | 0.557317 | 3.508277 | 0.538405 | 0.659771 |
| Release | PB1 | Protein PB1-F2, RNA-directed RNA polymerase catalytic subunit | 7.405237 | 0.528279 | 0.508942 | ||
| Removal of aminoterminal propeptides from gamma-carboxylated proteins | F7 | Coagulation factor VII | 8.562762 | 0.751371 | 2.676759 | 0.467047 | 0.519515 |
| Removal of aminoterminal propeptides from gamma-carboxylated proteins | F9 | Christmas factor, Coagulation factor IX | 8.562762 | 0.751371 | 2.676759 | 0.467047 | 0.632191 |
| Removal of aminoterminal propeptides from gamma-carboxylated proteins | FURIN | Furin, Furin, paired basic amino acid cleaving enzyme, Trans Golgi network protease furin | 8.562762 | 0.751371 | 2.676759 | 0.467047 | 0.715659 |
| Removal of aminoterminal propeptides from gamma-carboxylated proteins | F2 | Activation peptide fragment 1, Prothrombin | 8.562762 | 0.751371 | 2.676759 | 0.467047 | 0.5402 |
| Removal of aminoterminal propeptides from gamma-carboxylated proteins | F10 | Coagulation factor VII, Coagulation factor X, MGC107878 protein, Uncharacterized protein | 8.562762 | 0.751371 | 2.676759 | 0.467047 | 0.588553 |
| Resolution of Sister Chromatid Cohesion | RPS27 | 40S ribosomal protein S27 | 9.08431 | 0.530281 | 4 | 0.61429 | 0.549907 |
| Resolution of Sister Chromatid Cohesion | CENPE | Centromere protein E, Centromere-associated protein E, Kinesin motor domain-containing protein | 9.08431 | 0.530281 | 4 | 0.61429 | 0.564477 |
| Resolution of Sister Chromatid Cohesion | CCNB1 | Cyclin B1, G2/mitotic-specific cyclin-B1, Uncharacterized protein | 9.08431 | 0.530281 | 4 | 0.61429 | 0.513205 |
| Resolution of Sister Chromatid Cohesion | CDK1 | Cell division control protein 2, Cyclin dependent kinase 1, Cyclin-dependent kinase 1 | 9.08431 | 0.530281 | 4 | 0.61429 | 0.514008 |
| Resolution of Sister Chromatid Cohesion | KIF2C | Kinesin-like protein, Kinesin-like protein KIF2C | 9.08431 | 0.530281 | 4 | 0.61429 | 0.611637 |
| Resolution of Sister Chromatid Cohesion | HDAC8 | Histone deacetylase, Histone deacetylase 8, WD repeat and SOCS box containing 2 | 9.08431 | 0.530281 | 4 | 0.61429 | 0.535733 |
| Resolution of Sister Chromatid Cohesion | PPP2R5A | Serine/threonine protein phosphatase 2A regulatory subunit, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform | 9.08431 | 0.530281 | 4 | 0.61429 | 0.559184 |
| Resolution of Sister Chromatid Cohesion | AURKB | Aurora kinase, Aurora kinase B | 9.08431 | 0.530281 | 4 | 0.61429 | 0.524412 |
| Resolution of Sister Chromatid Cohesion | PLK1 | Serine/threonine-protein kinase PLK, Serine/threonine-protein kinase PLK1 | 9.08431 | 0.530281 | 4 | 0.61429 | 0.504187 |
| Resolution of Sister Chromatid Cohesion | PPP1CC | Serine/threonine-protein phosphatase, Serine/threonine-protein phosphatase PP1-gamma catalytic subunit | 9.08431 | 0.530281 | 4 | 0.61429 | 0.503303 |
| Resolution of Sister Chromatid Cohesion | INCENP | Inner centromere protein, Inner centromere protein, isoform A, Uncharacterized protein | 9.08431 | 0.530281 | 4 | 0.61429 | 0.527883 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL22 | 60S ribosomal protein L22, 60S ribosomal protein L22 1, Ribosomal protein L22, Uncharacterized protein | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL13A | 60S ribosomal protein L13a | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL18 | 60S ribosomal protein L18, Ribosomal protein L18 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS14 | 40S ribosomal protein S14, Ribosomal protein S14, Uncharacterized protein | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS10 | 40S ribosomal protein S10, Ribosomal protein S10, S10_plectin domain-containing protein | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL31 | 60S ribosomal protein L31 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL29 | 60S ribosomal protein L29 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPLP0 | 60S acidic ribosomal protein P0 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL32 | 60S ribosomal protein L32, Hypothetical LOC496894, Ribosomal protein L32, Uncharacterized protein | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL7A | 60S ribosomal protein L7-A, 60S ribosomal protein L7a | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS12 | 40S ribosomal protein S12 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL41 | 60S ribosomal protein L41 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS25 | 40S ribosomal protein S25 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL14 | 60S ribosomal protein L14 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL37 | 60S ribosomal protein L37, Ribosomal protein L37 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL26 | 60S ribosomal protein L26, GEO07453p1, KOW domain-containing protein, Ribosomal protein L26 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS13 | 40S ribosomal protein S13 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL23A | 60S ribosomal protein L23-A, 60S ribosomal protein L23a, FI01658p, MGC89958 protein, Ribosomal protein L23a, Ribosomal_L23eN domain-containing protein | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL8 | 60S ribosomal protein L8 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS27 | 40S ribosomal protein S27 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.549907 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL11 | 60S ribosomal protein L11 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL34 | 60S ribosomal protein L34 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS29 | 40S ribosomal protein S29 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS11 | 40S ribosomal protein S11 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS4X | 40S ribosomal protein S4, 40S ribosomal protein S4, X isoform | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL3 | 60S ribosomal protein L3, LOC100145083 protein, Ribosomal protein L3, Uncharacterized protein | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL24 | 60S ribosomal protein L24, Ribosomal protein L24, TRASH domain-containing protein | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS24 | 40S ribosomal protein S24 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS20 | 40S ribosomal protein S20 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS16 | 40S ribosomal protein S16, Ribosomal protein S16, Uncharacterized protein | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS6 | 40S ribosomal protein S6 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.566528 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL27 | 60S ribosomal protein L27 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL17 | 60S ribosomal protein L17 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS3 | 40S ribosomal protein S3, DNA-(apurinic or apyrimidinic site) lyase | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS23 | 40S ribosomal protein S23, 40S ribosomal protein S23-A | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL5 | 60S ribosomal protein L5, Ribosomal protein L5 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL21 | 60S ribosomal protein L21 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL7 | 60S ribosomal protein L7, MGC79754 protein, Ribosomal protein L7, Uncharacterized protein | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL28 | 60S ribosomal protein L28 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL35 | 60S ribosomal protein L35 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL6 | 60S ribosomal protein L6 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS15A | 40S ribosomal protein S15a | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL30 | 60S ribosomal protein L30 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL15 | 60S ribosomal protein L15, Ribosomal protein L15 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS21 | 40S ribosomal protein S21 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS28 | 40S ribosomal protein S28 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL36A | 60S ribosomal protein L36-A, 60S ribosomal protein L36a, IP17351p, ribosomal protein L36a, Uncharacterized protein | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS3A | 40S ribosomal protein S3a | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS26 | 40S ribosomal protein S26 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS15 | 40S ribosomal protein S15 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL10A | 60S ribosomal protein L10a, Ribosomal protein | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS9 | 40S ribosomal protein S9 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS2 | 40S ribosomal protein S2 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL23 | 60S ribosomal protein L23 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL35A | 60S ribosomal protein L35-A, 60S ribosomal protein L35a | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL12 | 60S ribosomal protein L12 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS5 | 40S ribosomal protein S5, 40S ribosomal protein S5 [Cleaved into: 40S ribosomal protein S5, N-terminally processed], 40S ribosomal protein S5-A, Ribosomal protein S5 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL18A | 60S ribosomal protein L18-A, 60S ribosomal protein L18a | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPLP1 | 60S acidic ribosomal protein P1, Ribosomal protein, large, P1 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL4 | 60S ribosomal protein L4, Ribos_L4_asso_C domain-containing protein, Ribosomal protein L4 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL27A | 60S ribosomal protein L27-A, 60S ribosomal protein L27a | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS4Y1 | 40S ribosomal protein S4, 40S ribosomal protein S4, Y isoform 1 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS18 | 40S ribosomal protein S18 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS19 | 40S ribosomal protein S19, Uncharacterized protein | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL37A | 60S ribosomal protein L37-A, 60S ribosomal protein L37a, MGC89854 protein, Probable 60S ribosomal protein L37-A, Ribosomal protein L37a | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPSA | 40S ribosomal protein SA | 11.531418 | 0.884186 | 4 | 0.917487 | 0.521531 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL39 | 60S ribosomal protein L39, LOC100135132 protein, Ribosomal protein L39 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS7 | 40S ribosomal protein S7 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL13 | 60S ribosomal protein L13 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS8 | 40S ribosomal protein S8 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPS17 | 40S ribosomal protein S17 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL36 | 60S ribosomal protein L36 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL38 | 60S ribosomal protein L38 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL19 | 60S ribosomal protein L19, Ribosomal protein L19 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | FAU | 40S ribosomal protein S30 | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | RPL10 | 60S ribosomal protein L10, MGC89303 protein | 11.531418 | 0.884186 | 4 | 0.917487 | 0.579609 |
| RET signaling | PRKCA | Protein kinase C, Protein kinase C alpha type, Protein kinase C, alpha | 9.242127 | 0.523266 | 2.848725 | 0.440786 | 0.52202 |
| RET signaling | RET | Proto-oncogene tyrosine-protein kinase receptor Ret | 9.242127 | 0.523266 | 2.848725 | 0.440786 | 0.515319 |
| RET signaling | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.242127 | 0.523266 | 2.848725 | 0.440786 | 0.660919 |
| RET signaling | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 9.242127 | 0.523266 | 2.848725 | 0.440786 | 0.594807 |
| RET signaling | PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.242127 | 0.523266 | 2.848725 | 0.440786 | 0.589385 |
| RET signaling | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 9.242127 | 0.523266 | 2.848725 | 0.440786 | 0.646924 |
| RET signaling | SHC1 | SHC-transforming protein 1 | 9.242127 | 0.523266 | 2.848725 | 0.440786 | 0.521858 |
| RET signaling | P2RY1 | P2Y purinoceptor 1 | 9.242127 | 0.523266 | 2.848725 | 0.440786 | 0.550301 |
| RET signaling | PIK3CD | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.242127 | 0.523266 | 2.848725 | 0.440786 | 0.632077 |
| RHO GTPases activate IQGAPs | CTNNB1 | Catenin, Catenin beta-1, Uncharacterized protein | 8.886429 | 0.534318 | 3.266166 | 0.494906 | 0.657916 |
| RHO GTPases activate IQGAPs | MEN1 | Actin like 6A, Menin | 8.886429 | 0.534318 | 3.266166 | 0.494906 | 0.517021 |
| Ribosomal scanning and start codon recognition | RPS15A | 40S ribosomal protein S15a | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS19 | 40S ribosomal protein S19, Uncharacterized protein | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS28 | 40S ribosomal protein S28 | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS24 | 40S ribosomal protein S24 | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS16 | 40S ribosomal protein S16, Ribosomal protein S16, Uncharacterized protein | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS9 | 40S ribosomal protein S9 | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS11 | 40S ribosomal protein S11 | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS4X | 40S ribosomal protein S4, 40S ribosomal protein S4, X isoform | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS7 | 40S ribosomal protein S7 | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | FAU | 40S ribosomal protein S30 | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS13 | 40S ribosomal protein S13 | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS21 | 40S ribosomal protein S21 | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS3A | 40S ribosomal protein S3a | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS27 | 40S ribosomal protein S27 | 10.454788 | 0.799662 | 4 | 0.819683 | 0.549907 |
| Ribosomal scanning and start codon recognition | RPS2 | 40S ribosomal protein S2 | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS10 | 40S ribosomal protein S10, Ribosomal protein S10, S10_plectin domain-containing protein | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS3 | 40S ribosomal protein S3, DNA-(apurinic or apyrimidinic site) lyase | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS6 | 40S ribosomal protein S6 | 10.454788 | 0.799662 | 4 | 0.819683 | 0.566528 |
| Ribosomal scanning and start codon recognition | RPS12 | 40S ribosomal protein S12 | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS25 | 40S ribosomal protein S25 | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS29 | 40S ribosomal protein S29 | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS17 | 40S ribosomal protein S17 | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS15 | 40S ribosomal protein S15 | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS4Y1 | 40S ribosomal protein S4, 40S ribosomal protein S4, Y isoform 1 | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS20 | 40S ribosomal protein S20 | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS14 | 40S ribosomal protein S14, Ribosomal protein S14, Uncharacterized protein | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS8 | 40S ribosomal protein S8 | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS26 | 40S ribosomal protein S26 | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS18 | 40S ribosomal protein S18 | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPSA | 40S ribosomal protein SA | 10.454788 | 0.799662 | 4 | 0.819683 | 0.521531 |
| Ribosomal scanning and start codon recognition | RPS5 | 40S ribosomal protein S5, 40S ribosomal protein S5 [Cleaved into: 40S ribosomal protein S5, N-terminally processed], 40S ribosomal protein S5-A, Ribosomal protein S5 | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Ribosomal scanning and start codon recognition | RPS23 | 40S ribosomal protein S23, 40S ribosomal protein S23-A | 10.454788 | 0.799662 | 4 | 0.819683 | 0.579609 |
| Role of LAT2/NTAL/LAB on calcium mobilization | SHC | DShc protein | 9.503864 | 0.625154 | 2.856471 | 0.488007 | 0.605 |
| Role of LAT2/NTAL/LAB on calcium mobilization | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 9.503864 | 0.625154 | 2.856471 | 0.488007 | 0.646924 |
| Role of LAT2/NTAL/LAB on calcium mobilization | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.503864 | 0.625154 | 2.856471 | 0.488007 | 0.660919 |
| Role of LAT2/NTAL/LAB on calcium mobilization | SHC1 | SHC-transforming protein 1 | 9.503864 | 0.625154 | 2.856471 | 0.488007 | 0.521858 |
| Role of LAT2/NTAL/LAB on calcium mobilization | PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.503864 | 0.625154 | 2.856471 | 0.488007 | 0.589385 |
| Role of LAT2/NTAL/LAB on calcium mobilization | SYK | Tyrosine-protein kinase, Tyrosine-protein kinase SYK | 9.503864 | 0.625154 | 2.856471 | 0.488007 | 0.557587 |
| rRNA modification in the nucleus and cytosol | RPS14 | 40S ribosomal protein S14, Ribosomal protein S14, Uncharacterized protein | 10.600973 | 0.867526 | 4 | 0.861156 | 0.579609 |
| rRNA modification in the nucleus and cytosol | RPS2 | 40S ribosomal protein S2 | 10.600973 | 0.867526 | 4 | 0.861156 | 0.579609 |
| rRNA modification in the nucleus and cytosol | RPS7 | 40S ribosomal protein S7 | 10.600973 | 0.867526 | 4 | 0.861156 | 0.579609 |
| rRNA modification in the nucleus and cytosol | RPS9 | 40S ribosomal protein S9 | 10.600973 | 0.867526 | 4 | 0.861156 | 0.579609 |
| rRNA modification in the nucleus and cytosol | RPS6 | 40S ribosomal protein S6 | 10.600973 | 0.867526 | 4 | 0.861156 | 0.566528 |
| rRNA modification in the nucleus and cytosol | NAT10 | Chaperonin containing TCP1 subunit 3, RNA cytidine acetyltransferase | 10.600973 | 0.867526 | 4 | 0.861156 | 0.524821 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.824931 | 0.69856 | 4 | 0.736638 | 0.568645 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.54735 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.547953 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | ABL1 | Tyrosine-protein kinase, Tyrosine-protein kinase ABL1 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.590046 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.577481 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.515416 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.515416 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.515416 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.546881 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.573417 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.54735 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.515416 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.515416 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.565601 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.515416 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.515416 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | KMT2A | Histone-lysine N-methyltransferase, Histone-lysine N-methyltransferase 2A | 9.824931 | 0.69856 | 4 | 0.736638 | 0.518598 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.54735 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.824931 | 0.69856 | 4 | 0.736638 | 0.579609 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.577847 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.54735 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.572294 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.54735 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.824931 | 0.69856 | 4 | 0.736638 | 0.515416 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.54735 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.57244 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.515416 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.54735 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | YAP1 | Transcriptional coactivator YAP1, Yes1 associated transcriptional regulator | 9.824931 | 0.69856 | 4 | 0.736638 | 0.628833 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.824931 | 0.69856 | 4 | 0.736638 | 0.515416 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.5197 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.54735 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.54735 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.515416 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.58858 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.515416 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.515416 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.515416 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.515416 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.824931 | 0.69856 | 4 | 0.736638 | 0.54735 |
| RUNX3 regulates YAP1-mediated transcription | YAP1 | Transcriptional coactivator YAP1, Yes1 associated transcriptional regulator | 10.482821 | 0.986849 | 3.460322 | 0.791309 | 0.628833 |
| RUNX3 regulates YAP1-mediated transcription | TEAD1 | TEA domain transcription factor 1, TEA domain-containing protein, Transcriptional enhancer factor TEF-1 | 10.482821 | 0.986849 | 3.460322 | 0.791309 | 0.632738 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.515416 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.515416 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.565601 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.546881 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.54735 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.515416 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.54735 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.515416 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.54735 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.58858 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.572294 |
| SCF-beta-TrCP mediated degradation of Emi1 | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.527109 | 0.806612 | 4 | 0.775299 | 0.579609 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.527109 | 0.806612 | 4 | 0.775299 | 0.568645 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.515416 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.515416 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.54735 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.54735 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.54735 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.515416 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.54735 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.527109 | 0.806612 | 4 | 0.775299 | 0.515416 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.54735 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.515416 |
| SCF-beta-TrCP mediated degradation of Emi1 | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.515416 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.527109 | 0.806612 | 4 | 0.775299 | 0.515416 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.5197 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.515416 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.515416 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.547953 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.515416 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.577847 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.515416 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.54735 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.57244 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.573417 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.577481 |
| SCF-beta-TrCP mediated degradation of Emi1 | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.527109 | 0.806612 | 4 | 0.775299 | 0.54735 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.515416 |
| SCF(Skp2)-mediated degradation of p27/p21 | CCNA2 | Cyclin-A2 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.527977 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.389193 | 0.731182 | 4 | 0.730469 | 0.515416 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.515416 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.54735 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.515416 |
| SCF(Skp2)-mediated degradation of p27/p21 | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.515416 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.546881 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.515416 |
| SCF(Skp2)-mediated degradation of p27/p21 | CCND1 | Cyclin D1, G1/S-specific cyclin-D1 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.525654 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.57244 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.515416 |
| SCF(Skp2)-mediated degradation of p27/p21 | CDK2 | Cyclin dependent kinase 2, Cyclin-dependent kinase 2, Protein kinase domain-containing protein, Uncharacterized protein | 9.389193 | 0.731182 | 4 | 0.730469 | 0.532011 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.515416 |
| SCF(Skp2)-mediated degradation of p27/p21 | CCNE1 | Cyclin E1, Cyclin N-terminal domain-containing protein, G1/S-specific cyclin-E1 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.526741 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.565601 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.5197 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.54735 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.515416 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.54735 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.389193 | 0.731182 | 4 | 0.730469 | 0.568645 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.572294 |
| SCF(Skp2)-mediated degradation of p27/p21 | CCNE2 | Cyclin E2, Cyclin N-terminal domain-containing protein, G1/S-specific cyclin-E2 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.503983 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.577847 |
| SCF(Skp2)-mediated degradation of p27/p21 | CCNA1 | Cyclin A1, Cyclin-A1, Cyclin-A2 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.506777 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.515416 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.389193 | 0.731182 | 4 | 0.730469 | 0.515416 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.515416 |
| SCF(Skp2)-mediated degradation of p27/p21 | CDK1 | Cell division control protein 2, Cyclin dependent kinase 1, Cyclin-dependent kinase 1 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.514008 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.54735 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.515416 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.547953 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.515416 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.54735 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.58858 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.54735 |
| SCF(Skp2)-mediated degradation of p27/p21 | CDK4 | Cyclin dependent kinase 4, Cyclin-dependent kinase 4, Cyclin-dependent kinase 4, isoform A | 9.389193 | 0.731182 | 4 | 0.730469 | 0.511296 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.54735 |
| SCF(Skp2)-mediated degradation of p27/p21 | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.389193 | 0.731182 | 4 | 0.730469 | 0.579609 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.54735 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.573417 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.577481 |
| SCF(Skp2)-mediated degradation of p27/p21 | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.389193 | 0.731182 | 4 | 0.730469 | 0.515416 |
| Selenoamino acid metabolism | RPS10 | 40S ribosomal protein S10, Ribosomal protein S10, S10_plectin domain-containing protein | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL36 | 60S ribosomal protein L36 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPS3 | 40S ribosomal protein S3, DNA-(apurinic or apyrimidinic site) lyase | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL13 | 60S ribosomal protein L13 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL35 | 60S ribosomal protein L35 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPS19 | 40S ribosomal protein S19, Uncharacterized protein | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL27A | 60S ribosomal protein L27-A, 60S ribosomal protein L27a | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPS15 | 40S ribosomal protein S15 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL39 | 60S ribosomal protein L39, LOC100135132 protein, Ribosomal protein L39 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL13A | 60S ribosomal protein L13a | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL15 | 60S ribosomal protein L15, Ribosomal protein L15 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL23 | 60S ribosomal protein L23 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPS23 | 40S ribosomal protein S23, 40S ribosomal protein S23-A | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPS3A | 40S ribosomal protein S3a | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL7A | 60S ribosomal protein L7-A, 60S ribosomal protein L7a | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPS27 | 40S ribosomal protein S27 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.549907 |
| Selenoamino acid metabolism | RPS7 | 40S ribosomal protein S7 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL18A | 60S ribosomal protein L18-A, 60S ribosomal protein L18a | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPS13 | 40S ribosomal protein S13 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL10 | 60S ribosomal protein L10, MGC89303 protein | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPS8 | 40S ribosomal protein S8 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL37 | 60S ribosomal protein L37, Ribosomal protein L37 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL32 | 60S ribosomal protein L32, Hypothetical LOC496894, Ribosomal protein L32, Uncharacterized protein | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPS4Y1 | 40S ribosomal protein S4, 40S ribosomal protein S4, Y isoform 1 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL24 | 60S ribosomal protein L24, Ribosomal protein L24, TRASH domain-containing protein | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPS5 | 40S ribosomal protein S5, 40S ribosomal protein S5 [Cleaved into: 40S ribosomal protein S5, N-terminally processed], 40S ribosomal protein S5-A, Ribosomal protein S5 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPSA | 40S ribosomal protein SA | 6.808357 | 0.847061 | 4 | 0.655266 | 0.521531 |
| Selenoamino acid metabolism | RPS17 | 40S ribosomal protein S17 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL30 | 60S ribosomal protein L30 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL36A | 60S ribosomal protein L36-A, 60S ribosomal protein L36a, IP17351p, ribosomal protein L36a, Uncharacterized protein | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPS15A | 40S ribosomal protein S15a | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL12 | 60S ribosomal protein L12 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPS12 | 40S ribosomal protein S12 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPS9 | 40S ribosomal protein S9 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPS25 | 40S ribosomal protein S25 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPS14 | 40S ribosomal protein S14, Ribosomal protein S14, Uncharacterized protein | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | FAU | 40S ribosomal protein S30 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPS16 | 40S ribosomal protein S16, Ribosomal protein S16, Uncharacterized protein | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPS4X | 40S ribosomal protein S4, 40S ribosomal protein S4, X isoform | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL22 | 60S ribosomal protein L22, 60S ribosomal protein L22 1, Ribosomal protein L22, Uncharacterized protein | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL8 | 60S ribosomal protein L8 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL19 | 60S ribosomal protein L19, Ribosomal protein L19 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL18 | 60S ribosomal protein L18, Ribosomal protein L18 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL4 | 60S ribosomal protein L4, Ribos_L4_asso_C domain-containing protein, Ribosomal protein L4 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL21 | 60S ribosomal protein L21 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL31 | 60S ribosomal protein L31 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPS20 | 40S ribosomal protein S20 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPS28 | 40S ribosomal protein S28 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL34 | 60S ribosomal protein L34 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL14 | 60S ribosomal protein L14 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL17 | 60S ribosomal protein L17 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL5 | 60S ribosomal protein L5, Ribosomal protein L5 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL41 | 60S ribosomal protein L41 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPS18 | 40S ribosomal protein S18 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL23A | 60S ribosomal protein L23-A, 60S ribosomal protein L23a, FI01658p, MGC89958 protein, Ribosomal protein L23a, Ribosomal_L23eN domain-containing protein | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPLP0 | 60S acidic ribosomal protein P0 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL38 | 60S ribosomal protein L38 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPS21 | 40S ribosomal protein S21 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL7 | 60S ribosomal protein L7, MGC79754 protein, Ribosomal protein L7, Uncharacterized protein | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL6 | 60S ribosomal protein L6 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPS29 | 40S ribosomal protein S29 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL29 | 60S ribosomal protein L29 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL28 | 60S ribosomal protein L28 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL37A | 60S ribosomal protein L37-A, 60S ribosomal protein L37a, MGC89854 protein, Probable 60S ribosomal protein L37-A, Ribosomal protein L37a | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL27 | 60S ribosomal protein L27 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL10A | 60S ribosomal protein L10a, Ribosomal protein | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPLP1 | 60S acidic ribosomal protein P1, Ribosomal protein, large, P1 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPS6 | 40S ribosomal protein S6 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.566528 |
| Selenoamino acid metabolism | RPS11 | 40S ribosomal protein S11 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPS2 | 40S ribosomal protein S2 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL35A | 60S ribosomal protein L35-A, 60S ribosomal protein L35a | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL3 | 60S ribosomal protein L3, LOC100145083 protein, Ribosomal protein L3, Uncharacterized protein | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPS26 | 40S ribosomal protein S26 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | HNMT | Histamine N-methyltransferase | 6.808357 | 0.847061 | 4 | 0.655266 | 0.532374 |
| Selenoamino acid metabolism | RPS24 | 40S ribosomal protein S24 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL26 | 60S ribosomal protein L26, GEO07453p1, KOW domain-containing protein, Ribosomal protein L26 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPL11 | 60S ribosomal protein L11 | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenoamino acid metabolism | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 6.808357 | 0.847061 | 4 | 0.655266 | 0.579609 |
| Selenocysteine synthesis | RPL39 | 60S ribosomal protein L39, LOC100135132 protein, Ribosomal protein L39 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL12 | 60S ribosomal protein L12 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL21 | 60S ribosomal protein L21 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL11 | 60S ribosomal protein L11 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS4X | 40S ribosomal protein S4, 40S ribosomal protein S4, X isoform | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS17 | 40S ribosomal protein S17 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPSA | 40S ribosomal protein SA | 11.801728 | 0.914845 | 4 | 0.946761 | 0.521531 |
| Selenocysteine synthesis | RPL24 | 60S ribosomal protein L24, Ribosomal protein L24, TRASH domain-containing protein | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS18 | 40S ribosomal protein S18 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL22 | 60S ribosomal protein L22, 60S ribosomal protein L22 1, Ribosomal protein L22, Uncharacterized protein | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL7A | 60S ribosomal protein L7-A, 60S ribosomal protein L7a | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS19 | 40S ribosomal protein S19, Uncharacterized protein | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS23 | 40S ribosomal protein S23, 40S ribosomal protein S23-A | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS7 | 40S ribosomal protein S7 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL27A | 60S ribosomal protein L27-A, 60S ribosomal protein L27a | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL34 | 60S ribosomal protein L34 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL26 | 60S ribosomal protein L26, GEO07453p1, KOW domain-containing protein, Ribosomal protein L26 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL6 | 60S ribosomal protein L6 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL23 | 60S ribosomal protein L23 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL23A | 60S ribosomal protein L23-A, 60S ribosomal protein L23a, FI01658p, MGC89958 protein, Ribosomal protein L23a, Ribosomal_L23eN domain-containing protein | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL36 | 60S ribosomal protein L36 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL8 | 60S ribosomal protein L8 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS8 | 40S ribosomal protein S8 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL7 | 60S ribosomal protein L7, MGC79754 protein, Ribosomal protein L7, Uncharacterized protein | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL17 | 60S ribosomal protein L17 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS21 | 40S ribosomal protein S21 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL30 | 60S ribosomal protein L30 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS16 | 40S ribosomal protein S16, Ribosomal protein S16, Uncharacterized protein | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS6 | 40S ribosomal protein S6 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.566528 |
| Selenocysteine synthesis | RPL31 | 60S ribosomal protein L31 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL37A | 60S ribosomal protein L37-A, 60S ribosomal protein L37a, MGC89854 protein, Probable 60S ribosomal protein L37-A, Ribosomal protein L37a | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL18A | 60S ribosomal protein L18-A, 60S ribosomal protein L18a | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL10A | 60S ribosomal protein L10a, Ribosomal protein | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL35A | 60S ribosomal protein L35-A, 60S ribosomal protein L35a | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL10 | 60S ribosomal protein L10, MGC89303 protein | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS29 | 40S ribosomal protein S29 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL28 | 60S ribosomal protein L28 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS27 | 40S ribosomal protein S27 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.549907 |
| Selenocysteine synthesis | RPS28 | 40S ribosomal protein S28 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS20 | 40S ribosomal protein S20 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS24 | 40S ribosomal protein S24 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL19 | 60S ribosomal protein L19, Ribosomal protein L19 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS9 | 40S ribosomal protein S9 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS25 | 40S ribosomal protein S25 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPLP0 | 60S acidic ribosomal protein P0 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS12 | 40S ribosomal protein S12 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL3 | 60S ribosomal protein L3, LOC100145083 protein, Ribosomal protein L3, Uncharacterized protein | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL15 | 60S ribosomal protein L15, Ribosomal protein L15 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL18 | 60S ribosomal protein L18, Ribosomal protein L18 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL36A | 60S ribosomal protein L36-A, 60S ribosomal protein L36a, IP17351p, ribosomal protein L36a, Uncharacterized protein | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS10 | 40S ribosomal protein S10, Ribosomal protein S10, S10_plectin domain-containing protein | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS14 | 40S ribosomal protein S14, Ribosomal protein S14, Uncharacterized protein | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL27 | 60S ribosomal protein L27 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS26 | 40S ribosomal protein S26 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL37 | 60S ribosomal protein L37, Ribosomal protein L37 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL13 | 60S ribosomal protein L13 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS11 | 40S ribosomal protein S11 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL4 | 60S ribosomal protein L4, Ribos_L4_asso_C domain-containing protein, Ribosomal protein L4 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS3 | 40S ribosomal protein S3, DNA-(apurinic or apyrimidinic site) lyase | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL35 | 60S ribosomal protein L35 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL5 | 60S ribosomal protein L5, Ribosomal protein L5 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL32 | 60S ribosomal protein L32, Hypothetical LOC496894, Ribosomal protein L32, Uncharacterized protein | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL14 | 60S ribosomal protein L14 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL13A | 60S ribosomal protein L13a | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL29 | 60S ribosomal protein L29 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPLP1 | 60S acidic ribosomal protein P1, Ribosomal protein, large, P1 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | FAU | 40S ribosomal protein S30 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS5 | 40S ribosomal protein S5, 40S ribosomal protein S5 [Cleaved into: 40S ribosomal protein S5, N-terminally processed], 40S ribosomal protein S5-A, Ribosomal protein S5 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS15 | 40S ribosomal protein S15 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS4Y1 | 40S ribosomal protein S4, 40S ribosomal protein S4, Y isoform 1 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS15A | 40S ribosomal protein S15a | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS13 | 40S ribosomal protein S13 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS2 | 40S ribosomal protein S2 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPS3A | 40S ribosomal protein S3a | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL38 | 60S ribosomal protein L38 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Selenocysteine synthesis | RPL41 | 60S ribosomal protein L41 | 11.801728 | 0.914845 | 4 | 0.946761 | 0.579609 |
| Separation of Sister Chromatids | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.515416 |
| Separation of Sister Chromatids | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.573417 |
| Separation of Sister Chromatids | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.515416 |
| Separation of Sister Chromatids | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.515416 |
| Separation of Sister Chromatids | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.54735 |
| Separation of Sister Chromatids | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.515416 |
| Separation of Sister Chromatids | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.57244 |
| Separation of Sister Chromatids | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.515416 |
| Separation of Sister Chromatids | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.515416 |
| Separation of Sister Chromatids | PPP1CC | Serine/threonine-protein phosphatase, Serine/threonine-protein phosphatase PP1-gamma catalytic subunit | 9.651117 | 0.666167 | 4 | 0.711474 | 0.503303 |
| Separation of Sister Chromatids | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.547953 |
| Separation of Sister Chromatids | KIF2C | Kinesin-like protein, Kinesin-like protein KIF2C | 9.651117 | 0.666167 | 4 | 0.711474 | 0.611637 |
| Separation of Sister Chromatids | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.54735 |
| Separation of Sister Chromatids | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.54735 |
| Separation of Sister Chromatids | RPS27 | 40S ribosomal protein S27 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.549907 |
| Separation of Sister Chromatids | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.54735 |
| Separation of Sister Chromatids | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.54735 |
| Separation of Sister Chromatids | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.58858 |
| Separation of Sister Chromatids | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.54735 |
| Separation of Sister Chromatids | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.54735 |
| Separation of Sister Chromatids | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.651117 | 0.666167 | 4 | 0.711474 | 0.579609 |
| Separation of Sister Chromatids | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.565601 |
| Separation of Sister Chromatids | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.5197 |
| Separation of Sister Chromatids | CENPE | Centromere protein E, Centromere-associated protein E, Kinesin motor domain-containing protein | 9.651117 | 0.666167 | 4 | 0.711474 | 0.564477 |
| Separation of Sister Chromatids | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.515416 |
| Separation of Sister Chromatids | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.515416 |
| Separation of Sister Chromatids | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.651117 | 0.666167 | 4 | 0.711474 | 0.568645 |
| Separation of Sister Chromatids | INCENP | Inner centromere protein, Inner centromere protein, isoform A, Uncharacterized protein | 9.651117 | 0.666167 | 4 | 0.711474 | 0.527883 |
| Separation of Sister Chromatids | AURKB | Aurora kinase, Aurora kinase B | 9.651117 | 0.666167 | 4 | 0.711474 | 0.524412 |
| Separation of Sister Chromatids | PPP2R5A | Serine/threonine protein phosphatase 2A regulatory subunit, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform | 9.651117 | 0.666167 | 4 | 0.711474 | 0.559184 |
| Separation of Sister Chromatids | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.515416 |
| Separation of Sister Chromatids | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.572294 |
| Separation of Sister Chromatids | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.54735 |
| Separation of Sister Chromatids | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.54735 |
| Separation of Sister Chromatids | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.515416 |
| Separation of Sister Chromatids | HDAC8 | Histone deacetylase, Histone deacetylase 8, WD repeat and SOCS box containing 2 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.535733 |
| Separation of Sister Chromatids | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.577481 |
| Separation of Sister Chromatids | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.651117 | 0.666167 | 4 | 0.711474 | 0.515416 |
| Separation of Sister Chromatids | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.54735 |
| Separation of Sister Chromatids | PLK1 | Serine/threonine-protein kinase PLK, Serine/threonine-protein kinase PLK1 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.504187 |
| Separation of Sister Chromatids | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.546881 |
| Separation of Sister Chromatids | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.515416 |
| Separation of Sister Chromatids | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.577847 |
| Separation of Sister Chromatids | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.515416 |
| Separation of Sister Chromatids | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.651117 | 0.666167 | 4 | 0.711474 | 0.515416 |
| Separation of Sister Chromatids | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.651117 | 0.666167 | 4 | 0.711474 | 0.515416 |
| SHC1 events in ERBB2 signaling | SHC1 | SHC-transforming protein 1 | 9.065746 | 0.579299 | 3.396816 | 0.541657 | 0.521858 |
| SHC1 events in ERBB2 signaling | EGFR | Epidermal growth factor receptor, Receptor protein-tyrosine kinase | 9.065746 | 0.579299 | 3.396816 | 0.541657 | 0.625817 |
| SHC1 events in ERBB2 signaling | PRKCA | Protein kinase C, Protein kinase C alpha type, Protein kinase C, alpha | 9.065746 | 0.579299 | 3.396816 | 0.541657 | 0.52202 |
| SHC1 events in ERBB2 signaling | ERBB2 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-2 | 9.065746 | 0.579299 | 3.396816 | 0.541657 | 0.589497 |
| SHC1 events in ERBB2 signaling | PRKCE | Protein kinase C, Protein kinase C epsilon type | 9.065746 | 0.579299 | 3.396816 | 0.541657 | 0.512211 |
| SHC1 events in ERBB2 signaling | ERBB4 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-4 | 9.065746 | 0.579299 | 3.396816 | 0.541657 | 0.550399 |
| SHC1 events in ERBB2 signaling | PRKCD | Protein kinase C, Protein kinase C delta type | 9.065746 | 0.579299 | 3.396816 | 0.541657 | 0.520528 |
| SHC1 events in ERBB2 signaling | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 9.065746 | 0.579299 | 3.396816 | 0.541657 | 0.505988 |
| SHC1 events in ERBB2 signaling | SHC | DShc protein | 9.065746 | 0.579299 | 3.396816 | 0.541657 | 0.605 |
| SHC1 events in ERBB2 signaling | ERBB3 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-3 | 9.065746 | 0.579299 | 3.396816 | 0.541657 | 0.551662 |
| Signaling by BRAF and RAF fusions | RAF1 | Non-specific serine/threonine protein kinase, RAF proto-oncogene serine/threonine-protein kinase | 8.838917 | 0.483022 | 3.920849 | 0.566564 | 0.573556 |
| Signaling by BRAF and RAF fusions | YWHAB | 14-3-3 protein beta/alpha, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta | 8.838917 | 0.483022 | 3.920849 | 0.566564 | 0.513843 |
| Signaling by BRAF and RAF fusions | MAP2K1 | Dual specificity mitogen-activated protein kinase kinase 1, MAP2K1 protein, Mitogen-activated protein kinase kinase 1, Protein kinase domain-containing protein | 8.838917 | 0.483022 | 3.920849 | 0.566564 | 0.545374 |
| Signaling by BRAF and RAF fusions | BRAF | Non-specific serine/threonine protein kinase, Serine/threonine-protein kinase B-raf | 8.838917 | 0.483022 | 3.920849 | 0.566564 | 0.590074 |
| Signaling by BRAF and RAF fusions | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 8.838917 | 0.483022 | 3.920849 | 0.566564 | 0.505988 |
| Signaling by BRAF and RAF fusions | LMNA | Lamin A, Lamin A/C, Prelamin-A/C, Prelamin-A/C [Cleaved into: Lamin-A/C, Prelamin-A/C [Cleaved into: Lamin-A/C], Uncharacterized protein | 8.838917 | 0.483022 | 3.920849 | 0.566564 | 0.5357 |
| Signaling by BRAF and RAF fusions | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 8.838917 | 0.483022 | 3.920849 | 0.566564 | 0.541254 |
| Signaling by BRAF and RAF fusions | VWF | von Willebrand factor | 8.838917 | 0.483022 | 3.920849 | 0.566564 | 0.548181 |
| Signaling by BRAF and RAF fusions | JAK2 | Janus kinase 2, Tyrosine-protein kinase, Tyrosine-protein kinase JAK2 | 8.838917 | 0.483022 | 3.920849 | 0.566564 | 0.573235 |
| Signaling by BRAF and RAF fusions | FN1 | Fibronectin | 8.838917 | 0.483022 | 3.920849 | 0.566564 | 0.578096 |
| Signaling by BRAF and RAF fusions | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 8.838917 | 0.483022 | 3.920849 | 0.566564 | 0.594807 |
| Signaling by BRAF and RAF fusions | RAP1A | RAP1A, member of RAS oncogene family, Ras-related protein Rap-1A, Uncharacterized protein | 8.838917 | 0.483022 | 3.920849 | 0.566564 | 0.530823 |
| Signaling by ERBB2 | HSP90AA1 | Heat shock protein 90 alpha family class A member 1, Heat shock protein HSP 90-alpha | 9.068875 | 0.486529 | 3.738892 | 0.552999 | 0.624325 |
| Signaling by ERBB2 | PRKCE | Protein kinase C, Protein kinase C epsilon type | 9.068875 | 0.486529 | 3.738892 | 0.552999 | 0.512211 |
| Signaling by ERBB2 | SHC | DShc protein | 9.068875 | 0.486529 | 3.738892 | 0.552999 | 0.605 |
| Signaling by ERBB2 | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 9.068875 | 0.486529 | 3.738892 | 0.552999 | 0.505988 |
| Signaling by ERBB2 | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 9.068875 | 0.486529 | 3.738892 | 0.552999 | 0.646924 |
| Signaling by ERBB2 | YES1 | Tyrosine-protein kinase, Tyrosine-protein kinase yes, Tyrosine-protein kinase Yes | 9.068875 | 0.486529 | 3.738892 | 0.552999 | 0.506641 |
| Signaling by ERBB2 | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 9.068875 | 0.486529 | 3.738892 | 0.552999 | 0.589663 |
| Signaling by ERBB2 | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 9.068875 | 0.486529 | 3.738892 | 0.552999 | 0.552041 |
| Signaling by ERBB2 | SHC1 | SHC-transforming protein 1 | 9.068875 | 0.486529 | 3.738892 | 0.552999 | 0.521858 |
| Signaling by ERBB2 | ERBB3 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-3 | 9.068875 | 0.486529 | 3.738892 | 0.552999 | 0.551662 |
| Signaling by ERBB2 | ERBB2 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-2 | 9.068875 | 0.486529 | 3.738892 | 0.552999 | 0.589497 |
| Signaling by ERBB2 | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 9.068875 | 0.486529 | 3.738892 | 0.552999 | 0.594807 |
| Signaling by ERBB2 | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.068875 | 0.486529 | 3.738892 | 0.552999 | 0.660919 |
| Signaling by ERBB2 | PRKCD | Protein kinase C, Protein kinase C delta type | 9.068875 | 0.486529 | 3.738892 | 0.552999 | 0.520528 |
| Signaling by ERBB2 | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.068875 | 0.486529 | 3.738892 | 0.552999 | 0.579609 |
| Signaling by ERBB2 | ERBB4 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-4 | 9.068875 | 0.486529 | 3.738892 | 0.552999 | 0.550399 |
| Signaling by ERBB2 | EGFR | Epidermal growth factor receptor, Receptor protein-tyrosine kinase | 9.068875 | 0.486529 | 3.738892 | 0.552999 | 0.625817 |
| Signaling by ERBB2 | PRKCA | Protein kinase C, Protein kinase C alpha type, Protein kinase C, alpha | 9.068875 | 0.486529 | 3.738892 | 0.552999 | 0.52202 |
| Signaling by ERBB2 KD Mutants | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.521281 | 0.548592 | 2.889588 | 0.46666 | 0.660919 |
| Signaling by ERBB2 KD Mutants | ERBB3 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-3 | 9.521281 | 0.548592 | 2.889588 | 0.46666 | 0.551662 |
| Signaling by ERBB2 KD Mutants | ERBB2 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-2 | 9.521281 | 0.548592 | 2.889588 | 0.46666 | 0.589497 |
| Signaling by ERBB2 KD Mutants | EGFR | Epidermal growth factor receptor, Receptor protein-tyrosine kinase | 9.521281 | 0.548592 | 2.889588 | 0.46666 | 0.625817 |
| Signaling by ERBB2 KD Mutants | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 9.521281 | 0.548592 | 2.889588 | 0.46666 | 0.505988 |
| Signaling by ERBB2 KD Mutants | SHC1 | SHC-transforming protein 1 | 9.521281 | 0.548592 | 2.889588 | 0.46666 | 0.521858 |
| Signaling by ERBB2 KD Mutants | ERBB4 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-4 | 9.521281 | 0.548592 | 2.889588 | 0.46666 | 0.550399 |
| Signaling by ERBB2 KD Mutants | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 9.521281 | 0.548592 | 2.889588 | 0.46666 | 0.646924 |
| Signaling by ERBB2 KD Mutants | HSP90AA1 | Heat shock protein 90 alpha family class A member 1, Heat shock protein HSP 90-alpha | 9.521281 | 0.548592 | 2.889588 | 0.46666 | 0.624325 |
| Signaling by ERBB4 | APH1A | Anterior pharynx defective 1 homolog A, Anterior pharynx defective 1a homolog, Aph-1 homolog A, gamma-secretase subunit, Gamma-secretase subunit APH-1A, Uncharacterized protein | 7.724622 | 0.604724 | 4 | 0.581367 | 0.60749 |
| Signaling by ERBB4 | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 7.724622 | 0.604724 | 4 | 0.581367 | 0.660919 |
| Signaling by ERBB4 | ERBB4 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-4 | 7.724622 | 0.604724 | 4 | 0.581367 | 0.550399 |
| Signaling by ERBB4 | SHC | DShc protein | 7.724622 | 0.604724 | 4 | 0.581367 | 0.605 |
| Signaling by ERBB4 | NCSTN | Nicastrin | 7.724622 | 0.604724 | 4 | 0.581367 | 0.60749 |
| Signaling by ERBB4 | GABRA1 | GABA(A) receptor subunit alpha-1, Gamma-aminobutyric acid receptor subunit alpha-1 | 7.724622 | 0.604724 | 4 | 0.581367 | 0.508013 |
| Signaling by ERBB4 | GABRB3 | Gabrb3 protein, Gamma-aminobutyric acid receptor subunit beta-3, Gamma-aminobutyric acid type A receptor beta3 subunit, Gamma-aminobutyric acid type A receptor subunit beta3 | 7.724622 | 0.604724 | 4 | 0.581367 | 0.53034 |
| Signaling by ERBB4 | CSN2 | Beta-casein, COP9 signalosome complex subunit 2 | 7.724622 | 0.604724 | 4 | 0.581367 | 0.537553 |
| Signaling by ERBB4 | EGFR | Epidermal growth factor receptor, Receptor protein-tyrosine kinase | 7.724622 | 0.604724 | 4 | 0.581367 | 0.625817 |
| Signaling by ERBB4 | ERBB3 | Receptor protein-tyrosine kinase, Receptor tyrosine-protein kinase erbB-3 | 7.724622 | 0.604724 | 4 | 0.581367 | 0.551662 |
| Signaling by ERBB4 | PSEN2 | Presenilin, Presenilin-2 | 7.724622 | 0.604724 | 4 | 0.581367 | 0.607393 |
| Signaling by ERBB4 | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 7.724622 | 0.604724 | 4 | 0.581367 | 0.594807 |
| Signaling by ERBB4 | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 7.724622 | 0.604724 | 4 | 0.581367 | 0.579609 |
| Signaling by ERBB4 | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 7.724622 | 0.604724 | 4 | 0.581367 | 0.646924 |
| Signaling by ERBB4 | YAP1 | Transcriptional coactivator YAP1, Yes1 associated transcriptional regulator | 7.724622 | 0.604724 | 4 | 0.581367 | 0.628833 |
| Signaling by ERBB4 | GABRG2 | GABA(A) receptor subunit gamma-2, Gamma-aminobutyric acid, Gamma-aminobutyric acid receptor subunit gamma-2, Gamma-aminobutyric acid type A receptor subunit gamma2 | 7.724622 | 0.604724 | 4 | 0.581367 | 0.522585 |
| Signaling by ERBB4 | ESR1 | Estrogen receptor | 7.724622 | 0.604724 | 4 | 0.581367 | 0.561602 |
| Signaling by ERBB4 | APH1B | Aph-1 homolog B, gamma-secretase subunit, APH-1B', Gamma-secretase subunit APH-1B, Uncharacterized protein | 7.724622 | 0.604724 | 4 | 0.581367 | 0.60749 |
| Signaling by ERBB4 | PSENEN | Gamma-secretase subunit PEN-2 | 7.724622 | 0.604724 | 4 | 0.581367 | 0.60725 |
| Signaling by ERBB4 | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 7.724622 | 0.604724 | 4 | 0.581367 | 0.505988 |
| Signaling by ERBB4 | PGR | Nuclear receptor subfamily 3 group C member 3, Progesterone receptor | 7.724622 | 0.604724 | 4 | 0.581367 | 0.5833 |
| Signaling by ERBB4 | SHC1 | SHC-transforming protein 1 | 7.724622 | 0.604724 | 4 | 0.581367 | 0.521858 |
| Signaling by ERBB4 | PSEN1 | Presenilin, Presenilin-1 | 7.724622 | 0.604724 | 4 | 0.581367 | 0.611222 |
| Signaling by high-kinase activity BRAF mutants | BRAF | Non-specific serine/threonine protein kinase, Serine/threonine-protein kinase B-raf | 8.906294 | 0.495804 | 2.839452 | 0.417306 | 0.590074 |
| Signaling by high-kinase activity BRAF mutants | RAP1A | RAP1A, member of RAS oncogene family, Ras-related protein Rap-1A, Uncharacterized protein | 8.906294 | 0.495804 | 2.839452 | 0.417306 | 0.530823 |
| Signaling by high-kinase activity BRAF mutants | YWHAB | 14-3-3 protein beta/alpha, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta | 8.906294 | 0.495804 | 2.839452 | 0.417306 | 0.513843 |
| Signaling by high-kinase activity BRAF mutants | RAF1 | Non-specific serine/threonine protein kinase, RAF proto-oncogene serine/threonine-protein kinase | 8.906294 | 0.495804 | 2.839452 | 0.417306 | 0.573556 |
| Signaling by high-kinase activity BRAF mutants | FN1 | Fibronectin | 8.906294 | 0.495804 | 2.839452 | 0.417306 | 0.578096 |
| Signaling by high-kinase activity BRAF mutants | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 8.906294 | 0.495804 | 2.839452 | 0.417306 | 0.505988 |
| Signaling by high-kinase activity BRAF mutants | MAP2K1 | Dual specificity mitogen-activated protein kinase kinase 1, MAP2K1 protein, Mitogen-activated protein kinase kinase 1, Protein kinase domain-containing protein | 8.906294 | 0.495804 | 2.839452 | 0.417306 | 0.545374 |
| Signaling by high-kinase activity BRAF mutants | VWF | von Willebrand factor | 8.906294 | 0.495804 | 2.839452 | 0.417306 | 0.548181 |
| Signaling by high-kinase activity BRAF mutants | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 8.906294 | 0.495804 | 2.839452 | 0.417306 | 0.541254 |
| Signaling by high-kinase activity BRAF mutants | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 8.906294 | 0.495804 | 2.839452 | 0.417306 | 0.594807 |
| Signaling by moderate kinase activity BRAF mutants | VWF | von Willebrand factor | 8.880042 | 0.512627 | 4 | 0.594925 | 0.548181 |
| Signaling by moderate kinase activity BRAF mutants | MAP2K1 | Dual specificity mitogen-activated protein kinase kinase 1, MAP2K1 protein, Mitogen-activated protein kinase kinase 1, Protein kinase domain-containing protein | 8.880042 | 0.512627 | 4 | 0.594925 | 0.545374 |
| Signaling by moderate kinase activity BRAF mutants | FN1 | Fibronectin | 8.880042 | 0.512627 | 4 | 0.594925 | 0.578096 |
| Signaling by moderate kinase activity BRAF mutants | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 8.880042 | 0.512627 | 4 | 0.594925 | 0.505988 |
| Signaling by moderate kinase activity BRAF mutants | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 8.880042 | 0.512627 | 4 | 0.594925 | 0.541254 |
| Signaling by moderate kinase activity BRAF mutants | RAF1 | Non-specific serine/threonine protein kinase, RAF proto-oncogene serine/threonine-protein kinase | 8.880042 | 0.512627 | 4 | 0.594925 | 0.573556 |
| Signaling by moderate kinase activity BRAF mutants | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 8.880042 | 0.512627 | 4 | 0.594925 | 0.594807 |
| Signaling by moderate kinase activity BRAF mutants | JAK2 | Janus kinase 2, Tyrosine-protein kinase, Tyrosine-protein kinase JAK2 | 8.880042 | 0.512627 | 4 | 0.594925 | 0.573235 |
| Signaling by moderate kinase activity BRAF mutants | RAP1A | RAP1A, member of RAS oncogene family, Ras-related protein Rap-1A, Uncharacterized protein | 8.880042 | 0.512627 | 4 | 0.594925 | 0.530823 |
| Signaling by moderate kinase activity BRAF mutants | BRAF | Non-specific serine/threonine protein kinase, Serine/threonine-protein kinase B-raf | 8.880042 | 0.512627 | 4 | 0.594925 | 0.590074 |
| Signaling by moderate kinase activity BRAF mutants | YWHAB | 14-3-3 protein beta/alpha, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta | 8.880042 | 0.512627 | 4 | 0.594925 | 0.513843 |
| Signaling by phosphorylated juxtamembrane, extracellular and kinase domain KIT mutants | YES1 | Tyrosine-protein kinase, Tyrosine-protein kinase yes, Tyrosine-protein kinase Yes | 8.401489 | 0.521679 | 2.837065 | 0.40766 | 0.506641 |
| Signaling by phosphorylated juxtamembrane, extracellular and kinase domain KIT mutants | JAK2 | Janus kinase 2, Tyrosine-protein kinase, Tyrosine-protein kinase JAK2 | 8.401489 | 0.521679 | 2.837065 | 0.40766 | 0.573235 |
| Signaling by phosphorylated juxtamembrane, extracellular and kinase domain KIT mutants | KIT | Mast/stem cell growth factor receptor, Mast/stem cell growth factor receptor Kit | 8.401489 | 0.521679 | 2.837065 | 0.40766 | 0.562353 |
| Signaling by phosphorylated juxtamembrane, extracellular and kinase domain KIT mutants | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 8.401489 | 0.521679 | 2.837065 | 0.40766 | 0.505988 |
| Signaling by phosphorylated juxtamembrane, extracellular and kinase domain KIT mutants | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 8.401489 | 0.521679 | 2.837065 | 0.40766 | 0.594807 |
| Signaling by phosphorylated juxtamembrane, extracellular and kinase domain KIT mutants | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 8.401489 | 0.521679 | 2.837065 | 0.40766 | 0.646924 |
| Signaling by phosphorylated juxtamembrane, extracellular and kinase domain KIT mutants | LCK | Proto-oncogene tyrosine-protein kinase LCK, Tyrosine-protein kinase, Tyrosine-protein kinase Lck | 8.401489 | 0.521679 | 2.837065 | 0.40766 | 0.543278 |
| Signaling by phosphorylated juxtamembrane, extracellular and kinase domain KIT mutants | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 8.401489 | 0.521679 | 2.837065 | 0.40766 | 0.660919 |
| Signaling by RAF1 mutants | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 8.926693 | 0.548719 | 4 | 0.615378 | 0.594807 |
| Signaling by RAF1 mutants | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 8.926693 | 0.548719 | 4 | 0.615378 | 0.505988 |
| Signaling by RAF1 mutants | YWHAB | 14-3-3 protein beta/alpha, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta | 8.926693 | 0.548719 | 4 | 0.615378 | 0.513843 |
| Signaling by RAF1 mutants | RAP1A | RAP1A, member of RAS oncogene family, Ras-related protein Rap-1A, Uncharacterized protein | 8.926693 | 0.548719 | 4 | 0.615378 | 0.530823 |
| Signaling by RAF1 mutants | BRAF | Non-specific serine/threonine protein kinase, Serine/threonine-protein kinase B-raf | 8.926693 | 0.548719 | 4 | 0.615378 | 0.590074 |
| Signaling by RAF1 mutants | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 8.926693 | 0.548719 | 4 | 0.615378 | 0.541254 |
| Signaling by RAF1 mutants | MAP2K1 | Dual specificity mitogen-activated protein kinase kinase 1, MAP2K1 protein, Mitogen-activated protein kinase kinase 1, Protein kinase domain-containing protein | 8.926693 | 0.548719 | 4 | 0.615378 | 0.545374 |
| Signaling by RAF1 mutants | RAF1 | Non-specific serine/threonine protein kinase, RAF proto-oncogene serine/threonine-protein kinase | 8.926693 | 0.548719 | 4 | 0.615378 | 0.573556 |
| Signaling by RAF1 mutants | FN1 | Fibronectin | 8.926693 | 0.548719 | 4 | 0.615378 | 0.578096 |
| Signaling by RAF1 mutants | VWF | von Willebrand factor | 8.926693 | 0.548719 | 4 | 0.615378 | 0.548181 |
| Signaling by RAF1 mutants | JAK2 | Janus kinase 2, Tyrosine-protein kinase, Tyrosine-protein kinase JAK2 | 8.926693 | 0.548719 | 4 | 0.615378 | 0.573235 |
| Signaling by ROBO receptors | RPS13 | 40S ribosomal protein S13 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPS20 | 40S ribosomal protein S20 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPS5 | 40S ribosomal protein S5, 40S ribosomal protein S5 [Cleaved into: 40S ribosomal protein S5, N-terminally processed], 40S ribosomal protein S5-A, Ribosomal protein S5 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPS26 | 40S ribosomal protein S26 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPS11 | 40S ribosomal protein S11 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL27 | 60S ribosomal protein L27 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.54735 |
| Signaling by ROBO receptors | RPL13 | 60S ribosomal protein L13 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.515416 |
| Signaling by ROBO receptors | RPL27A | 60S ribosomal protein L27-A, 60S ribosomal protein L27a | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPLP0 | 60S acidic ribosomal protein P0 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.58858 |
| Signaling by ROBO receptors | RPS2 | 40S ribosomal protein S2 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.54735 |
| Signaling by ROBO receptors | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.577847 |
| Signaling by ROBO receptors | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.54735 |
| Signaling by ROBO receptors | RPS16 | 40S ribosomal protein S16, Ribosomal protein S16, Uncharacterized protein | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPS10 | 40S ribosomal protein S10, Ribosomal protein S10, S10_plectin domain-containing protein | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL7A | 60S ribosomal protein L7-A, 60S ribosomal protein L7a | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPS27 | 40S ribosomal protein S27 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.549907 |
| Signaling by ROBO receptors | RPL21 | 60S ribosomal protein L21 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL31 | 60S ribosomal protein L31 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.515416 |
| Signaling by ROBO receptors | RPL14 | 60S ribosomal protein L14 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL7 | 60S ribosomal protein L7, MGC79754 protein, Ribosomal protein L7, Uncharacterized protein | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL10A | 60S ribosomal protein L10a, Ribosomal protein | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL39 | 60S ribosomal protein L39, LOC100135132 protein, Ribosomal protein L39 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPS3 | 40S ribosomal protein S3, DNA-(apurinic or apyrimidinic site) lyase | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | ABL1 | Tyrosine-protein kinase, Tyrosine-protein kinase ABL1 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.590046 |
| Signaling by ROBO receptors | RPS21 | 40S ribosomal protein S21 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.515416 |
| Signaling by ROBO receptors | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.54735 |
| Signaling by ROBO receptors | RPS17 | 40S ribosomal protein S17 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPS28 | 40S ribosomal protein S28 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL12 | 60S ribosomal protein L12 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL4 | 60S ribosomal protein L4, Ribos_L4_asso_C domain-containing protein, Ribosomal protein L4 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.54735 |
| Signaling by ROBO receptors | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.54735 |
| Signaling by ROBO receptors | RPL18 | 60S ribosomal protein L18, Ribosomal protein L18 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL29 | 60S ribosomal protein L29 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPS29 | 40S ribosomal protein S29 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL23 | 60S ribosomal protein L23 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL17 | 60S ribosomal protein L17 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL22 | 60S ribosomal protein L22, 60S ribosomal protein L22 1, Ribosomal protein L22, Uncharacterized protein | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL6 | 60S ribosomal protein L6 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPS25 | 40S ribosomal protein S25 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL35 | 60S ribosomal protein L35 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL26 | 60S ribosomal protein L26, GEO07453p1, KOW domain-containing protein, Ribosomal protein L26 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.515416 |
| Signaling by ROBO receptors | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 6.413861 | 0.737935 | 4 | 0.580351 | 0.594807 |
| Signaling by ROBO receptors | RPS7 | 40S ribosomal protein S7 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.5197 |
| Signaling by ROBO receptors | RPS24 | 40S ribosomal protein S24 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL35A | 60S ribosomal protein L35-A, 60S ribosomal protein L35a | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPS12 | 40S ribosomal protein S12 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL15 | 60S ribosomal protein L15, Ribosomal protein L15 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL23A | 60S ribosomal protein L23-A, 60S ribosomal protein L23a, FI01658p, MGC89958 protein, Ribosomal protein L23a, Ribosomal_L23eN domain-containing protein | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 6.413861 | 0.737935 | 4 | 0.580351 | 0.515416 |
| Signaling by ROBO receptors | RPL5 | 60S ribosomal protein L5, Ribosomal protein L5 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL30 | 60S ribosomal protein L30 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL36 | 60S ribosomal protein L36 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | ABL2 | Tyrosine-protein kinase, Tyrosine-protein kinase ABL2 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.52908 |
| Signaling by ROBO receptors | RPS15 | 40S ribosomal protein S15 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPS8 | 40S ribosomal protein S8 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.515416 |
| Signaling by ROBO receptors | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.515416 |
| Signaling by ROBO receptors | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.577481 |
| Signaling by ROBO receptors | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.547953 |
| Signaling by ROBO receptors | RPL8 | 60S ribosomal protein L8 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL37 | 60S ribosomal protein L37, Ribosomal protein L37 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPLP1 | 60S acidic ribosomal protein P1, Ribosomal protein, large, P1 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL37A | 60S ribosomal protein L37-A, 60S ribosomal protein L37a, MGC89854 protein, Probable 60S ribosomal protein L37-A, Ribosomal protein L37a | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.565601 |
| Signaling by ROBO receptors | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.515416 |
| Signaling by ROBO receptors | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.515416 |
| Signaling by ROBO receptors | RPS23 | 40S ribosomal protein S23, 40S ribosomal protein S23-A | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.54735 |
| Signaling by ROBO receptors | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.573417 |
| Signaling by ROBO receptors | RPS19 | 40S ribosomal protein S19, Uncharacterized protein | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL24 | 60S ribosomal protein L24, Ribosomal protein L24, TRASH domain-containing protein | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPSA | 40S ribosomal protein SA | 6.413861 | 0.737935 | 4 | 0.580351 | 0.521531 |
| Signaling by ROBO receptors | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.515416 |
| Signaling by ROBO receptors | RPL41 | 60S ribosomal protein L41 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 6.413861 | 0.737935 | 4 | 0.580351 | 0.515416 |
| Signaling by ROBO receptors | RPS4Y1 | 40S ribosomal protein S4, 40S ribosomal protein S4, Y isoform 1 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | FAU | 40S ribosomal protein S30 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL28 | 60S ribosomal protein L28 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL36A | 60S ribosomal protein L36-A, 60S ribosomal protein L36a, IP17351p, ribosomal protein L36a, Uncharacterized protein | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.54735 |
| Signaling by ROBO receptors | RPS14 | 40S ribosomal protein S14, Ribosomal protein S14, Uncharacterized protein | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.546881 |
| Signaling by ROBO receptors | PRKCA | Protein kinase C, Protein kinase C alpha type, Protein kinase C, alpha | 6.413861 | 0.737935 | 4 | 0.580351 | 0.52202 |
| Signaling by ROBO receptors | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.57244 |
| Signaling by ROBO receptors | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.515416 |
| Signaling by ROBO receptors | RPS6 | 40S ribosomal protein S6 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.566528 |
| Signaling by ROBO receptors | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.515416 |
| Signaling by ROBO receptors | CXCR4 | C-X-C chemokine receptor type 4, G_PROTEIN_RECEP_F1_2 domain-containing protein | 6.413861 | 0.737935 | 4 | 0.580351 | 0.512583 |
| Signaling by ROBO receptors | RPS4X | 40S ribosomal protein S4, 40S ribosomal protein S4, X isoform | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPS15A | 40S ribosomal protein S15a | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.572294 |
| Signaling by ROBO receptors | RPL10 | 60S ribosomal protein L10, MGC89303 protein | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL32 | 60S ribosomal protein L32, Hypothetical LOC496894, Ribosomal protein L32, Uncharacterized protein | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 6.413861 | 0.737935 | 4 | 0.580351 | 0.568645 |
| Signaling by ROBO receptors | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.515416 |
| Signaling by ROBO receptors | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.54735 |
| Signaling by ROBO receptors | RPL13A | 60S ribosomal protein L13a | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPS18 | 40S ribosomal protein S18 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL19 | 60S ribosomal protein L19, Ribosomal protein L19 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.54735 |
| Signaling by ROBO receptors | RPL34 | 60S ribosomal protein L34 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL3 | 60S ribosomal protein L3, LOC100145083 protein, Ribosomal protein L3, Uncharacterized protein | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.515416 |
| Signaling by ROBO receptors | RPL38 | 60S ribosomal protein L38 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | PRKAR2A | cAMP-dependent protein kinase type II-alpha regulatory subunit, Protein kinase cAMP-dependent type II regulatory subunit alpha, Protein kinase, cAMP-dependent, regulatory subunit type II alpha, Uncharacterized protein | 6.413861 | 0.737935 | 4 | 0.580351 | 0.527065 |
| Signaling by ROBO receptors | RPL11 | 60S ribosomal protein L11 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPL18A | 60S ribosomal protein L18-A, 60S ribosomal protein L18a | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPS9 | 40S ribosomal protein S9 | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by ROBO receptors | RPS3A | 40S ribosomal protein S3a | 6.413861 | 0.737935 | 4 | 0.580351 | 0.579609 |
| Signaling by VEGF | FLT1 | Receptor protein-tyrosine kinase, Vascular endothelial growth factor receptor 1 | 6.352699 | 0.447974 | 4 | 0.432215 | 0.518548 |
| Signaling by VEGF | MAPK13 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 13 | 6.352699 | 0.447974 | 4 | 0.432215 | 0.573201 |
| Signaling by VEGF | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 6.352699 | 0.447974 | 4 | 0.432215 | 0.594807 |
| Signaling by VEGF | PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 6.352699 | 0.447974 | 4 | 0.432215 | 0.589385 |
| Signaling by VEGF | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 6.352699 | 0.447974 | 4 | 0.432215 | 0.552041 |
| Signaling by VEGF | KDR | Receptor protein-tyrosine kinase, Vascular endothelial growth factor receptor 2 | 6.352699 | 0.447974 | 4 | 0.432215 | 0.609728 |
| Signaling by VEGF | PRKCA | Protein kinase C, Protein kinase C alpha type, Protein kinase C, alpha | 6.352699 | 0.447974 | 4 | 0.432215 | 0.52202 |
| Signaling by VEGF | CRK | 14_3_3 domain-containing protein, Adapter molecule crk, Adapter molecule Crk, CRK proto-oncogene, adaptor protein, Uncharacterized protein, V-crk avian sarcoma virus CT10 oncogene homolog | 6.352699 | 0.447974 | 4 | 0.432215 | 0.513369 |
| Signaling by VEGF | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 6.352699 | 0.447974 | 4 | 0.432215 | 0.660919 |
| Signaling by VEGF | MTOR | Nucleoprotein TPR, Serine/threonine-protein kinase mTOR, Serine/threonine-protein kinase TOR | 6.352699 | 0.447974 | 4 | 0.432215 | 0.619026 |
| Signaling by VEGF | MAPK11 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 11 | 6.352699 | 0.447974 | 4 | 0.432215 | 0.567539 |
| Signaling by VEGF | PRKCB | Protein kinase C, Protein kinase C beta type | 6.352699 | 0.447974 | 4 | 0.432215 | 0.520598 |
| Signaling by VEGF | PRKCZ | Protein kinase C, Protein kinase C zeta type | 6.352699 | 0.447974 | 4 | 0.432215 | 0.513845 |
| Signaling by VEGF | PTK2 | Focal adhesion kinase 1, Non-specific protein-tyrosine kinase, Serine/threonine-protein kinase PTK2/STK2 | 6.352699 | 0.447974 | 4 | 0.432215 | 0.503121 |
| Signaling by VEGF | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 6.352699 | 0.447974 | 4 | 0.432215 | 0.646924 |
| Signaling by VEGF | HSP90AA1 | Heat shock protein 90 alpha family class A member 1, Heat shock protein HSP 90-alpha | 6.352699 | 0.447974 | 4 | 0.432215 | 0.624325 |
| Signaling by VEGF | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 6.352699 | 0.447974 | 4 | 0.432215 | 0.589663 |
| Signaling by VEGF | MAPK12 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 12 | 6.352699 | 0.447974 | 4 | 0.432215 | 0.573393 |
| Signaling by VEGF | FLT4 | Vascular endothelial growth factor receptor 3 | 6.352699 | 0.447974 | 4 | 0.432215 | 0.501095 |
| Signaling by VEGF | PRKCD | Protein kinase C, Protein kinase C delta type | 6.352699 | 0.447974 | 4 | 0.432215 | 0.520528 |
| Signaling by VEGF | CTNNB1 | Catenin, Catenin beta-1, Uncharacterized protein | 6.352699 | 0.447974 | 4 | 0.432215 | 0.657916 |
| Signaling by VEGF | ROCK2 | Rho-associated protein kinase, Rho-associated protein kinase 2 | 6.352699 | 0.447974 | 4 | 0.432215 | 0.516942 |
| Signaling by VEGF | MAPK14 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 14 | 6.352699 | 0.447974 | 4 | 0.432215 | 0.591941 |
| Signaling by VEGF | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 6.352699 | 0.447974 | 4 | 0.432215 | 0.505988 |
| Signaling downstream of RAS mutants | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 8.880042 | 0.512627 | 4 | 0.594925 | 0.505988 |
| Signaling downstream of RAS mutants | BRAF | Non-specific serine/threonine protein kinase, Serine/threonine-protein kinase B-raf | 8.880042 | 0.512627 | 4 | 0.594925 | 0.590074 |
| Signaling downstream of RAS mutants | MAP2K1 | Dual specificity mitogen-activated protein kinase kinase 1, MAP2K1 protein, Mitogen-activated protein kinase kinase 1, Protein kinase domain-containing protein | 8.880042 | 0.512627 | 4 | 0.594925 | 0.545374 |
| Signaling downstream of RAS mutants | VWF | von Willebrand factor | 8.880042 | 0.512627 | 4 | 0.594925 | 0.548181 |
| Signaling downstream of RAS mutants | RAP1A | RAP1A, member of RAS oncogene family, Ras-related protein Rap-1A, Uncharacterized protein | 8.880042 | 0.512627 | 4 | 0.594925 | 0.530823 |
| Signaling downstream of RAS mutants | YWHAB | 14-3-3 protein beta/alpha, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta | 8.880042 | 0.512627 | 4 | 0.594925 | 0.513843 |
| Signaling downstream of RAS mutants | FN1 | Fibronectin | 8.880042 | 0.512627 | 4 | 0.594925 | 0.578096 |
| Signaling downstream of RAS mutants | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 8.880042 | 0.512627 | 4 | 0.594925 | 0.541254 |
| Signaling downstream of RAS mutants | RAF1 | Non-specific serine/threonine protein kinase, RAF proto-oncogene serine/threonine-protein kinase | 8.880042 | 0.512627 | 4 | 0.594925 | 0.573556 |
| Signaling downstream of RAS mutants | JAK2 | Janus kinase 2, Tyrosine-protein kinase, Tyrosine-protein kinase JAK2 | 8.880042 | 0.512627 | 4 | 0.594925 | 0.573235 |
| Signaling downstream of RAS mutants | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 8.880042 | 0.512627 | 4 | 0.594925 | 0.594807 |
| Signalling to RAS | NTRK1 | High affinity nerve growth factor receptor, Tyrosine-protein kinase receptor | 7.990193 | 0.647441 | 4 | 0.616426 | 0.527885 |
| Signalling to RAS | SHC1 | SHC-transforming protein 1 | 7.990193 | 0.647441 | 4 | 0.616426 | 0.521858 |
| Signalling to RAS | MAPK13 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 13 | 7.990193 | 0.647441 | 4 | 0.616426 | 0.573201 |
| Signalling to RAS | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 7.990193 | 0.647441 | 4 | 0.616426 | 0.594807 |
| Signalling to RAS | MAPK11 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 11 | 7.990193 | 0.647441 | 4 | 0.616426 | 0.567539 |
| Signalling to RAS | MAPK12 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 12 | 7.990193 | 0.647441 | 4 | 0.616426 | 0.573393 |
| Signalling to RAS | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 7.990193 | 0.647441 | 4 | 0.616426 | 0.505988 |
| Signalling to RAS | SHC | DShc protein | 7.990193 | 0.647441 | 4 | 0.616426 | 0.605 |
| Signalling to RAS | MAPK14 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 14 | 7.990193 | 0.647441 | 4 | 0.616426 | 0.591941 |
| SRP-dependent cotranslational protein targeting to membrane | RPS5 | 40S ribosomal protein S5, 40S ribosomal protein S5 [Cleaved into: 40S ribosomal protein S5, N-terminally processed], 40S ribosomal protein S5-A, Ribosomal protein S5 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS24 | 40S ribosomal protein S24 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL31 | 60S ribosomal protein L31 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL37 | 60S ribosomal protein L37, Ribosomal protein L37 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS23 | 40S ribosomal protein S23, 40S ribosomal protein S23-A | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL18A | 60S ribosomal protein L18-A, 60S ribosomal protein L18a | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS8 | 40S ribosomal protein S8 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL38 | 60S ribosomal protein L38 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL36A | 60S ribosomal protein L36-A, 60S ribosomal protein L36a, IP17351p, ribosomal protein L36a, Uncharacterized protein | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS20 | 40S ribosomal protein S20 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL10A | 60S ribosomal protein L10a, Ribosomal protein | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS16 | 40S ribosomal protein S16, Ribosomal protein S16, Uncharacterized protein | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL7 | 60S ribosomal protein L7, MGC79754 protein, Ribosomal protein L7, Uncharacterized protein | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL28 | 60S ribosomal protein L28 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL27A | 60S ribosomal protein L27-A, 60S ribosomal protein L27a | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS25 | 40S ribosomal protein S25 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL39 | 60S ribosomal protein L39, LOC100135132 protein, Ribosomal protein L39 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL29 | 60S ribosomal protein L29 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL14 | 60S ribosomal protein L14 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS26 | 40S ribosomal protein S26 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL34 | 60S ribosomal protein L34 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL23 | 60S ribosomal protein L23 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL19 | 60S ribosomal protein L19, Ribosomal protein L19 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS9 | 40S ribosomal protein S9 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL13A | 60S ribosomal protein L13a | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS13 | 40S ribosomal protein S13 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS15 | 40S ribosomal protein S15 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPSA | 40S ribosomal protein SA | 12.008381 | 0.90445 | 4 | 0.952225 | 0.521531 |
| SRP-dependent cotranslational protein targeting to membrane | RPS14 | 40S ribosomal protein S14, Ribosomal protein S14, Uncharacterized protein | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL27 | 60S ribosomal protein L27 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS29 | 40S ribosomal protein S29 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL23A | 60S ribosomal protein L23-A, 60S ribosomal protein L23a, FI01658p, MGC89958 protein, Ribosomal protein L23a, Ribosomal_L23eN domain-containing protein | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL17 | 60S ribosomal protein L17 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL5 | 60S ribosomal protein L5, Ribosomal protein L5 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS27 | 40S ribosomal protein S27 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.549907 |
| SRP-dependent cotranslational protein targeting to membrane | RPS7 | 40S ribosomal protein S7 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL30 | 60S ribosomal protein L30 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL36 | 60S ribosomal protein L36 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS17 | 40S ribosomal protein S17 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS6 | 40S ribosomal protein S6 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.566528 |
| SRP-dependent cotranslational protein targeting to membrane | RPS18 | 40S ribosomal protein S18 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS12 | 40S ribosomal protein S12 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL8 | 60S ribosomal protein L8 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL9 | 60S ribosomal protein L9 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL37A | 60S ribosomal protein L37-A, 60S ribosomal protein L37a, MGC89854 protein, Probable 60S ribosomal protein L37-A, Ribosomal protein L37a | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL13 | 60S ribosomal protein L13 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPLP0 | 60S acidic ribosomal protein P0 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL32 | 60S ribosomal protein L32, Hypothetical LOC496894, Ribosomal protein L32, Uncharacterized protein | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS3 | 40S ribosomal protein S3, DNA-(apurinic or apyrimidinic site) lyase | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL15 | 60S ribosomal protein L15, Ribosomal protein L15 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS28 | 40S ribosomal protein S28 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS3A | 40S ribosomal protein S3a | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL41 | 60S ribosomal protein L41 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS10 | 40S ribosomal protein S10, Ribosomal protein S10, S10_plectin domain-containing protein | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL35 | 60S ribosomal protein L35 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL35A | 60S ribosomal protein L35-A, 60S ribosomal protein L35a | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS4Y1 | 40S ribosomal protein S4, 40S ribosomal protein S4, Y isoform 1 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL10 | 60S ribosomal protein L10, MGC89303 protein | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPLP1 | 60S acidic ribosomal protein P1, Ribosomal protein, large, P1 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL24 | 60S ribosomal protein L24, Ribosomal protein L24, TRASH domain-containing protein | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL26 | 60S ribosomal protein L26, GEO07453p1, KOW domain-containing protein, Ribosomal protein L26 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL11 | 60S ribosomal protein L11 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS21 | 40S ribosomal protein S21 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS19 | 40S ribosomal protein S19, Uncharacterized protein | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL12 | 60S ribosomal protein L12 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL3 | 60S ribosomal protein L3, LOC100145083 protein, Ribosomal protein L3, Uncharacterized protein | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | FAU | 40S ribosomal protein S30 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS4X | 40S ribosomal protein S4, 40S ribosomal protein S4, X isoform | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL22 | 60S ribosomal protein L22, 60S ribosomal protein L22 1, Ribosomal protein L22, Uncharacterized protein | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL21 | 60S ribosomal protein L21 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL7A | 60S ribosomal protein L7-A, 60S ribosomal protein L7a | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS11 | 40S ribosomal protein S11 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL18 | 60S ribosomal protein L18, Ribosomal protein L18 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL6 | 60S ribosomal protein L6 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS15A | 40S ribosomal protein S15a | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPS2 | 40S ribosomal protein S2 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| SRP-dependent cotranslational protein targeting to membrane | RPL4 | 60S ribosomal protein L4, Ribos_L4_asso_C domain-containing protein, Ribosomal protein L4 | 12.008381 | 0.90445 | 4 | 0.952225 | 0.579609 |
| Stabilization of p53 | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.577847 |
| Stabilization of p53 | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.546881 |
| Stabilization of p53 | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.515416 |
| Stabilization of p53 | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.54735 |
| Stabilization of p53 | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.54735 |
| Stabilization of p53 | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.515416 |
| Stabilization of p53 | MDM4 | Double minute 4 protein, Protein Mdm4 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.510741 |
| Stabilization of p53 | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.573417 |
| Stabilization of p53 | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.515416 |
| Stabilization of p53 | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.54735 |
| Stabilization of p53 | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.54735 |
| Stabilization of p53 | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.58858 |
| Stabilization of p53 | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.515416 |
| Stabilization of p53 | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.54735 |
| Stabilization of p53 | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.547953 |
| Stabilization of p53 | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.572294 |
| Stabilization of p53 | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.515416 |
| Stabilization of p53 | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.515416 |
| Stabilization of p53 | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 8.673342 | 0.777853 | 4 | 0.716875 | 0.515416 |
| Stabilization of p53 | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.577481 |
| Stabilization of p53 | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.54735 |
| Stabilization of p53 | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 8.673342 | 0.777853 | 4 | 0.716875 | 0.579609 |
| Stabilization of p53 | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.57244 |
| Stabilization of p53 | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.54735 |
| Stabilization of p53 | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.515416 |
| Stabilization of p53 | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.515416 |
| Stabilization of p53 | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.5197 |
| Stabilization of p53 | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.565601 |
| Stabilization of p53 | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.54735 |
| Stabilization of p53 | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.515416 |
| Stabilization of p53 | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 8.673342 | 0.777853 | 4 | 0.716875 | 0.515416 |
| Stabilization of p53 | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 8.673342 | 0.777853 | 4 | 0.716875 | 0.568645 |
| Stabilization of p53 | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.54735 |
| Stabilization of p53 | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.515416 |
| Stabilization of p53 | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.515416 |
| Stabilization of p53 | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.515416 |
| Stabilization of p53 | TP53 | Cellular tumor antigen p53 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.525937 |
| Stabilization of p53 | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.54735 |
| Stabilization of p53 | MDM2 | E3 ubiquitin-protein ligase Mdm2 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.659771 |
| Stabilization of p53 | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 8.673342 | 0.777853 | 4 | 0.716875 | 0.515416 |
| Switching of origins to a post-replicative state | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.54735 |
| Switching of origins to a post-replicative state | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.515416 |
| Switching of origins to a post-replicative state | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.515416 |
| Switching of origins to a post-replicative state | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.515416 |
| Switching of origins to a post-replicative state | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.577481 |
| Switching of origins to a post-replicative state | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.54735 |
| Switching of origins to a post-replicative state | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 8.757997 | 0.744394 | 4 | 0.704512 | 0.515416 |
| Switching of origins to a post-replicative state | CDK2 | Cyclin dependent kinase 2, Cyclin-dependent kinase 2, Protein kinase domain-containing protein, Uncharacterized protein | 8.757997 | 0.744394 | 4 | 0.704512 | 0.532011 |
| Switching of origins to a post-replicative state | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.515416 |
| Switching of origins to a post-replicative state | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.54735 |
| Switching of origins to a post-replicative state | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.57244 |
| Switching of origins to a post-replicative state | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.515416 |
| Switching of origins to a post-replicative state | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.54735 |
| Switching of origins to a post-replicative state | CCNA1 | Cyclin A1, Cyclin-A1, Cyclin-A2 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.506777 |
| Switching of origins to a post-replicative state | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.54735 |
| Switching of origins to a post-replicative state | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.54735 |
| Switching of origins to a post-replicative state | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 8.757997 | 0.744394 | 4 | 0.704512 | 0.515416 |
| Switching of origins to a post-replicative state | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.515416 |
| Switching of origins to a post-replicative state | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.577847 |
| Switching of origins to a post-replicative state | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.5197 |
| Switching of origins to a post-replicative state | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 8.757997 | 0.744394 | 4 | 0.704512 | 0.568645 |
| Switching of origins to a post-replicative state | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.515416 |
| Switching of origins to a post-replicative state | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.54735 |
| Switching of origins to a post-replicative state | CCNE2 | Cyclin E2, Cyclin N-terminal domain-containing protein, G1/S-specific cyclin-E2 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.503983 |
| Switching of origins to a post-replicative state | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.54735 |
| Switching of origins to a post-replicative state | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.565601 |
| Switching of origins to a post-replicative state | CCNE1 | Cyclin E1, Cyclin N-terminal domain-containing protein, G1/S-specific cyclin-E1 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.526741 |
| Switching of origins to a post-replicative state | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.573417 |
| Switching of origins to a post-replicative state | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.58858 |
| Switching of origins to a post-replicative state | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.546881 |
| Switching of origins to a post-replicative state | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.547953 |
| Switching of origins to a post-replicative state | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.572294 |
| Switching of origins to a post-replicative state | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 8.757997 | 0.744394 | 4 | 0.704512 | 0.579609 |
| Switching of origins to a post-replicative state | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.54735 |
| Switching of origins to a post-replicative state | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.515416 |
| Switching of origins to a post-replicative state | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.54735 |
| Switching of origins to a post-replicative state | CDK1 | Cell division control protein 2, Cyclin dependent kinase 1, Cyclin-dependent kinase 1 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.514008 |
| Switching of origins to a post-replicative state | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.515416 |
| Switching of origins to a post-replicative state | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.515416 |
| Switching of origins to a post-replicative state | CCNA2 | Cyclin-A2 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.527977 |
| Switching of origins to a post-replicative state | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.515416 |
| Switching of origins to a post-replicative state | MCM4 | DNA helicase, DNA replication licensing factor mcm4, DNA replication licensing factor MCM4 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.515269 |
| Switching of origins to a post-replicative state | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.515416 |
| Switching of origins to a post-replicative state | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 8.757997 | 0.744394 | 4 | 0.704512 | 0.515416 |
| Synthesis and processing of ENV and VPU | ENV | Envelope glycoprotein gp160, Truncated surface protein | 8.772297 | 0.997261 | 2.000945 | 0.416038 | 0.538307 |
| Synthesis and processing of ENV and VPU | FURIN | Furin, Furin, paired basic amino acid cleaving enzyme, Trans Golgi network protease furin | 8.772297 | 0.997261 | 2.000945 | 0.416038 | 0.715659 |
| Tachykinin receptors bind tachykinins | TACR3 | Neuromedin-K receptor, NK-3 receptor | 9.89071 | 0.806464 | 2.416758 | 0.479716 | 0.602621 |
| Tachykinin receptors bind tachykinins | TACR1 | NK-1 receptor, Substance-P receptor | 9.89071 | 0.806464 | 2.416758 | 0.479716 | 0.738091 |
| Tachykinin receptors bind tachykinins | TACR2 | NK-2 receptor, Substance-K receptor | 9.89071 | 0.806464 | 2.416758 | 0.479716 | 0.601846 |
| TCF dependent signaling in response to WNT | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.515416 |
| TCF dependent signaling in response to WNT | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.589663 |
| TCF dependent signaling in response to WNT | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.515416 |
| TCF dependent signaling in response to WNT | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 7.909926 | 0.539611 | 4 | 0.55837 | 0.568645 |
| TCF dependent signaling in response to WNT | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.58858 |
| TCF dependent signaling in response to WNT | YWHAZ | 14_3_3 domain-containing protein, 14-3-3 protein zeta, 14-3-3 protein zeta/delta, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta | 7.909926 | 0.539611 | 4 | 0.55837 | 0.602005 |
| TCF dependent signaling in response to WNT | CTNNB1 | Catenin, Catenin beta-1, Uncharacterized protein | 7.909926 | 0.539611 | 4 | 0.55837 | 0.657916 |
| TCF dependent signaling in response to WNT | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.54735 |
| TCF dependent signaling in response to WNT | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.54735 |
| TCF dependent signaling in response to WNT | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.515416 |
| TCF dependent signaling in response to WNT | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.54735 |
| TCF dependent signaling in response to WNT | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.565601 |
| TCF dependent signaling in response to WNT | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.577847 |
| TCF dependent signaling in response to WNT | CSNK2B | Casein kinase II subunit beta | 7.909926 | 0.539611 | 4 | 0.55837 | 0.537688 |
| TCF dependent signaling in response to WNT | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.515416 |
| TCF dependent signaling in response to WNT | PDE10A | cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A, Histone acetyltransferase, Phosphodiesterase | 7.909926 | 0.539611 | 4 | 0.55837 | 0.660672 |
| TCF dependent signaling in response to WNT | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.54735 |
| TCF dependent signaling in response to WNT | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.577481 |
| TCF dependent signaling in response to WNT | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.54735 |
| TCF dependent signaling in response to WNT | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.515416 |
| TCF dependent signaling in response to WNT | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.515416 |
| TCF dependent signaling in response to WNT | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.572294 |
| TCF dependent signaling in response to WNT | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.515416 |
| TCF dependent signaling in response to WNT | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.5197 |
| TCF dependent signaling in response to WNT | XIAP | E3 ubiquitin-protein ligase XIAP, RING-type domain-containing protein, X-linked inhibitor of apoptosis | 7.909926 | 0.539611 | 4 | 0.55837 | 0.589894 |
| TCF dependent signaling in response to WNT | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 7.909926 | 0.539611 | 4 | 0.55837 | 0.515416 |
| TCF dependent signaling in response to WNT | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.54735 |
| TCF dependent signaling in response to WNT | DVL2 | Dishevelled segment polarity protein 2, Segment polarity protein dishevelled homolog DVL-2, Uncharacterized protein | 7.909926 | 0.539611 | 4 | 0.55837 | 0.529242 |
| TCF dependent signaling in response to WNT | MEN1 | Actin like 6A, Menin | 7.909926 | 0.539611 | 4 | 0.55837 | 0.517021 |
| TCF dependent signaling in response to WNT | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 7.909926 | 0.539611 | 4 | 0.55837 | 0.579609 |
| TCF dependent signaling in response to WNT | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.54735 |
| TCF dependent signaling in response to WNT | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.515416 |
| TCF dependent signaling in response to WNT | PPP2R5A | Serine/threonine protein phosphatase 2A regulatory subunit, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform | 7.909926 | 0.539611 | 4 | 0.55837 | 0.559184 |
| TCF dependent signaling in response to WNT | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.515416 |
| TCF dependent signaling in response to WNT | CSNK2A1 | Casein kinase 2, alpha 1 polypeptide, Casein kinase II subunit alpha, Protein kinase domain-containing protein | 7.909926 | 0.539611 | 4 | 0.55837 | 0.538737 |
| TCF dependent signaling in response to WNT | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.54735 |
| TCF dependent signaling in response to WNT | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.546881 |
| TCF dependent signaling in response to WNT | HDAC1 | Histone deacetylase 1, Histone deacetylase HDAC1 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.540105 |
| TCF dependent signaling in response to WNT | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.515416 |
| TCF dependent signaling in response to WNT | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.515416 |
| TCF dependent signaling in response to WNT | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.573417 |
| TCF dependent signaling in response to WNT | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.54735 |
| TCF dependent signaling in response to WNT | TNKS | Poly [ADP-ribose] polymerase, Poly [ADP-ribose] polymerase tankyrase, Poly [ADP-ribose] polymerase tankyrase-1 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.506213 |
| TCF dependent signaling in response to WNT | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 7.909926 | 0.539611 | 4 | 0.55837 | 0.552041 |
| TCF dependent signaling in response to WNT | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.547953 |
| TCF dependent signaling in response to WNT | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.54735 |
| TCF dependent signaling in response to WNT | TCF7L2 | HMG box domain-containing protein, Transcription factor 7 like 2, Transcription factor 7-like 2 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.688556 |
| TCF dependent signaling in response to WNT | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.515416 |
| TCF dependent signaling in response to WNT | CSNK2A2 | Casein kinase 2 alpha 2, Casein kinase II subunit alpha' | 7.909926 | 0.539611 | 4 | 0.55837 | 0.53622 |
| TCF dependent signaling in response to WNT | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.515416 |
| TCF dependent signaling in response to WNT | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 7.909926 | 0.539611 | 4 | 0.55837 | 0.57244 |
| TCF dependent signaling in response to WNT | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 7.909926 | 0.539611 | 4 | 0.55837 | 0.515416 |
| The NLRP1 inflammasome | BCL2L1 | Apoptosis regulator Bcl-X, Bcl-2-like protein 1, BCL2-like 1 | 11.623954 | 0.913099 | 2.8597 | 0.669683 | 0.865729 |
| The NLRP1 inflammasome | BCL2 | Apoptosis regulator Bcl-2 | 11.623954 | 0.913099 | 2.8597 | 0.669683 | 1 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.776258 | 0.740758 | 4 | 0.755226 | 0.579609 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.54735 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.515416 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.565601 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.515416 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.572294 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | HSP90AA1 | Heat shock protein 90 alpha family class A member 1, Heat shock protein HSP 90-alpha | 9.776258 | 0.740758 | 4 | 0.755226 | 0.624325 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.515416 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.515416 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.776258 | 0.740758 | 4 | 0.755226 | 0.515416 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.515416 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.54735 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.577481 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.54735 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.515416 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.515416 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.5197 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.776258 | 0.740758 | 4 | 0.755226 | 0.568645 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | TP53 | Cellular tumor antigen p53 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.525937 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | HSP90AB1 | Heat shock protein 90 alpha family class B member 1, Heat shock protein 90kDa alpha, Heat shock protein HSP 90-beta, Heat shock protein HSP 90-beta isoform a | 9.776258 | 0.740758 | 4 | 0.755226 | 0.569354 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.546881 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.54735 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PLK1 | Serine/threonine-protein kinase PLK, Serine/threonine-protein kinase PLK1 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.504187 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.57244 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.776258 | 0.740758 | 4 | 0.755226 | 0.515416 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.515416 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.54735 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.54735 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.573417 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.58858 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.54735 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.54735 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.54735 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.515416 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | CCNB1 | Cyclin B1, G2/mitotic-specific cyclin-B1, Uncharacterized protein | 9.776258 | 0.740758 | 4 | 0.755226 | 0.513205 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.547953 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.515416 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.577847 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.54735 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.515416 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.515416 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.515416 |
| The role of GTSE1 in G2/M progression after G2 checkpoint | CDK1 | Cell division control protein 2, Cyclin dependent kinase 1, Cyclin-dependent kinase 1 | 9.776258 | 0.740758 | 4 | 0.755226 | 0.514008 |
| TNFR2 non-canonical NF-kB pathway | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.515416 |
| TNFR2 non-canonical NF-kB pathway | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.515416 |
| TNFR2 non-canonical NF-kB pathway | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.54735 |
| TNFR2 non-canonical NF-kB pathway | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.58858 |
| TNFR2 non-canonical NF-kB pathway | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.515416 |
| TNFR2 non-canonical NF-kB pathway | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.54735 |
| TNFR2 non-canonical NF-kB pathway | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.54735 |
| TNFR2 non-canonical NF-kB pathway | MAP3K14 | Mitogen-activated protein kinase kinase kinase 14 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.556123 |
| TNFR2 non-canonical NF-kB pathway | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.658667 | 0.670185 | 4 | 0.713873 | 0.515416 |
| TNFR2 non-canonical NF-kB pathway | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.54735 |
| TNFR2 non-canonical NF-kB pathway | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.515416 |
| TNFR2 non-canonical NF-kB pathway | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.54735 |
| TNFR2 non-canonical NF-kB pathway | BIRC2 | Baculoviral IAP repeat containing 2, Baculoviral IAP repeat-containing 2, Baculoviral IAP repeat-containing protein 2 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.565892 |
| TNFR2 non-canonical NF-kB pathway | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.515416 |
| TNFR2 non-canonical NF-kB pathway | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.57244 |
| TNFR2 non-canonical NF-kB pathway | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.515416 |
| TNFR2 non-canonical NF-kB pathway | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.572294 |
| TNFR2 non-canonical NF-kB pathway | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.515416 |
| TNFR2 non-canonical NF-kB pathway | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.515416 |
| TNFR2 non-canonical NF-kB pathway | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.515416 |
| TNFR2 non-canonical NF-kB pathway | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.658667 | 0.670185 | 4 | 0.713873 | 0.568645 |
| TNFR2 non-canonical NF-kB pathway | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.546881 |
| TNFR2 non-canonical NF-kB pathway | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.577481 |
| TNFR2 non-canonical NF-kB pathway | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.54735 |
| TNFR2 non-canonical NF-kB pathway | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.515416 |
| TNFR2 non-canonical NF-kB pathway | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.515416 |
| TNFR2 non-canonical NF-kB pathway | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.565601 |
| TNFR2 non-canonical NF-kB pathway | XIAP | E3 ubiquitin-protein ligase XIAP, RING-type domain-containing protein, X-linked inhibitor of apoptosis | 9.658667 | 0.670185 | 4 | 0.713873 | 0.589894 |
| TNFR2 non-canonical NF-kB pathway | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.515416 |
| TNFR2 non-canonical NF-kB pathway | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.54735 |
| TNFR2 non-canonical NF-kB pathway | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.658667 | 0.670185 | 4 | 0.713873 | 0.515416 |
| TNFR2 non-canonical NF-kB pathway | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.54735 |
| TNFR2 non-canonical NF-kB pathway | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.54735 |
| TNFR2 non-canonical NF-kB pathway | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.54735 |
| TNFR2 non-canonical NF-kB pathway | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.547953 |
| TNFR2 non-canonical NF-kB pathway | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.577847 |
| TNFR2 non-canonical NF-kB pathway | BIRC3 | Baculoviral IAP repeat-containing 3, Baculoviral IAP repeat-containing protein 3, Uncharacterized protein | 9.658667 | 0.670185 | 4 | 0.713873 | 0.605019 |
| TNFR2 non-canonical NF-kB pathway | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.515416 |
| TNFR2 non-canonical NF-kB pathway | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.5197 |
| TNFR2 non-canonical NF-kB pathway | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.658667 | 0.670185 | 4 | 0.713873 | 0.573417 |
| TNFR2 non-canonical NF-kB pathway | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.658667 | 0.670185 | 4 | 0.713873 | 0.579609 |
| Toll Like Receptor TLR1:TLR2 Cascade | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 10.161622 | 0.393425 | 3.531766 | 0.531036 | 0.579609 |
| Toll Like Receptor TLR1:TLR2 Cascade | MDM2 | E3 ubiquitin-protein ligase Mdm2 | 10.161622 | 0.393425 | 3.531766 | 0.531036 | 0.659771 |
| Toll Like Receptor TLR1:TLR2 Cascade | BTK | Tyrosine-protein kinase, Tyrosine-protein kinase BTK | 10.161622 | 0.393425 | 3.531766 | 0.531036 | 0.602174 |
| Toll Like Receptor TLR1:TLR2 Cascade | UBE2V1 | Ubiquitin conjugating enzyme E2 V1, Ubiquitin-conjugating enzyme E2 variant 1 | 10.161622 | 0.393425 | 3.531766 | 0.531036 | 0.593364 |
| Toll Like Receptor TLR1:TLR2 Cascade | MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | 10.161622 | 0.393425 | 3.531766 | 0.531036 | 0.519182 |
| Toll Like Receptor TLR1:TLR2 Cascade | MAPK14 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 14 | 10.161622 | 0.393425 | 3.531766 | 0.531036 | 0.591941 |
| Toll Like Receptor TLR1:TLR2 Cascade | IKBKB | I-kappa-B kinase, Inhibitor of nuclear factor kappa-B kinase subunit beta | 10.161622 | 0.393425 | 3.531766 | 0.531036 | 0.518113 |
| Toll Like Receptor TLR1:TLR2 Cascade | APP | ABPP, Amyloid-beta A4 protein, Amyloid-beta precursor protein | 10.161622 | 0.393425 | 3.531766 | 0.531036 | 0.506343 |
| Toll Like Receptor TLR1:TLR2 Cascade | MAP2K1 | Dual specificity mitogen-activated protein kinase kinase 1, MAP2K1 protein, Mitogen-activated protein kinase kinase 1, Protein kinase domain-containing protein | 10.161622 | 0.393425 | 3.531766 | 0.531036 | 0.545374 |
| Toll Like Receptor TLR1:TLR2 Cascade | MAPK11 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 11 | 10.161622 | 0.393425 | 3.531766 | 0.531036 | 0.567539 |
| Toll Like Receptor TLR1:TLR2 Cascade | MAPK10 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 10 | 10.161622 | 0.393425 | 3.531766 | 0.531036 | 0.506109 |
| Toll Like Receptor TLR1:TLR2 Cascade | RIPK2 | Receptor-interacting serine-threonine kinase 2, Receptor-interacting serine/threonine-protein kinase 2 | 10.161622 | 0.393425 | 3.531766 | 0.531036 | 0.530344 |
| Toll Like Receptor TLR1:TLR2 Cascade | IRAK4 | Interleukin-1 receptor-associated kinase 4 | 10.161622 | 0.393425 | 3.531766 | 0.531036 | 0.50686 |
| Toll Like Receptor TLR1:TLR2 Cascade | MAPK8 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 8 | 10.161622 | 0.393425 | 3.531766 | 0.531036 | 0.503967 |
| Toll Like Receptor TLR1:TLR2 Cascade | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 10.161622 | 0.393425 | 3.531766 | 0.531036 | 0.541254 |
| Toll Like Receptor TLR6:TLR2 Cascade | IRAK4 | Interleukin-1 receptor-associated kinase 4 | 7.70401 | 0.39878 | 3.38468 | 0.403903 | 0.50686 |
| Toll Like Receptor TLR6:TLR2 Cascade | MAPK11 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 11 | 7.70401 | 0.39878 | 3.38468 | 0.403903 | 0.567539 |
| Toll Like Receptor TLR6:TLR2 Cascade | MAPK10 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 10 | 7.70401 | 0.39878 | 3.38468 | 0.403903 | 0.506109 |
| Toll Like Receptor TLR6:TLR2 Cascade | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 7.70401 | 0.39878 | 3.38468 | 0.403903 | 0.579609 |
| Toll Like Receptor TLR6:TLR2 Cascade | MAPK14 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 14 | 7.70401 | 0.39878 | 3.38468 | 0.403903 | 0.591941 |
| Toll Like Receptor TLR6:TLR2 Cascade | UBE2V1 | Ubiquitin conjugating enzyme E2 V1, Ubiquitin-conjugating enzyme E2 variant 1 | 7.70401 | 0.39878 | 3.38468 | 0.403903 | 0.593364 |
| Toll Like Receptor TLR6:TLR2 Cascade | MAP2K1 | Dual specificity mitogen-activated protein kinase kinase 1, MAP2K1 protein, Mitogen-activated protein kinase kinase 1, Protein kinase domain-containing protein | 7.70401 | 0.39878 | 3.38468 | 0.403903 | 0.545374 |
| Toll Like Receptor TLR6:TLR2 Cascade | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 7.70401 | 0.39878 | 3.38468 | 0.403903 | 0.541254 |
| Toll Like Receptor TLR6:TLR2 Cascade | RIPK2 | Receptor-interacting serine-threonine kinase 2, Receptor-interacting serine/threonine-protein kinase 2 | 7.70401 | 0.39878 | 3.38468 | 0.403903 | 0.530344 |
| Toll Like Receptor TLR6:TLR2 Cascade | BTK | Tyrosine-protein kinase, Tyrosine-protein kinase BTK | 7.70401 | 0.39878 | 3.38468 | 0.403903 | 0.602174 |
| Toll Like Receptor TLR6:TLR2 Cascade | IKBKB | I-kappa-B kinase, Inhibitor of nuclear factor kappa-B kinase subunit beta | 7.70401 | 0.39878 | 3.38468 | 0.403903 | 0.518113 |
| Toll Like Receptor TLR6:TLR2 Cascade | MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | 7.70401 | 0.39878 | 3.38468 | 0.403903 | 0.519182 |
| Toll Like Receptor TLR6:TLR2 Cascade | APP | ABPP, Amyloid-beta A4 protein, Amyloid-beta precursor protein | 7.70401 | 0.39878 | 3.38468 | 0.403903 | 0.506343 |
| Toll Like Receptor TLR6:TLR2 Cascade | MDM2 | E3 ubiquitin-protein ligase Mdm2 | 7.70401 | 0.39878 | 3.38468 | 0.403903 | 0.659771 |
| Toll Like Receptor TLR6:TLR2 Cascade | MAPK8 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 8 | 7.70401 | 0.39878 | 3.38468 | 0.403903 | 0.503967 |
| TP53 Regulates Metabolic Genes | TP53 | Cellular tumor antigen p53 | 9.063348 | 0.479922 | 3.561571 | 0.523577 | 0.525937 |
| TP53 Regulates Metabolic Genes | PRKAB1 | 5'-AMP-activated protein kinase subunit beta-1, 5'AMP-activated protein kinase beta-1 non-catalytic subunit, Protein kinase AMP-activated non-catalytic subunit beta 1, Protein kinase, AMP-activated, beta 1 non-catalytic subunit | 9.063348 | 0.479922 | 3.561571 | 0.523577 | 0.500481 |
| TP53 Regulates Metabolic Genes | MTOR | Nucleoprotein TPR, Serine/threonine-protein kinase mTOR, Serine/threonine-protein kinase TOR | 9.063348 | 0.479922 | 3.561571 | 0.523577 | 0.619026 |
| TP53 Regulates Metabolic Genes | PRKAG2 | 5'-AMP-activated protein kinase subunit gamma-2, 5'-AMP-activated protein kinase subunit gamma-2 isoform X8, Protein kinase AMP-activated non-catalytic subunit gamma 2, Protein kinase, AMP-activated, gamma 2 non-catalytic subunit, Uncharacterized protein | 9.063348 | 0.479922 | 3.561571 | 0.523577 | 0.510193 |
| TP53 Regulates Metabolic Genes | CRK | 14_3_3 domain-containing protein, Adapter molecule crk, Adapter molecule Crk, CRK proto-oncogene, adaptor protein, Uncharacterized protein, V-crk avian sarcoma virus CT10 oncogene homolog | 9.063348 | 0.479922 | 3.561571 | 0.523577 | 0.513369 |
| TP53 Regulates Metabolic Genes | PRKAG1 | 5'-AMP-activated protein kinase subunit gamma-1, Protein kinase, AMP-activated, gamma 1 non-catalytic subunit | 9.063348 | 0.479922 | 3.561571 | 0.523577 | 0.515113 |
| TP53 Regulates Metabolic Genes | RPTOR | Regulatory associated protein of MTOR complex 1, Regulatory associated protein of MTOR, complex 1, Regulatory-associated protein of mTOR, Regulatory-associated protein of MTOR, complex 1, WD_REPEATS_REGION domain-containing protein | 9.063348 | 0.479922 | 3.561571 | 0.523577 | 0.543357 |
| TP53 Regulates Metabolic Genes | YWHAZ | 14_3_3 domain-containing protein, 14-3-3 protein zeta, 14-3-3 protein zeta/delta, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta | 9.063348 | 0.479922 | 3.561571 | 0.523577 | 0.602005 |
| TP53 Regulates Metabolic Genes | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 9.063348 | 0.479922 | 3.561571 | 0.523577 | 0.552041 |
| TP53 Regulates Metabolic Genes | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 9.063348 | 0.479922 | 3.561571 | 0.523577 | 0.589663 |
| TP53 Regulates Metabolic Genes | YWHAB | 14-3-3 protein beta/alpha, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta | 9.063348 | 0.479922 | 3.561571 | 0.523577 | 0.513843 |
| TP53 Regulates Transcription of Genes Involved in G1 Cell Cycle Arrest | CCNE2 | Cyclin E2, Cyclin N-terminal domain-containing protein, G1/S-specific cyclin-E2 | 7.466186 | 0.675665 | 2.806881 | 0.423491 | 0.503983 |
| TP53 Regulates Transcription of Genes Involved in G1 Cell Cycle Arrest | CCNA2 | Cyclin-A2 | 7.466186 | 0.675665 | 2.806881 | 0.423491 | 0.527977 |
| TP53 Regulates Transcription of Genes Involved in G1 Cell Cycle Arrest | TP53 | Cellular tumor antigen p53 | 7.466186 | 0.675665 | 2.806881 | 0.423491 | 0.525937 |
| TP53 Regulates Transcription of Genes Involved in G1 Cell Cycle Arrest | CCNE1 | Cyclin E1, Cyclin N-terminal domain-containing protein, G1/S-specific cyclin-E1 | 7.466186 | 0.675665 | 2.806881 | 0.423491 | 0.526741 |
| TP53 Regulates Transcription of Genes Involved in G1 Cell Cycle Arrest | CCNA1 | Cyclin A1, Cyclin-A1, Cyclin-A2 | 7.466186 | 0.675665 | 2.806881 | 0.423491 | 0.506777 |
| TP53 Regulates Transcription of Genes Involved in G1 Cell Cycle Arrest | CDK2 | Cyclin dependent kinase 2, Cyclin-dependent kinase 2, Protein kinase domain-containing protein, Uncharacterized protein | 7.466186 | 0.675665 | 2.806881 | 0.423491 | 0.532011 |
| Transcription of SARS-CoV-1 sgRNAs | 1A | Replicase polyprotein 1a | 12.767545 | 1 | 0.701304 | ||
| Transcription of SARS-CoV-1 sgRNAs | REP | Rab proteins geranylgeranyltransferase component A, Replicase polyprotein 1ab | 12.767545 | 1 | 0.746505 | ||
| Transcriptional regulation by RUNX2 | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.54735 |
| Transcriptional regulation by RUNX2 | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.515416 |
| Transcriptional regulation by RUNX2 | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.546881 |
| Transcriptional regulation by RUNX2 | MMP13 | Collagenase 3 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.515873 |
| Transcriptional regulation by RUNX2 | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.589663 |
| Transcriptional regulation by RUNX2 | NR3C1 | Glucocorticoid receptor | 7.957408 | 0.611292 | 4 | 0.59666 | 0.59851 |
| Transcriptional regulation by RUNX2 | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.577847 |
| Transcriptional regulation by RUNX2 | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 7.957408 | 0.611292 | 4 | 0.59666 | 0.579609 |
| Transcriptional regulation by RUNX2 | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.515416 |
| Transcriptional regulation by RUNX2 | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.515416 |
| Transcriptional regulation by RUNX2 | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.54735 |
| Transcriptional regulation by RUNX2 | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.547953 |
| Transcriptional regulation by RUNX2 | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.565601 |
| Transcriptional regulation by RUNX2 | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.577481 |
| Transcriptional regulation by RUNX2 | CDK4 | Cyclin dependent kinase 4, Cyclin-dependent kinase 4, Cyclin-dependent kinase 4, isoform A | 7.957408 | 0.611292 | 4 | 0.59666 | 0.511296 |
| Transcriptional regulation by RUNX2 | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.54735 |
| Transcriptional regulation by RUNX2 | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.58858 |
| Transcriptional regulation by RUNX2 | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.54735 |
| Transcriptional regulation by RUNX2 | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.515416 |
| Transcriptional regulation by RUNX2 | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.515416 |
| Transcriptional regulation by RUNX2 | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.515416 |
| Transcriptional regulation by RUNX2 | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.5197 |
| Transcriptional regulation by RUNX2 | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 7.957408 | 0.611292 | 4 | 0.59666 | 0.515416 |
| Transcriptional regulation by RUNX2 | HDAC6 | Histone deacetylase 6, Histone deacetylase 6, isoform G | 7.957408 | 0.611292 | 4 | 0.59666 | 0.531859 |
| Transcriptional regulation by RUNX2 | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.54735 |
| Transcriptional regulation by RUNX2 | ABL1 | Tyrosine-protein kinase, Tyrosine-protein kinase ABL1 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.590046 |
| Transcriptional regulation by RUNX2 | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.54735 |
| Transcriptional regulation by RUNX2 | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.515416 |
| Transcriptional regulation by RUNX2 | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.54735 |
| Transcriptional regulation by RUNX2 | MAPK1 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 1 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.541254 |
| Transcriptional regulation by RUNX2 | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.515416 |
| Transcriptional regulation by RUNX2 | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 7.957408 | 0.611292 | 4 | 0.59666 | 0.552041 |
| Transcriptional regulation by RUNX2 | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 7.957408 | 0.611292 | 4 | 0.59666 | 0.594807 |
| Transcriptional regulation by RUNX2 | HDAC4 | Histone deacetylase, Histone deacetylase 4 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.528897 |
| Transcriptional regulation by RUNX2 | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.515416 |
| Transcriptional regulation by RUNX2 | PPARGC1A | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | 7.957408 | 0.611292 | 4 | 0.59666 | 0.539093 |
| Transcriptional regulation by RUNX2 | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.515416 |
| Transcriptional regulation by RUNX2 | YES1 | Tyrosine-protein kinase, Tyrosine-protein kinase yes, Tyrosine-protein kinase Yes | 7.957408 | 0.611292 | 4 | 0.59666 | 0.506641 |
| Transcriptional regulation by RUNX2 | YAP1 | Transcriptional coactivator YAP1, Yes1 associated transcriptional regulator | 7.957408 | 0.611292 | 4 | 0.59666 | 0.628833 |
| Transcriptional regulation by RUNX2 | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.572294 |
| Transcriptional regulation by RUNX2 | ESR1 | Estrogen receptor | 7.957408 | 0.611292 | 4 | 0.59666 | 0.561602 |
| Transcriptional regulation by RUNX2 | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.515416 |
| Transcriptional regulation by RUNX2 | HDAC3 | Histone deacetylase, Histone deacetylase 3 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.528637 |
| Transcriptional regulation by RUNX2 | CCND1 | Cyclin D1, G1/S-specific cyclin-D1 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.525654 |
| Transcriptional regulation by RUNX2 | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.573417 |
| Transcriptional regulation by RUNX2 | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.515416 |
| Transcriptional regulation by RUNX2 | CDK1 | Cell division control protein 2, Cyclin dependent kinase 1, Cyclin-dependent kinase 1 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.514008 |
| Transcriptional regulation by RUNX2 | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 7.957408 | 0.611292 | 4 | 0.59666 | 0.515416 |
| Transcriptional regulation by RUNX2 | CCNB1 | Cyclin B1, G2/mitotic-specific cyclin-B1, Uncharacterized protein | 7.957408 | 0.611292 | 4 | 0.59666 | 0.513205 |
| Transcriptional regulation by RUNX2 | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.54735 |
| Transcriptional regulation by RUNX2 | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.54735 |
| Transcriptional regulation by RUNX2 | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 7.957408 | 0.611292 | 4 | 0.59666 | 0.568645 |
| Transcriptional regulation by RUNX2 | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.54735 |
| Transcriptional regulation by RUNX2 | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.57244 |
| Transcriptional regulation by RUNX2 | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 7.957408 | 0.611292 | 4 | 0.59666 | 0.515416 |
| Translation initiation complex formation | RPS13 | 40S ribosomal protein S13 | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS4X | 40S ribosomal protein S4, 40S ribosomal protein S4, X isoform | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS24 | 40S ribosomal protein S24 | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS23 | 40S ribosomal protein S23, 40S ribosomal protein S23-A | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS6 | 40S ribosomal protein S6 | 10.379304 | 0.787218 | 4 | 0.809566 | 0.566528 |
| Translation initiation complex formation | FAU | 40S ribosomal protein S30 | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS4Y1 | 40S ribosomal protein S4, 40S ribosomal protein S4, Y isoform 1 | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS11 | 40S ribosomal protein S11 | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS16 | 40S ribosomal protein S16, Ribosomal protein S16, Uncharacterized protein | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS3A | 40S ribosomal protein S3a | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS9 | 40S ribosomal protein S9 | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS8 | 40S ribosomal protein S8 | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS2 | 40S ribosomal protein S2 | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPSA | 40S ribosomal protein SA | 10.379304 | 0.787218 | 4 | 0.809566 | 0.521531 |
| Translation initiation complex formation | RPS28 | 40S ribosomal protein S28 | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS26 | 40S ribosomal protein S26 | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS20 | 40S ribosomal protein S20 | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS29 | 40S ribosomal protein S29 | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS10 | 40S ribosomal protein S10, Ribosomal protein S10, S10_plectin domain-containing protein | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS7 | 40S ribosomal protein S7 | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS27 | 40S ribosomal protein S27 | 10.379304 | 0.787218 | 4 | 0.809566 | 0.549907 |
| Translation initiation complex formation | RPS5 | 40S ribosomal protein S5, 40S ribosomal protein S5 [Cleaved into: 40S ribosomal protein S5, N-terminally processed], 40S ribosomal protein S5-A, Ribosomal protein S5 | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS12 | 40S ribosomal protein S12 | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS25 | 40S ribosomal protein S25 | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS19 | 40S ribosomal protein S19, Uncharacterized protein | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS17 | 40S ribosomal protein S17 | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS15A | 40S ribosomal protein S15a | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS15 | 40S ribosomal protein S15 | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS14 | 40S ribosomal protein S14, Ribosomal protein S14, Uncharacterized protein | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS21 | 40S ribosomal protein S21 | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS3 | 40S ribosomal protein S3, DNA-(apurinic or apyrimidinic site) lyase | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translation initiation complex formation | RPS18 | 40S ribosomal protein S18 | 10.379304 | 0.787218 | 4 | 0.809566 | 0.579609 |
| Translocation of SLC2A4 (GLUT4) to the plasma membrane | PRKAB1 | 5'-AMP-activated protein kinase subunit beta-1, 5'AMP-activated protein kinase beta-1 non-catalytic subunit, Protein kinase AMP-activated non-catalytic subunit beta 1, Protein kinase, AMP-activated, beta 1 non-catalytic subunit | 8.949121 | 0.549709 | 4 | 0.61703 | 0.500481 |
| Translocation of SLC2A4 (GLUT4) to the plasma membrane | PRKAG2 | 5'-AMP-activated protein kinase subunit gamma-2, 5'-AMP-activated protein kinase subunit gamma-2 isoform X8, Protein kinase AMP-activated non-catalytic subunit gamma 2, Protein kinase, AMP-activated, gamma 2 non-catalytic subunit, Uncharacterized protein | 8.949121 | 0.549709 | 4 | 0.61703 | 0.510193 |
| Translocation of SLC2A4 (GLUT4) to the plasma membrane | YWHAZ | 14_3_3 domain-containing protein, 14-3-3 protein zeta, 14-3-3 protein zeta/delta, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta | 8.949121 | 0.549709 | 4 | 0.61703 | 0.602005 |
| Translocation of SLC2A4 (GLUT4) to the plasma membrane | YWHAB | 14-3-3 protein beta/alpha, Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta | 8.949121 | 0.549709 | 4 | 0.61703 | 0.513843 |
| Translocation of SLC2A4 (GLUT4) to the plasma membrane | RAB10 | LD39986p, RAB10 protein, RAB10, member RAS oncogene family, Ras-related protein Rab-10 | 8.949121 | 0.549709 | 4 | 0.61703 | 0.527339 |
| Translocation of SLC2A4 (GLUT4) to the plasma membrane | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 8.949121 | 0.549709 | 4 | 0.61703 | 0.589663 |
| Translocation of SLC2A4 (GLUT4) to the plasma membrane | PRKAG1 | 5'-AMP-activated protein kinase subunit gamma-1, Protein kinase, AMP-activated, gamma 1 non-catalytic subunit | 8.949121 | 0.549709 | 4 | 0.61703 | 0.515113 |
| Translocation of SLC2A4 (GLUT4) to the plasma membrane | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 8.949121 | 0.549709 | 4 | 0.61703 | 0.552041 |
| Ub-specific processing proteases | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.462575 | 0.506149 | 4 | 0.621738 | 0.515416 |
| Ub-specific processing proteases | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.577847 |
| Ub-specific processing proteases | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.515416 |
| Ub-specific processing proteases | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.462575 | 0.506149 | 4 | 0.621738 | 0.568645 |
| Ub-specific processing proteases | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.54735 |
| Ub-specific processing proteases | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.546881 |
| Ub-specific processing proteases | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.462575 | 0.506149 | 4 | 0.621738 | 0.515416 |
| Ub-specific processing proteases | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.515416 |
| Ub-specific processing proteases | CCNA1 | Cyclin A1, Cyclin-A1, Cyclin-A2 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.506777 |
| Ub-specific processing proteases | BIRC3 | Baculoviral IAP repeat-containing 3, Baculoviral IAP repeat-containing protein 3, Uncharacterized protein | 9.462575 | 0.506149 | 4 | 0.621738 | 0.605019 |
| Ub-specific processing proteases | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.547953 |
| Ub-specific processing proteases | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.54735 |
| Ub-specific processing proteases | TGFBR1 | Receptor protein serine/threonine kinase, Serine/threonine-protein kinase receptor, TGF-beta receptor type-1 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.504115 |
| Ub-specific processing proteases | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.577481 |
| Ub-specific processing proteases | USP13 | Ubiquitin carboxyl-terminal hydrolase, Ubiquitin carboxyl-terminal hydrolase 13 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.538123 |
| Ub-specific processing proteases | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.515416 |
| Ub-specific processing proteases | MDM2 | E3 ubiquitin-protein ligase Mdm2 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.659771 |
| Ub-specific processing proteases | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.54735 |
| Ub-specific processing proteases | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.54735 |
| Ub-specific processing proteases | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.58858 |
| Ub-specific processing proteases | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.572294 |
| Ub-specific processing proteases | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.515416 |
| Ub-specific processing proteases | BIRC2 | Baculoviral IAP repeat containing 2, Baculoviral IAP repeat-containing 2, Baculoviral IAP repeat-containing protein 2 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.565892 |
| Ub-specific processing proteases | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.573417 |
| Ub-specific processing proteases | CCNA2 | Cyclin-A2 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.527977 |
| Ub-specific processing proteases | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.54735 |
| Ub-specific processing proteases | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.462575 | 0.506149 | 4 | 0.621738 | 0.579609 |
| Ub-specific processing proteases | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.515416 |
| Ub-specific processing proteases | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.54735 |
| Ub-specific processing proteases | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.54735 |
| Ub-specific processing proteases | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.515416 |
| Ub-specific processing proteases | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.515416 |
| Ub-specific processing proteases | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.515416 |
| Ub-specific processing proteases | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.515416 |
| Ub-specific processing proteases | WDR48 | USP1-associated factor 1, WD repeat-containing protein 48 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.519063 |
| Ub-specific processing proteases | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.515416 |
| Ub-specific processing proteases | SMAD3 | Mothers against decapentaplegic homolog, Mothers against decapentaplegic homolog 3 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.502216 |
| Ub-specific processing proteases | TNKS | Poly [ADP-ribose] polymerase, Poly [ADP-ribose] polymerase tankyrase, Poly [ADP-ribose] polymerase tankyrase-1 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.506213 |
| Ub-specific processing proteases | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.54735 |
| Ub-specific processing proteases | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.57244 |
| Ub-specific processing proteases | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.515416 |
| Ub-specific processing proteases | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.515416 |
| Ub-specific processing proteases | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.54735 |
| Ub-specific processing proteases | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.565601 |
| Ub-specific processing proteases | MDM4 | Double minute 4 protein, Protein Mdm4 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.510741 |
| Ub-specific processing proteases | MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.519182 |
| Ub-specific processing proteases | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.54735 |
| Ub-specific processing proteases | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.5197 |
| Ub-specific processing proteases | ADRM1 | Adhesion regulating molecule 1, Oxysterol-binding protein, Proteasomal ubiquitin receptor ADRM1 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.515416 |
| Ub-specific processing proteases | RPS3A | 40S ribosomal protein S3a | 9.462575 | 0.506149 | 4 | 0.621738 | 0.579609 |
| Ub-specific processing proteases | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.515416 |
| Ub-specific processing proteases | TP53 | Cellular tumor antigen p53 | 9.462575 | 0.506149 | 4 | 0.621738 | 0.525937 |
| Ubiquitin-dependent degradation of Cyclin D | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.54735 |
| Ubiquitin-dependent degradation of Cyclin D | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.515416 |
| Ubiquitin-dependent degradation of Cyclin D | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.577481 |
| Ubiquitin-dependent degradation of Cyclin D | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.529994 | 0.793073 | 4 | 0.768679 | 0.568645 |
| Ubiquitin-dependent degradation of Cyclin D | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.529994 | 0.793073 | 4 | 0.768679 | 0.515416 |
| Ubiquitin-dependent degradation of Cyclin D | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.515416 |
| Ubiquitin-dependent degradation of Cyclin D | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.529994 | 0.793073 | 4 | 0.768679 | 0.515416 |
| Ubiquitin-dependent degradation of Cyclin D | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.515416 |
| Ubiquitin-dependent degradation of Cyclin D | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.572294 |
| Ubiquitin-dependent degradation of Cyclin D | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.515416 |
| Ubiquitin-dependent degradation of Cyclin D | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.54735 |
| Ubiquitin-dependent degradation of Cyclin D | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.565601 |
| Ubiquitin-dependent degradation of Cyclin D | CCND1 | Cyclin D1, G1/S-specific cyclin-D1 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.525654 |
| Ubiquitin-dependent degradation of Cyclin D | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.54735 |
| Ubiquitin-dependent degradation of Cyclin D | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.515416 |
| Ubiquitin-dependent degradation of Cyclin D | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.515416 |
| Ubiquitin-dependent degradation of Cyclin D | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.57244 |
| Ubiquitin-dependent degradation of Cyclin D | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.54735 |
| Ubiquitin-dependent degradation of Cyclin D | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.5197 |
| Ubiquitin-dependent degradation of Cyclin D | CDK4 | Cyclin dependent kinase 4, Cyclin-dependent kinase 4, Cyclin-dependent kinase 4, isoform A | 9.529994 | 0.793073 | 4 | 0.768679 | 0.511296 |
| Ubiquitin-dependent degradation of Cyclin D | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.547953 |
| Ubiquitin-dependent degradation of Cyclin D | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.577847 |
| Ubiquitin-dependent degradation of Cyclin D | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.54735 |
| Ubiquitin-dependent degradation of Cyclin D | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.58858 |
| Ubiquitin-dependent degradation of Cyclin D | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.54735 |
| Ubiquitin-dependent degradation of Cyclin D | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.54735 |
| Ubiquitin-dependent degradation of Cyclin D | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.515416 |
| Ubiquitin-dependent degradation of Cyclin D | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.515416 |
| Ubiquitin-dependent degradation of Cyclin D | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.515416 |
| Ubiquitin-dependent degradation of Cyclin D | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.54735 |
| Ubiquitin-dependent degradation of Cyclin D | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.529994 | 0.793073 | 4 | 0.768679 | 0.579609 |
| Ubiquitin-dependent degradation of Cyclin D | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.515416 |
| Ubiquitin-dependent degradation of Cyclin D | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.573417 |
| Ubiquitin-dependent degradation of Cyclin D | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.515416 |
| Ubiquitin-dependent degradation of Cyclin D | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.54735 |
| Ubiquitin-dependent degradation of Cyclin D | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.515416 |
| Ubiquitin-dependent degradation of Cyclin D | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.54735 |
| Ubiquitin-dependent degradation of Cyclin D | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.515416 |
| Ubiquitin-dependent degradation of Cyclin D | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.529994 | 0.793073 | 4 | 0.768679 | 0.546881 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.567097 | 0.765606 | 4 | 0.756859 | 0.515416 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.546881 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.54735 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.54735 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.54735 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.515416 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.515416 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.515416 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.58858 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.515416 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.54735 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.54735 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.567097 | 0.765606 | 4 | 0.756859 | 0.515416 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.57244 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.54735 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.515416 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.54735 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.5197 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.547953 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.515416 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.515416 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.577847 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.515416 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.515416 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.54735 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.577481 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.515416 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PTK2 | Focal adhesion kinase 1, Non-specific protein-tyrosine kinase, Serine/threonine-protein kinase PTK2/STK2 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.503121 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.515416 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.565601 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.54735 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.572294 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.567097 | 0.765606 | 4 | 0.756859 | 0.568645 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.573417 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.515416 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.515416 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.567097 | 0.765606 | 4 | 0.756859 | 0.54735 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.567097 | 0.765606 | 4 | 0.756859 | 0.579609 |
| UCH proteinases | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.54735 |
| UCH proteinases | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.515416 |
| UCH proteinases | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.547953 |
| UCH proteinases | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.54735 |
| UCH proteinases | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.54735 |
| UCH proteinases | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.515416 |
| UCH proteinases | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.54735 |
| UCH proteinases | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.515416 |
| UCH proteinases | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.262568 | 0.689501 | 4 | 0.703096 | 0.568645 |
| UCH proteinases | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.5197 |
| UCH proteinases | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.54735 |
| UCH proteinases | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.54735 |
| UCH proteinases | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.515416 |
| UCH proteinases | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.515416 |
| UCH proteinases | TGFBR1 | Receptor protein serine/threonine kinase, Serine/threonine-protein kinase receptor, TGF-beta receptor type-1 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.504115 |
| UCH proteinases | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.54735 |
| UCH proteinases | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.577847 |
| UCH proteinases | MEN1 | Actin like 6A, Menin | 9.262568 | 0.689501 | 4 | 0.703096 | 0.517021 |
| UCH proteinases | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.565601 |
| UCH proteinases | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.58858 |
| UCH proteinases | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.54735 |
| UCH proteinases | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.54735 |
| UCH proteinases | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.262568 | 0.689501 | 4 | 0.703096 | 0.579609 |
| UCH proteinases | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.515416 |
| UCH proteinases | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.515416 |
| UCH proteinases | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.262568 | 0.689501 | 4 | 0.703096 | 0.515416 |
| UCH proteinases | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.572294 |
| UCH proteinases | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.515416 |
| UCH proteinases | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.515416 |
| UCH proteinases | ADRM1 | Adhesion regulating molecule 1, Oxysterol-binding protein, Proteasomal ubiquitin receptor ADRM1 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.515416 |
| UCH proteinases | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.515416 |
| UCH proteinases | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.577481 |
| UCH proteinases | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.54735 |
| UCH proteinases | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.515416 |
| UCH proteinases | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.57244 |
| UCH proteinases | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.515416 |
| UCH proteinases | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.546881 |
| UCH proteinases | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.573417 |
| UCH proteinases | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.262568 | 0.689501 | 4 | 0.703096 | 0.515416 |
| UCH proteinases | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.262568 | 0.689501 | 4 | 0.703096 | 0.515416 |
| Uncoating of the Influenza Virion | PB1 | Protein PB1-F2, RNA-directed RNA polymerase catalytic subunit | 7.405237 | 0.528279 | 0.508942 | ||
| Vasopressin-like receptors | OXTR | Isotocin receptor-like 2, Oxytocin receptor, Oxytocin-like receptor | 8.505077 | 0.842654 | 2.855126 | 0.528622 | 0.526619 |
| Vasopressin-like receptors | AVPR1A | Arginine vasopressin receptor 1A, G_PROTEIN_RECEP_F1_2 domain-containing protein, Vasopressin V1a receptor | 8.505077 | 0.842654 | 2.855126 | 0.528622 | 0.54747 |
| Vasopressin-like receptors | AVPR1B | G_PROTEIN_RECEP_F1_2 domain-containing protein, Vasopressin V1b receptor | 8.505077 | 0.842654 | 2.855126 | 0.528622 | 0.514485 |
| Vasopressin-like receptors | AVPR2 | AVPR V2, Vasopressin V2 receptor | 8.505077 | 0.842654 | 2.855126 | 0.528622 | 0.569629 |
| Vasopressin regulates renal water homeostasis via Aquaporins | ADCY9 | Adenylate cyclase 9, Adenylate cyclase type 9 | 8.957262 | 0.545229 | 3.056762 | 0.470137 | 0.550534 |
| Vasopressin regulates renal water homeostasis via Aquaporins | ADCY7 | Adenylate cyclase type 7 | 8.957262 | 0.545229 | 3.056762 | 0.470137 | 0.550655 |
| Vasopressin regulates renal water homeostasis via Aquaporins | AVPR2 | AVPR V2, Vasopressin V2 receptor | 8.957262 | 0.545229 | 3.056762 | 0.470137 | 0.569629 |
| Vasopressin regulates renal water homeostasis via Aquaporins | PRKAR2A | cAMP-dependent protein kinase type II-alpha regulatory subunit, Protein kinase cAMP-dependent type II regulatory subunit alpha, Protein kinase, cAMP-dependent, regulatory subunit type II alpha, Uncharacterized protein | 8.957262 | 0.545229 | 3.056762 | 0.470137 | 0.527065 |
| VEGFA-VEGFR2 Pathway | ROCK2 | Rho-associated protein kinase, Rho-associated protein kinase 2 | 9.380837 | 0.450954 | 4 | 0.589924 | 0.516942 |
| VEGFA-VEGFR2 Pathway | SRC | Neuronal proto-oncogene tyrosine-protein kinase Src, Proto-oncogene tyrosine-protein kinase Src, Tyrosine-protein kinase | 9.380837 | 0.450954 | 4 | 0.589924 | 0.594807 |
| VEGFA-VEGFR2 Pathway | HRAS | GTPase HRas, GTPase HRas isoform 1, Harvey rat sarcoma viral oncogene homolog, HRas proto-oncogene, GTPase, Uncharacterized protein | 9.380837 | 0.450954 | 4 | 0.589924 | 0.505988 |
| VEGFA-VEGFR2 Pathway | PRKCA | Protein kinase C, Protein kinase C alpha type, Protein kinase C, alpha | 9.380837 | 0.450954 | 4 | 0.589924 | 0.52202 |
| VEGFA-VEGFR2 Pathway | PRKCZ | Protein kinase C, Protein kinase C zeta type | 9.380837 | 0.450954 | 4 | 0.589924 | 0.513845 |
| VEGFA-VEGFR2 Pathway | PTK2 | Focal adhesion kinase 1, Non-specific protein-tyrosine kinase, Serine/threonine-protein kinase PTK2/STK2 | 9.380837 | 0.450954 | 4 | 0.589924 | 0.503121 |
| VEGFA-VEGFR2 Pathway | AKT2 | AKT serine/threonine kinase 2, Non-specific serine/threonine protein kinase, RAC-beta serine/threonine-protein kinase | 9.380837 | 0.450954 | 4 | 0.589924 | 0.552041 |
| VEGFA-VEGFR2 Pathway | PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.380837 | 0.450954 | 4 | 0.589924 | 0.589385 |
| VEGFA-VEGFR2 Pathway | CRK | 14_3_3 domain-containing protein, Adapter molecule crk, Adapter molecule Crk, CRK proto-oncogene, adaptor protein, Uncharacterized protein, V-crk avian sarcoma virus CT10 oncogene homolog | 9.380837 | 0.450954 | 4 | 0.589924 | 0.513369 |
| VEGFA-VEGFR2 Pathway | AKT1 | Non-specific serine/threonine protein kinase, RAC serine/threonine-protein kinase, RAC-alpha serine/threonine-protein kinase, RAC-PK-alpha, Serine protease 2 | 9.380837 | 0.450954 | 4 | 0.589924 | 0.589663 |
| VEGFA-VEGFR2 Pathway | MAPK13 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 13 | 9.380837 | 0.450954 | 4 | 0.589924 | 0.573201 |
| VEGFA-VEGFR2 Pathway | PRKCD | Protein kinase C, Protein kinase C delta type | 9.380837 | 0.450954 | 4 | 0.589924 | 0.520528 |
| VEGFA-VEGFR2 Pathway | PIK3R1 | Phosphatidylinositol 3-kinase 85 kDa regulatory subunit alpha, Phosphatidylinositol 3-kinase regulatory subunit alpha | 9.380837 | 0.450954 | 4 | 0.589924 | 0.646924 |
| VEGFA-VEGFR2 Pathway | HSP90AA1 | Heat shock protein 90 alpha family class A member 1, Heat shock protein HSP 90-alpha | 9.380837 | 0.450954 | 4 | 0.589924 | 0.624325 |
| VEGFA-VEGFR2 Pathway | MAPK14 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 14 | 9.380837 | 0.450954 | 4 | 0.589924 | 0.591941 |
| VEGFA-VEGFR2 Pathway | CTNNB1 | Catenin, Catenin beta-1, Uncharacterized protein | 9.380837 | 0.450954 | 4 | 0.589924 | 0.657916 |
| VEGFA-VEGFR2 Pathway | MAPK11 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 11 | 9.380837 | 0.450954 | 4 | 0.589924 | 0.567539 |
| VEGFA-VEGFR2 Pathway | MAPK12 | Mitogen-activated protein kinase, Mitogen-activated protein kinase 12 | 9.380837 | 0.450954 | 4 | 0.589924 | 0.573393 |
| VEGFA-VEGFR2 Pathway | PRKCB | Protein kinase C, Protein kinase C beta type | 9.380837 | 0.450954 | 4 | 0.589924 | 0.520598 |
| VEGFA-VEGFR2 Pathway | KDR | Receptor protein-tyrosine kinase, Vascular endothelial growth factor receptor 2 | 9.380837 | 0.450954 | 4 | 0.589924 | 0.609728 |
| VEGFA-VEGFR2 Pathway | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, Phosphatidylinositol-4,5-bisphosphate 3-kinase | 9.380837 | 0.450954 | 4 | 0.589924 | 0.660919 |
| VEGFA-VEGFR2 Pathway | MTOR | Nucleoprotein TPR, Serine/threonine-protein kinase mTOR, Serine/threonine-protein kinase TOR | 9.380837 | 0.450954 | 4 | 0.589924 | 0.619026 |
| Vif-mediated degradation of APOBEC3G | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.636578 | 0.763053 | 4 | 0.759167 | 0.579609 |
| Vif-mediated degradation of APOBEC3G | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.54735 |
| Vif-mediated degradation of APOBEC3G | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.515416 |
| Vif-mediated degradation of APOBEC3G | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.577847 |
| Vif-mediated degradation of APOBEC3G | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.54735 |
| Vif-mediated degradation of APOBEC3G | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.54735 |
| Vif-mediated degradation of APOBEC3G | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.515416 |
| Vif-mediated degradation of APOBEC3G | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.515416 |
| Vif-mediated degradation of APOBEC3G | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.54735 |
| Vif-mediated degradation of APOBEC3G | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.54735 |
| Vif-mediated degradation of APOBEC3G | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.515416 |
| Vif-mediated degradation of APOBEC3G | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.515416 |
| Vif-mediated degradation of APOBEC3G | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.515416 |
| Vif-mediated degradation of APOBEC3G | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.515416 |
| Vif-mediated degradation of APOBEC3G | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.573417 |
| Vif-mediated degradation of APOBEC3G | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.636578 | 0.763053 | 4 | 0.759167 | 0.515416 |
| Vif-mediated degradation of APOBEC3G | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.565601 |
| Vif-mediated degradation of APOBEC3G | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.546881 |
| Vif-mediated degradation of APOBEC3G | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.515416 |
| Vif-mediated degradation of APOBEC3G | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.636578 | 0.763053 | 4 | 0.759167 | 0.568645 |
| Vif-mediated degradation of APOBEC3G | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.577481 |
| Vif-mediated degradation of APOBEC3G | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.54735 |
| Vif-mediated degradation of APOBEC3G | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.5197 |
| Vif-mediated degradation of APOBEC3G | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.572294 |
| Vif-mediated degradation of APOBEC3G | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.515416 |
| Vif-mediated degradation of APOBEC3G | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.54735 |
| Vif-mediated degradation of APOBEC3G | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.515416 |
| Vif-mediated degradation of APOBEC3G | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.54735 |
| Vif-mediated degradation of APOBEC3G | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.58858 |
| Vif-mediated degradation of APOBEC3G | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.636578 | 0.763053 | 4 | 0.759167 | 0.515416 |
| Vif-mediated degradation of APOBEC3G | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.54735 |
| Vif-mediated degradation of APOBEC3G | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.515416 |
| Vif-mediated degradation of APOBEC3G | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.547953 |
| Vif-mediated degradation of APOBEC3G | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.54735 |
| Vif-mediated degradation of APOBEC3G | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.515416 |
| Vif-mediated degradation of APOBEC3G | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.57244 |
| Vif-mediated degradation of APOBEC3G | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.636578 | 0.763053 | 4 | 0.759167 | 0.515416 |
| Viral mRNA Translation | RPLP1 | 60S acidic ribosomal protein P1, Ribosomal protein, large, P1 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS7 | 40S ribosomal protein S7 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS6 | 40S ribosomal protein S6 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.566528 |
| Viral mRNA Translation | RPL21 | 60S ribosomal protein L21 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL28 | 60S ribosomal protein L28 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL24 | 60S ribosomal protein L24, Ribosomal protein L24, TRASH domain-containing protein | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL22 | 60S ribosomal protein L22, 60S ribosomal protein L22 1, Ribosomal protein L22, Uncharacterized protein | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS21 | 40S ribosomal protein S21 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL41 | 60S ribosomal protein L41 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS9 | 40S ribosomal protein S9 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS19 | 40S ribosomal protein S19, Uncharacterized protein | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS15A | 40S ribosomal protein S15a | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL35A | 60S ribosomal protein L35-A, 60S ribosomal protein L35a | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS17 | 40S ribosomal protein S17 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS15 | 40S ribosomal protein S15 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL5 | 60S ribosomal protein L5, Ribosomal protein L5 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS3A | 40S ribosomal protein S3a | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS4Y1 | 40S ribosomal protein S4, 40S ribosomal protein S4, Y isoform 1 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL37 | 60S ribosomal protein L37, Ribosomal protein L37 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL19 | 60S ribosomal protein L19, Ribosomal protein L19 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | FAU | 40S ribosomal protein S30 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL36A | 60S ribosomal protein L36-A, 60S ribosomal protein L36a, IP17351p, ribosomal protein L36a, Uncharacterized protein | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL7A | 60S ribosomal protein L7-A, 60S ribosomal protein L7a | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS13 | 40S ribosomal protein S13 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS8 | 40S ribosomal protein S8 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL18 | 60S ribosomal protein L18, Ribosomal protein L18 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL12 | 60S ribosomal protein L12 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL29 | 60S ribosomal protein L29 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL13 | 60S ribosomal protein L13 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL18A | 60S ribosomal protein L18-A, 60S ribosomal protein L18a | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL10 | 60S ribosomal protein L10, MGC89303 protein | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS25 | 40S ribosomal protein S25 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS16 | 40S ribosomal protein S16, Ribosomal protein S16, Uncharacterized protein | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS28 | 40S ribosomal protein S28 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL35 | 60S ribosomal protein L35 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL10A | 60S ribosomal protein L10a, Ribosomal protein | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL38 | 60S ribosomal protein L38 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS20 | 40S ribosomal protein S20 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS27 | 40S ribosomal protein S27 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.549907 |
| Viral mRNA Translation | RPL27 | 60S ribosomal protein L27 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL4 | 60S ribosomal protein L4, Ribos_L4_asso_C domain-containing protein, Ribosomal protein L4 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS26 | 40S ribosomal protein S26 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL32 | 60S ribosomal protein L32, Hypothetical LOC496894, Ribosomal protein L32, Uncharacterized protein | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL7 | 60S ribosomal protein L7, MGC79754 protein, Ribosomal protein L7, Uncharacterized protein | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS2 | 40S ribosomal protein S2 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS12 | 40S ribosomal protein S12 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL36 | 60S ribosomal protein L36 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | PB1 | Protein PB1-F2, RNA-directed RNA polymerase catalytic subunit | 11.63711 | 0.918316 | 4 | 0.940004 | 0.508942 |
| Viral mRNA Translation | RPS10 | 40S ribosomal protein S10, Ribosomal protein S10, S10_plectin domain-containing protein | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL6 | 60S ribosomal protein L6 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS11 | 40S ribosomal protein S11 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL11 | 60S ribosomal protein L11 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS18 | 40S ribosomal protein S18 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS23 | 40S ribosomal protein S23, 40S ribosomal protein S23-A | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL3 | 60S ribosomal protein L3, LOC100145083 protein, Ribosomal protein L3, Uncharacterized protein | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS24 | 40S ribosomal protein S24 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPLP0 | 60S acidic ribosomal protein P0 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL39 | 60S ribosomal protein L39, LOC100135132 protein, Ribosomal protein L39 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL37A | 60S ribosomal protein L37-A, 60S ribosomal protein L37a, MGC89854 protein, Probable 60S ribosomal protein L37-A, Ribosomal protein L37a | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS29 | 40S ribosomal protein S29 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL17 | 60S ribosomal protein L17 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS4X | 40S ribosomal protein S4, 40S ribosomal protein S4, X isoform | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPSA | 40S ribosomal protein SA | 11.63711 | 0.918316 | 4 | 0.940004 | 0.521531 |
| Viral mRNA Translation | RPL23A | 60S ribosomal protein L23-A, 60S ribosomal protein L23a, FI01658p, MGC89958 protein, Ribosomal protein L23a, Ribosomal_L23eN domain-containing protein | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL26 | 60S ribosomal protein L26, GEO07453p1, KOW domain-containing protein, Ribosomal protein L26 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS3 | 40S ribosomal protein S3, DNA-(apurinic or apyrimidinic site) lyase | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL13A | 60S ribosomal protein L13a | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL27A | 60S ribosomal protein L27-A, 60S ribosomal protein L27a | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL14 | 60S ribosomal protein L14 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL8 | 60S ribosomal protein L8 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS5 | 40S ribosomal protein S5, 40S ribosomal protein S5 [Cleaved into: 40S ribosomal protein S5, N-terminally processed], 40S ribosomal protein S5-A, Ribosomal protein S5 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL23 | 60S ribosomal protein L23 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL30 | 60S ribosomal protein L30 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL31 | 60S ribosomal protein L31 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPS14 | 40S ribosomal protein S14, Ribosomal protein S14, Uncharacterized protein | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL15 | 60S ribosomal protein L15, Ribosomal protein L15 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Viral mRNA Translation | RPL34 | 60S ribosomal protein L34 | 11.63711 | 0.918316 | 4 | 0.940004 | 0.579609 |
| Vpu mediated degradation of CD4 | PSMB7 | Proteasome subunit beta, Proteasome subunit beta type-7 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.565601 |
| Vpu mediated degradation of CD4 | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.5197 |
| Vpu mediated degradation of CD4 | PSMA5 | Proteasome subunit alpha type, Proteasome subunit alpha type-5 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.54735 |
| Vpu mediated degradation of CD4 | PSMB1 | Proteasome subunit beta, Proteasome subunit beta type-1 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.577481 |
| Vpu mediated degradation of CD4 | PSMB5 | Proteasome subunit beta, Proteasome subunit beta type-5 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.58858 |
| Vpu mediated degradation of CD4 | PSMD13 | 26S proteasome non-ATPase regulatory subunit 13 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.515416 |
| Vpu mediated degradation of CD4 | PSMC3 | 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6A homolog, AAA domain-containing protein, Proteasome 26S subunit, ATPase 3, TPR_REGION domain-containing protein | 9.565729 | 0.793025 | 4 | 0.770498 | 0.515416 |
| Vpu mediated degradation of CD4 | PSMB6 | Proteasome subunit beta, Proteasome subunit beta type-6 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.546881 |
| Vpu mediated degradation of CD4 | PSMC1 | 26S protease regulatory subunit 4, 26S proteasome regulatory subunit 4, 26S proteasome regulatory subunit 4 homolog, Proteasome 26S subunit, ATPase 1 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.515416 |
| Vpu mediated degradation of CD4 | PSMB9 | Proteasome subunit beta, Proteasome subunit beta type-9 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.577847 |
| Vpu mediated degradation of CD4 | PSMC6 | 26S protease regulatory subunit S10B, 26S proteasome regulatory subunit 10B, Proteasome 26S subunit, ATPase 6 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.515416 |
| Vpu mediated degradation of CD4 | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.515416 |
| Vpu mediated degradation of CD4 | PSMA3 | Proteasome subunit alpha type, Proteasome subunit alpha type-3 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.54735 |
| Vpu mediated degradation of CD4 | PSMD4 | 26S proteasome non-ATPase regulatory subunit 4 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.515416 |
| Vpu mediated degradation of CD4 | SEM1 | 26S proteasome complex subunit SEM1, Probable 26S proteasome complex subunit sem1 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.515416 |
| Vpu mediated degradation of CD4 | PSMB10 | Proteasome subunit beta, Proteasome subunit beta type-10 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.572294 |
| Vpu mediated degradation of CD4 | RPS27A | 40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-40S ribosomal protein S27a [Cleaved into: Ubiquitin; 40S ribosomal protein S27a] | 9.565729 | 0.793025 | 4 | 0.770498 | 0.579609 |
| Vpu mediated degradation of CD4 | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2, Uncharacterized protein | 9.565729 | 0.793025 | 4 | 0.770498 | 0.515416 |
| Vpu mediated degradation of CD4 | PSMB4 | Proteasome subunit beta, Proteasome subunit beta type-4 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.54735 |
| Vpu mediated degradation of CD4 | PSMD8 | 26S proteasome non-ATPase regulatory subunit 8, Proteasome 26S subunit, non-ATPase 8 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.515416 |
| Vpu mediated degradation of CD4 | PSMC2 | 26S proteasome AAA-ATPase subunit RPT1, 26S proteasome regulatory subunit 7 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.515416 |
| Vpu mediated degradation of CD4 | PSMA1 | Proteasome subunit alpha type, Proteasome subunit alpha type-1 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.54735 |
| Vpu mediated degradation of CD4 | PSMA4 | Proteasome subunit alpha type, Proteasome subunit alpha type-4 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.54735 |
| Vpu mediated degradation of CD4 | PSMA2 | Proteasome subunit alpha type, Proteasome subunit alpha type-2 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.54735 |
| Vpu mediated degradation of CD4 | PSMA8 | Proteasome subunit alpha type, Proteasome subunit alpha type-8, Proteasome subunit alpha-type 8 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.54735 |
| Vpu mediated degradation of CD4 | PSMC5 | 26S proteasome regulatory subunit 8, AAA domain-containing protein, Proteasome, Proteasome 26S subunit, ATPase 5 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.515416 |
| Vpu mediated degradation of CD4 | PSMA7 | Proteasome subunit alpha 7, Proteasome subunit alpha type, Proteasome subunit alpha type-7 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.54735 |
| Vpu mediated degradation of CD4 | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 3 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.515416 |
| Vpu mediated degradation of CD4 | PSMB8 | Proteasome subunit beta, Proteasome subunit beta type-8 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.57244 |
| Vpu mediated degradation of CD4 | PSMB2 | Proteasome subunit beta, Proteasome subunit beta type-2 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.573417 |
| Vpu mediated degradation of CD4 | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12, PCI domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 12 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.515416 |
| Vpu mediated degradation of CD4 | PSMC4 | 26S proteasome AAA-ATPase subunit RPT3, 26S proteasome regulatory subunit 6B, 26S proteasome regulatory subunit 6B homolog | 9.565729 | 0.793025 | 4 | 0.770498 | 0.568645 |
| Vpu mediated degradation of CD4 | PSMB3 | Proteasome subunit beta, Proteasome subunit beta type-3 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.547953 |
| Vpu mediated degradation of CD4 | PSMB11 | Proteasome subunit beta, Proteasome subunit beta type-11 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.54735 |
| Vpu mediated degradation of CD4 | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7, MPN domain-containing protein, Proteasome, Proteasome 26S subunit, non-ATPase 7 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.515416 |
| Vpu mediated degradation of CD4 | PSMA6 | Proteasome subunit alpha type, Proteasome subunit alpha type-6 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.54735 |
| Vpu mediated degradation of CD4 | PSMD11 | 26S proteasome non-ATPase regulatory subunit 11, PCI domain-containing protein, Proteasome 26S subunit, non-ATPase 11 | 9.565729 | 0.793025 | 4 | 0.770498 | 0.515416 |
See all pathway scores
| pathwayname | aggregatedSarsScore | log10SumPositiveInteractions | combinedAggregationScore | enrichmentScore | -log10FDR | combinedPathwaysScore |
|---|---|---|---|---|---|---|
| 2-LTR circle formation | 2.352604517 | 1.77815125 | 6.483360285 | -0.764593039 | 1.401671157 | 0.209292402 |
| 5-Phosphoribose 1-diphosphate biosynthesis | 1.628445671 | 0 | 3.256891342 | 0.163350253 | ||
| A tetrasaccharide linker sequence is required for GAG synthesis | 4.843162618 | 0.84509804 | 10.53142328 | 0.713229252 | 0.275451609 | 0.05374196 |
| Abacavir metabolism | 2.002724514 | 0.301029996 | 4.306479023 | -0.477927063 | 0.696945318 | 0.059524163 |
| Abacavir transmembrane transport | 2.573399007 | 2.695481676 | 7.84227969 | 0.533040855 | 0.775952286 | 0.107002805 |
| Abacavir transport and metabolism | 0.306038913 | 0.205687834 | 0.01573712 | |||
| Abasic sugar-phosphate removal via the single-nucleotide replacement pathway | 2.720154873 | 2.507855872 | 7.948165619 | 0.671506849 | 0.606999218 | 0.095039435 |
| ABC-family proteins mediated transport | 3.060954534 | 3.318063335 | 9.439972402 | 0.656891409 | 4 | 0.695943665 |
| ABC transporter disorders | 0.709981986 | 4 | 0.709981986 | |||
| ABC transporters in lipid homeostasis | 0.7 | 0 | 1.4 | 0 | ||
| Aberrant regulation of mitotic cell cycle due to RB1 defects | 0.435967188 | 1.281079769 | 0.139627186 | |||
| Aberrant regulation of mitotic exit in cancer due to RB1 defects | 2.234536606 | 0.602059991 | 5.071133204 | -0.584864272 | 1.213453031 | 0.131825095 |
| Aberrant regulation of mitotic G1/S transition in cancer due to RB1 defects | 0.531181191 | 1.857708808 | 0.246694994 | |||
| Abnormal conversion of 2-oxoglutarate to 2-hydroxyglutarate | 2.269525144 | 2.456366033 | 6.995416321 | 0.894275541 | 0.659202877 | 0.113469108 |
| ABO blood group biosynthesis | 2.442992526 | 0 | 4.885985051 | -0.775068493 | 0.959949484 | 0.124816431 |
| Abortive elongation of HIV-1 transcript in the absence of Tat | 2.296017113 | 0 | 4.592034226 | 0.280802429 | ||
| Acetylation | 1.739316766 | 0 | 3.478633532 | -0.911506849 | 1.68808258 | 0.217640657 |
| Acetylcholine binding and downstream events | 0.305112699 | 0.272173256 | 0.020760879 | |||
| Acetylcholine Neurotransmitter Release Cycle | 2.502128579 | 2.021189299 | 7.025446457 | 0.278782895 | 0.013363962 | 0.001277348 |
| Acetylcholine regulates insulin secretion | 2.249575265 | 2.717670503 | 7.216821033 | 0.475172453 | 0.47988571 | 0.058833229 |
| Acrosome Reaction and Sperm:Oocyte Membrane Binding | 2.240410169 | 0.477121255 | 4.957941592 | 0.560942208 | 0.057142886 | 0.005953501 |
| Activated NOTCH1 Transmits Signal to the Nucleus | 3.070789304 | 3.148602655 | 9.290181263 | 0.7032472 | 4 | 0.711393961 |
| Activated NTRK2 signals through CDK5 | 2.592055372 | 2.247973266 | 7.43208401 | 0.450237449 | 0.40098188 | 0.049023184 |
| Activated NTRK2 signals through FRS2 and FRS3 | 2.690862115 | 2.397940009 | 7.77966424 | 0.510671894 | 0.886335463 | 0.119030422 |
| Activated NTRK2 signals through FYN | 2.459939639 | 2.785329835 | 7.705209113 | 0.569821613 | 0.972246931 | 0.136822801 |
| Activated NTRK2 signals through PI3K | 2.68252685 | 2.899820502 | 8.264874203 | 0.585188211 | 0.819543936 | 0.122822947 |
| Activated NTRK2 signals through PLCG1 | 2.108673596 | 2.235528447 | 6.452875638 | 0.410136986 | 0.038874218 | 0.004066871 |
| Activated NTRK2 signals through RAS | 2.836897398 | 1.924279286 | 7.598074082 | 0.56332124 | 1.216191785 | 0.168484093 |
| Activated NTRK3 signals through PI3K | 2.533756322 | 3.330616667 | 8.39812931 | 0.659796074 | 1.498586209 | 0.241140575 |
| Activated NTRK3 signals through PLCG1 | 2.146218374 | 1.959041392 | 6.25147814 | 0.410136986 | 0.038874218 | 0.003965896 |
| Activated NTRK3 signals through RAS | 2.737773079 | 1.755874856 | 7.231421014 | 0.511347732 | 0.890338205 | 0.113347737 |
| Activated PKN1 stimulates transcription of AR (androgen receptor) regulated genes KLK2 and KLK3 | 2.655134262 | 2.739572344 | 8.049840869 | 0.53093094 | 1.210727282 | 0.16987948 |
| Activated point mutants of FGFR2 | 2.801357858 | 2.170261715 | 7.772977432 | 0.477612545 | 0.632171666 | 0.082230577 |
| Activated TAK1 mediates Jun kinases (JNK) phosphorylation and activation | 3.817383417 | 1.176091259 | 8.810858093 | 0.651931278 | ||
| activated TAK1 mediates p38 MAPK activation | 2.805188561 | 3.266936911 | 8.877314033 | 0.575533255 | 3.406085947 | 0.5332547 |
| Activation and oligomerization of BAK protein | 3.463966703 | 1 | 7.927933406 | 0.481917808 | 0.115912307 | 0.01537148 |
| Activation of AKT2 | 2.49883118 | 2.472756449 | 7.470418809 | 0.798630137 | 1.059296889 | 0.176162507 |
| Activation of AMPA receptors | 1.892089004 | 2.08278537 | 5.866963378 | 0.541940789 | 0.63226851 | 0.071784577 |
| Activation of AMPK downstream of NMDARs | 2.756161933 | 3.108903128 | 8.621226993 | 0.650692743 | 4 | 0.650605944 |
| Activation of anterior HOX genes in hindbrain development during early embryogenesis | 2.341524101 | 2.580924976 | 7.263973178 | 0.241517431 | 0.175646968 | 0.016510758 |
| Activation of APC/C and APC/C:Cdc20 mediated degradation of mitotic proteins | 0.763032415 | 4 | 0.763032415 | |||
| Activation of ATR in response to replication stress | 2.533717359 | 2.664641976 | 7.732076694 | 0.276172735 | 0.172084172 | 0.017960228 |
| Activation of BH3-only proteins | 0.560141293 | 3.355210346 | 0.469847965 | |||
| Activation of BMF and translocation to mitochondria | 2.347791248 | 2.499687083 | 7.195269579 | 0.818679814 | 0.434263051 | 0.071765827 |
| Activation of C3 and C5 | 3.391477898 | 1.176091259 | 7.959047055 | 0.596581551 | 0.355528002 | 0.052386077 |
| Activation of Ca-permeable Kainate Receptor | 2.097023495 | 1.431363764 | 5.625410755 | 0.379526273 | 0.315235927 | 0.028408358 |
| Activation of caspases through apoptosome-mediated cleavage | 2.473860645 | 2.326335861 | 7.27405715 | 0.471749863 | 0.61901022 | 0.076081795 |
| Activation of G protein gated Potassium channels | 2.599434789 | 2.530199698 | 7.729069276 | 0.439770008 | 0.827485878 | 0.103253462 |
| Activation of GABAB receptors | 0.440581747 | 1.765694331 | 0.194483173 | |||
| Activation of gene expression by SREBF (SREBP) | 2.548688771 | 2.658011397 | 7.75538894 | 0.31096691 | 0.590710947 | 0.064398594 |
| Activation of HOX genes during differentiation | 0.241517431 | 0.175646968 | 0.010605451 | |||
| Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon | 3.070347088 | 2.556302501 | 8.696996676 | 0.578284228 | 1.545723167 | 0.238934247 |
| Activation of kainate receptors upon glutamate binding | -0.35319967 | 1.069593529 | 0.094445021 | |||
| Activation of Matrix Metalloproteinases | 2.593057293 | 3.269512944 | 8.455627531 | 0.381269584 | 1.284541312 | 0.162928403 |
| Activation of Na-permeable kainate receptors | 1.913478796 | 0.84509804 | 4.672055632 | -0.486027397 | 0.166558267 | 0.015179239 |
| Activation of NF-kappaB in B cells | 3.073280193 | 3.241297387 | 9.387857773 | 0.721213926 | 4 | 0.725416373 |
| Activation of NIMA Kinases NEK9, NEK6, NEK7 | 2.65348878 | 2.809559715 | 8.116537275 | 0.597256516 | 1.476603929 | 0.220697399 |
| Activation of NMDA receptors and postsynaptic events | 0.510178277 | 4 | 0.510178277 | |||
| Activation of NOXA and translocation to mitochondria | 2.487300291 | 2.387389826 | 7.361990409 | 0.453939455 | 0.20147299 | 0.024542752 |
| Activation of PPARGC1A (PGC-1alpha) by phosphorylation | 2.798789958 | 2.961895474 | 8.559475389 | 0.78830313 | 4 | 0.716225425 |
| Activation of PUMA and translocation to mitochondria | 3.173539487 | 2.389166084 | 8.736245059 | 0.59446922 | 0.84681802 | 0.133041011 |
| Activation of RAC1 | 2.645364539 | 2.392696953 | 7.683426032 | 0.492929518 | 1.03066882 | 0.134848578 |
| Activation of RAC1 downstream of NMDARs | 2.788958223 | 2.365487985 | 7.943404431 | 0.530573074 | 0.75980565 | 0.105532791 |
| Activation of RAS in B cells | 2.587260752 | 1.204119983 | 6.378641486 | 0.341861196 | 0.120551886 | 0.01146741 |
| Activation of SMO | 2.446306679 | 2.494154594 | 7.386767952 | 0.417483092 | 0.374431572 | 0.044025329 |
| Activation of the AP-1 family of transcription factors | 2.476053327 | 3.502973059 | 8.455079713 | 0.513666 | 1.304960289 | 0.187105589 |
| Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | 2.717003026 | 1.707570176 | 7.141576229 | 0.777154233 | 4 | 0.637502751 |
| Activation of the phototransduction cascade | 2.036933882 | 0.954242509 | 5.028110274 | -0.620680197 | 1.662489894 | 0.187127286 |
| Activation of the pre-replicative complex | 2.392260499 | 2.990338855 | 7.774859853 | 0.328890851 | 0.289524833 | 0.032285032 |
| Activation of the TFAP2 (AP-2) family of transcription factors | 2.36549144 | 1.568201724 | 6.299184603 | 0.570410959 | 0.282488127 | 0.034652313 |
| Activation of TRKA receptors | 0.682752887 | 1.899063194 | 0.32414772 | |||
| Activation, myristolyation of BID and translocation to mitochondria | 2.732946192 | 1.278753601 | 6.744645986 | 0.360197368 | 0.109389764 | 0.011172716 |
| Activation, translocation and oligomerization of BAX | 3.358755898 | 0.698970004 | 7.416481801 | -0.518356164 | 0.2388065 | 0.031181315 |
| Acyl chain remodeling of CL | 2.573319612 | 2.037426498 | 7.184065722 | 0.587512785 | 0.330792806 | 0.045060102 |
| Acyl chain remodeling of DAG and TAG | 1.938188665 | 1.84509804 | 5.72147537 | 0.390466485 | 0.233768386 | 0.021676002 |
| Acyl chain remodelling of PC | 2.801679292 | 2.45331834 | 8.056676925 | 0.353541537 | 0.608994058 | 0.071999187 |
| Acyl chain remodelling of PE | 2.801679292 | 2.45331834 | 8.056676925 | 0.319065724 | 0.405807039 | 0.046228297 |
| Acyl chain remodelling of PG | 2.87873805 | 2.451786436 | 8.209262536 | 0.442058935 | 0.920180789 | 0.120782043 |
| Acyl chain remodelling of PI | 2.934523929 | 2.450249108 | 8.319296966 | 0.502803153 | 1.360272912 | 0.190807125 |
| Acyl chain remodelling of PS | 2.866703927 | 2.44870632 | 8.182114173 | 0.354378137 | 0.496986484 | 0.05961293 |
| Adaptive Immune System | 0.354186256 | 4 | 0.354186256 | |||
| Adenosine P1 receptors | 2.269605648 | 3.070776463 | 7.60998776 | 0.68064693 | 1.403120521 | 0.215173463 |
| Adenylate cyclase activating pathway | 3.369402836 | 2.225309282 | 8.964114953 | 0.693110074 | 3.087337055 | 0.532182744 |
| Adenylate cyclase inhibitory pathway | 3.434938212 | 2.457881897 | 9.327758321 | 0.566700245 | 2.015495807 | 0.325028454 |
| Adherens junctions interactions | 2.435248791 | 2.371067862 | 7.241565445 | 0.364055389 | 0.168172435 | 0.018335491 |
| ADORA2B mediated anti-inflammatory cytokines production | 2.811043745 | 3.607025878 | 9.229113368 | 0.392462443 | 4 | 0.552851141 |
| ADP signalling through P2Y purinoceptor 1 | 2.565837472 | 3.349083169 | 8.480758113 | 0.571132257 | 1.251664103 | 0.188869551 |
| ADP signalling through P2Y purinoceptor 12 | 2.735192812 | 1.342422681 | 6.812808306 | 0.37136588 | 0.231124785 | 0.024132185 |
| Adrenaline signalling through Alpha-2 adrenergic receptor | 2.535888042 | 3.121887985 | 8.193664069 | 0.865442587 | 2.261877263 | 0.416142019 |
| Adrenaline,noradrenaline inhibits insulin secretion | 3.02812118 | 3.092720645 | 9.148963005 | 0.552695492 | 2.072753999 | 0.325853913 |
| Adrenoceptors | 2.485167174 | 3.740757323 | 8.711091672 | 0.643151249 | 2.395865641 | 0.39020939 |
| Advanced glycosylation endproduct receptor signaling | 2.659214881 | 2.850033258 | 8.16846302 | 0.408023592 | 0.484754043 | 0.061310923 |
| Aflatoxin activation and detoxification | 2.307859137 | 2.800717078 | 7.416435353 | 0.326559079 | 0.161834122 | 0.017250908 |
| Aggrephagy | 2.974426177 | 3.47099817 | 9.419850523 | 0.579865154 | 4 | 0.656392466 |
| AKT-mediated inactivation of FOXO1A | 2.621540024 | 3.028977705 | 8.272057753 | 0.612545622 | 0.935759104 | 0.143526506 |
| AKT phosphorylates targets in the cytosol | 2.541099981 | 3.369215857 | 8.451415819 | 0.567120013 | 2.015495807 | 0.302354182 |
| AKT phosphorylates targets in the nucleus | 2.501603117 | 3.034227261 | 8.037433495 | 0.301086679 | 0.132229426 | 0.014733184 |
| Alanine metabolism | 3.27872236 | 0.301029996 | 6.858474715 | 0.514927417 | 0.014693951 | 0.001806564 |
| ALKBH3 mediated reversal of alkylation damage | 1.949093606 | 0 | 3.898187212 | 0.219764886 | ||
| Alpha-defensins | 2.95619363 | 2.408239965 | 8.320627226 | 0.620661081 | 1.30546161 | 0.20237345 |
| alpha-linolenic (omega3) and linoleic (omega6) acid metabolism | -0.287125995 | 0.387389826 | 0.027807422 | |||
| alpha-linolenic acid (ALA) metabolism | 2.553901294 | 0.84509804 | 5.952900628 | -0.287125995 | 0.387389826 | 0.032072571 |
| Alpha-oxidation of phytanate | 2.743507359 | 0 | 5.487014718 | 0.359533632 | ||
| Alpha-protein kinase 1 signaling pathway | 2.822995183 | 1.86923172 | 7.515222086 | 0.613476483 | 0.940722523 | 0.135214741 |
| Alternative complement activation | 3.002086668 | 1.322219295 | 7.326392631 | 0.502189933 | 0.314512504 | 0.040065364 |
| Amine ligand-binding receptors | 1.988363537 | 1.113943352 | 5.090670427 | 0.562418669 | 4 | 0.424330464 |
| Amine Oxidase reactions | -0.327850877 | 0.084576278 | 0.006932102 | |||
| Amino acid transport across the plasma membrane | 2.573285091 | 1.62324929 | 6.769819473 | -0.443559462 | 1.951735888 | 0.220315239 |
| Amino acids regulate mTORC1 | 2.833126296 | 3.11991541 | 8.786168003 | 0.518878778 | 0.689700341 | 0.102283962 |
| AMPK inhibits chREBP transcriptional activation activity | 2.814664149 | 2.675778342 | 8.30510664 | 0.612317789 | 1.403692338 | 0.215856328 |
| Amplification of signal from unattached kinetochores via a MAD2 inhibitory signal | 2.981208715 | 2.756636108 | 8.719053539 | 0.459073468 | 1.489958479 | 0.208535742 |
| Amplification of signal from the kinetochores | 0.459073468 | 1.489958479 | 0.171000102 | |||
| Amyloid fiber formation | 2.580034607 | 3.153814864 | 8.313884078 | 0.467493824 | 1.974332372 | 0.268090112 |
| Anchoring fibril formation | 2.267528629 | 0.602059991 | 5.13711725 | 0.291852321 | 0.008296841 | 0.000604515 |
| Anchoring of the basal body to the plasma membrane | 2.538158693 | 3.311753861 | 8.388071247 | 0.445012018 | 1.639215947 | 0.21954727 |
| Androgen biosynthesis | 2.706537982 | 2.523746467 | 7.936822432 | 0.511019268 | 1.212088912 | 0.165286914 |
| Antagonism of Activin by Follistatin | 2.158539338 | 0 | 4.317078676 | 0.25661466 | ||
| Anti-inflammatory response favouring Leishmania parasite infection | 0.388842908 | 4 | 0.388842908 | |||
| Antigen activates B Cell Receptor (BCR) leading to generation of second messengers | 2.585809997 | 3.443262987 | 8.614882982 | 0.426357808 | 1.537819095 | 0.206879418 |
| Antigen Presentation: Folding, assembly and peptide loading of class I MHC | 2.298653956 | 1.908485019 | 6.505792932 | 0.553879901 | 0.879492878 | 0.108412022 |
| Antigen processing-Cross presentation | 0.589469215 | 4 | 0.589469215 | |||
| Antigen processing: Ubiquitination & Proteasome degradation | 3.135406117 | 2.995635195 | 9.266447428 | 0.58155531 | 4 | 0.649323607 |
| Antimicrobial action and antimicrobial resistance in Mtb | 1.895999547 | 0.698970004 | 4.490969098 | 0.271911756 | ||
| Antimicrobial peptides | 2.414955402 | 2.679427897 | 7.509338701 | 0.383614935 | 0.776748036 | 0.089268867 |
| Antiviral mechanism by IFN-stimulated genes | 0.37403909 | 0.963787827 | 0.09012358 | |||
| APC-Cdc20 mediated degradation of Nek2A | 2.693345551 | 1.792391689 | 7.179082792 | 0.382625862 | 0.21528498 | 0.023798328 |
| APC truncation mutants are not K63 polyubiquitinated | 3.966673517 | 0.77815125 | 8.711498285 | 0.808271706 | 0.413954674 | 0.07596611 |
| APC truncation mutants have impaired AXIN binding | 2.914767738 | 2.164352856 | 7.993888332 | 0.444696194 | 0.577408986 | 0.0743766 |
| APC/C-mediated degradation of cell cycle proteins | 0.723065546 | 4 | 0.723065546 | |||
| APC/C:Cdc20 mediated degradation of Cyclin B | 2.908264733 | 2.53529412 | 8.351823586 | 0.610577134 | 1.378138233 | 0.212457182 |
| APC/C:Cdc20 mediated degradation of mitotic proteins | 2.0418342 | 0 | 4.0836684 | 0.766670593 | 4 | 0.474506038 |
| APC/C:Cdc20 mediated degradation of Securin | 3.316512766 | 2.901458321 | 9.534483853 | 0.799276581 | 4 | 0.772012017 |
| APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | 3.218047557 | 3.211387553 | 9.647482667 | 0.755438637 | 4 | 0.755922558 |
| APC:Cdc20 mediated degradation of cell cycle proteins prior to satisfation of the cell cycle checkpoint | 0.764949115 | 4 | 0.764949115 | |||
| APEX1-Independent Resolution of AP Sites via the Single Nucleotide Replacement Pathway | 2.862017798 | 2.170261715 | 7.894297312 | 0.411806532 | 0.043502622 | 0.005368889 |
| APOBEC3G mediated resistance to HIV-1 infection | 3.114054267 | 2.133538908 | 8.361647441 | 0.502462425 | 0.314512504 | 0.044275442 |
| Apoptosis | 0.517189814 | 4 | 0.517189814 | |||
| Apoptosis induced DNA fragmentation | 2.463144927 | 1.973127854 | 6.899417708 | 0.460526316 | 0.337416349 | 0.039367718 |
| Apoptotic cleavage of cell adhesion proteins | 2.527385705 | 2.436162647 | 7.490934057 | 0.482116473 | 0.25717314 | 0.032661436 |
| Apoptotic cleavage of cellular proteins | 2.532448197 | 3.389874558 | 8.454770952 | 0.468617193 | 2.05345345 | 0.282853378 |
| Apoptotic execution phase | 2.072711275 | 1.556302501 | 5.701725051 | 0.443796259 | 2.024728731 | 0.200723028 |
| Apoptotic factor-mediated response | 0.472267985 | 1.01899404 | 0.120309565 | |||
| Aquaporin-mediated transport | 0.529029893 | 2.947854389 | 0.389875773 | |||
| Arachidonate production from DAG | 2.228178313 | 1.397940009 | 5.854296634 | -0.431861804 | 0.524233799 | 0.052219828 |
| Arachidonic acid metabolism | 2.649733661 | 2.602059991 | 7.901527314 | 0.312380042 | 0.864284172 | 0.096004879 |
| ARL13B-mediated ciliary trafficking of INPP5E | 2.213197668 | 1.612783857 | 6.039179193 | 0.634346754 | 0.165595473 | 0.021081433 |
| ARMS-mediated activation | 3.30393984 | 2.750508395 | 9.358388075 | 0.802272784 | 3.390943046 | 0.648030821 |
| Aromatic amines can be N-hydroxylated or N-dealkylated by CYP1A2 | 2.109151198 | 2.290034611 | 6.508337007 | 0.708572994 | 0.265941131 | 0.037932769 |
| Aryl hydrocarbon receptor signalling | 2.365551632 | 2.26245109 | 6.993554355 | 0.327505439 | 0.058073697 | 0.005880572 |
| Asparagine N-linked glycosylation | 2.302018214 | 0 | 4.604036429 | 0.225547223 | 0.286104318 | 0.016507483 |
| Aspartate and asparagine metabolism | 2.533320562 | 1.505149978 | 6.571791102 | -0.300520976 | 0.120898759 | 0.01117683 |
| Assembly and cell surface presentation of NMDA receptors | 2.515245843 | 3.697665163 | 8.728156849 | 0.547828699 | 4 | 0.604690344 |
| Assembly of active LPL and LIPC lipase complexes | 3.010706853 | 2.389166084 | 8.41057979 | 0.49563052 | 0.702098189 | 0.098681256 |
| Assembly of collagen fibrils and other multimeric structures | 2.521434721 | 2.889861721 | 7.932731163 | 0.293726878 | 0.292402848 | 0.03191607 |
| Assembly Of The HIV Virion | 3.430576143 | 2.235528447 | 9.096680734 | 0.634933313 | 1.340223717 | 0.223567572 |
| Assembly of the pre-replicative complex | 4.228483415 | 0.301029996 | 8.757996825 | 0.799396494 | 4 | 0.73201366 |
| Assembly of the SARS-CoV-1 Replication-Transcription Complex (RTC) | 5.18567279 | 2.396199347 | 12.76754493 | 1 | ||
| Assembly of Viral Components at the Budding Site | 3.395472905 | 1.113943352 | 7.904889162 | 0.881128458 | 0.618515789 | 0.112704174 |
| Association of TriC/CCT with target proteins during biosynthesis | 2.332529181 | 2.820201459 | 7.485259821 | 0.49631723 | 1.208050276 | 0.155480479 |
| Astrocytic Glutamate-Glutamine Uptake And Metabolism | 2.925248608 | 0.698970004 | 6.549467219 | -0.610852288 | 0.698970004 | 0.091531061 |
| Asymmetric localization of PCP proteins | 3.38571718 | 2.904174368 | 9.675608728 | 0.800354925 | 4 | 0.779831702 |
| ATF4 activates genes in response to endoplasmic reticulum stress | 2.279952446 | 1.204119983 | 5.764024876 | -0.248902305 | 0.144897739 | 0.010951011 |
| ATF6 (ATF6-alpha) activates chaperone genes | 2.366356027 | 1.73239376 | 6.465105813 | 0.426780367 | 0.342643191 | 0.036612905 |
| ATF6 (ATF6-alpha) activates chaperones | 2.749405589 | 1.579783597 | 7.078594775 | 0.445410807 | 0.497324641 | 0.058234462 |
| ATP sensitive Potassium channels | 2.199315969 | 0.954242509 | 5.352874448 | -0.438048246 | 0.364416975 | 0.034225326 |
| Attachment and Entry | 2.469631995 | 1.968482949 | 6.907746939 | 0.542699054 | 0.656592348 | 0.083422045 |
| Attachment of GPI anchor to uPAR | 2.329481659 | 0.84509804 | 5.504061358 | -0.66109589 | 0.5788774 | 0.071635436 |
| Attenuation phase | 2.415060968 | 2.953759692 | 7.783881629 | 0.248062545 | 0.083281157 | 0.008454972 |
| AUF1 (hnRNP D0) binds and destabilizes mRNA | 3.247671285 | 2.904715545 | 9.400058115 | 0.759684331 | 4 | 0.745280981 |
| AURKA Activation by TPX2 | 2.557599404 | 3.382377303 | 8.497576111 | 0.458061546 | 1.847252706 | 0.253032658 |
| Autodegradation of Cdh1 by Cdh1:APC/C | 3.318069574 | 2.884795364 | 9.520934512 | 0.795888088 | 4 | 0.769618771 |
| Autodegradation of the E3 ubiquitin ligase COP1 | 3.287193209 | 3.112269768 | 9.686656186 | 0.804352985 | 4 | 0.782400661 |
| Autointegration results in viral DNA circles | 2.884935624 | 1.77815125 | 7.548022499 | 0.540839956 | ||
| Autophagy | 0.492615927 | 4 | 0.492615927 | |||
| AXIN missense mutants destabilize the destruction complex | 2.914767738 | 2.164352856 | 7.993888332 | 0.444696194 | 0.577408986 | 0.0743766 |
| Axon guidance | 0.597959876 | 4 | 0.597959876 | |||
| Axonal growth inhibition (RHOA activation) | 2.657668582 | 0.903089987 | 6.218427151 | -0.455717028 | 0.606137904 | 0.065032485 |
| Axonal growth stimulation | 1.944454942 | 0.602059991 | 4.490969876 | -0.483004386 | 0.516374554 | 0.045658519 |
| B-WICH complex positively regulates rRNA expression | 2.327413119 | 2.638489257 | 7.293315495 | -0.24409665 | 0.243991495 | 0.02310612 |
| Base-Excision Repair, AP Site Formation | 0.428938957 | 0.42666194 | 0.045752982 | |||
| Base Excision Repair | 0.308943262 | 0.34186625 | 0.026404319 | |||
| Basigin interactions | 2.662799233 | 2.460897843 | 7.786496309 | 0.305146335 | 0.306214256 | 0.033283169 |
| BBSome-mediated cargo-targeting to cilium | 3.153458593 | 3.181557774 | 9.488474959 | 0.721692474 | 1.690308073 | 0.308838894 |
| BDNF activates NTRK2 (TRKB) signaling | 2.274010625 | 1.755874856 | 6.303896105 | 0.749931526 | 0.319254816 | 0.046345917 |
| Beta-catenin independent WNT signaling | 0.514372272 | 4 | 0.514372272 | |||
| Beta-catenin phosphorylation cascade | 2.485959763 | 2.79518459 | 7.767104116 | 0.46828187 | 0.791988351 | 0.102035231 |
| Beta-oxidation of pristanoyl-CoA | 2.821966568 | 0.477121255 | 6.12105439 | 0.306522933 | 0.067222445 | 0.005874221 |
| Beta-oxidation of very long chain fatty acids | 2.677834717 | 0.84509804 | 6.200767474 | -0.331140351 | 0.093350202 | 0.008540616 |
| Beta defensins | 3.156215034 | 2.465382851 | 8.777812919 | 0.589912281 | 0.823336925 | 0.129324381 |
| Beta oxidation of butanoyl-CoA to acetyl-CoA | 1.455863467 | 0 | 2.911726935 | -0.944094272 | 3.288066599 | 0.413275778 |
| Beta oxidation of decanoyl-CoA to octanoyl-CoA-CoA | 1.455863467 | 0 | 2.911726935 | -0.944094272 | 3.288066599 | 0.413275778 |
| Beta oxidation of hexanoyl-CoA to butanoyl-CoA | 1.455863467 | 0 | 2.911726935 | -0.944094272 | 3.288066599 | 0.413275778 |
| Beta oxidation of lauroyl-CoA to decanoyl-CoA-CoA | 2.502473001 | 0.301029996 | 5.305975997 | -0.533570841 | 0.481277214 | 0.050656109 |
| Beta oxidation of myristoyl-CoA to lauroyl-CoA | 3.549082534 | 0.301029996 | 7.399195064 | 0.527747645 | ||
| Beta oxidation of octanoyl-CoA to hexanoyl-CoA | 1.455863467 | 0 | 2.911726935 | -0.944094272 | 3.288066599 | 0.413275778 |
| Beta oxidation of palmitoyl-CoA to myristoyl-CoA | 2.847393016 | 0 | 5.694786032 | 0.75623117 | 0.328209658 | 0.045325961 |
| betaKlotho-mediated ligand binding | 2.946720461 | 2.257678575 | 8.151119497 | 0.911804985 | 0.726149667 | 0.137407416 |
| BH3-only proteins associate with and inactivate anti-apoptotic BCL-2 members | 3.842309241 | 3.729407797 | 11.41402628 | 0.563727897 | 1.216191785 | 0.22840133 |
| Bicarbonate transporters | 2.058652746 | 0 | 4.117305492 | 0.239040664 | ||
| Bile acid and bile salt metabolism | 0.213807453 | 0.095578501 | 0.005108849 | |||
| Binding and entry of HIV virion | 3.161921927 | 2.593286067 | 8.917129921 | 0.69627193 | 1.448878012 | 0.249446417 |
| Binding and Uptake of Ligands by Scavenger Receptors | 0.283977543 | 0.216542894 | 0.01537333 | |||
| Binding of TCF/LEF:CTNNB1 to target gene promoters | 3.107733196 | 2.359835482 | 8.575301875 | 0.543492651 | 0.95841845 | 0.142477687 |
| Biogenic amines are oxidatively deaminated to aldehydes by MAOA and MAOB | 1.767530948 | 2.593286067 | 6.128347962 | 0.561643836 | 0.264060748 | 0.031520665 |
| Biological oxidations | -0.131853517 | 0.82920964 | 0.027333552 | |||
| Biosynthesis of aspirin-triggered D-series resolvins | 2.196756256 | 2.507855872 | 6.901368384 | 0.528493151 | 0.201741048 | 0.025256975 |
| Biosynthesis of D-series resolvins | 2.215271454 | 2.636487896 | 7.067030804 | 0.528637983 | 0.397624043 | 0.050637312 |
| Biosynthesis of DHA-derived SPMs | 2.131329155 | 2.800717078 | 7.063375389 | 0.463843827 | 1.433252004 | 0.17084838 |
| Biosynthesis of DPA-derived SPMs | 0.691063596 | 1.417843778 | 0.244955055 | |||
| Biosynthesis of DPAn-3-derived 13-series resolvins | 2.240320111 | 2.482873584 | 6.963513806 | 0.82470556 | 0.448349512 | 0.073091319 |
| Biosynthesis of DPAn-3-derived maresins | 2.260388852 | 2.621176282 | 7.141953986 | 0.714520548 | 0.730992809 | 0.110782946 |
| Biosynthesis of DPAn-3-derived protectins and resolvins | 2.093093167 | 2.804139432 | 6.990325766 | 0.690684932 | 0.671116718 | 0.098396653 |
| Biosynthesis of DPAn-3 SPMs | 2.222634502 | 2.267171728 | 6.712440732 | 0.691063596 | 1.417843778 | 0.202864625 |
| Biosynthesis of DPAn-6 SPMs | 2.058284878 | 2.651278014 | 6.76784777 | 0.690684932 | 0.671116718 | 0.096470975 |
| Biosynthesis of E-series 18(R)-resolvins | 2.106766552 | 2.8162413 | 7.029774403 | 0.528637983 | 0.397624043 | 0.050446251 |
| Biosynthesis of E-series 18(S)-resolvins | 2.128413032 | 2.88422877 | 7.141054834 | 0.528782895 | 0.587985705 | 0.075451823 |
| Biosynthesis of electrophilic ω-3 PUFA oxo-derivatives | 0.783561644 | 0.99929896 | 0.195753084 | |||
| Biosynthesis of EPA-derived SPMs | 2.222634502 | 2.267171728 | 6.712440732 | 0.528927886 | 0.744237873 | 0.09140187 |
| Biosynthesis of maresin-like SPMs | 2.157155731 | 3.065579715 | 7.379891177 | 0.47368417 | 0.621176282 | 0.077346104 |
| Biosynthesis of maresin conjugates in tissue regeneration (MCTR) | 2.232720525 | 0.698970004 | 5.164411053 | 0.331154271 | ||
| Biosynthesis of maresins | 2.202156019 | 2.582063363 | 6.986375401 | 0.508457348 | 1.071680291 | 0.132659989 |
| Biosynthesis of protectin and resolvin conjugates in tissue regeneration (PCTR and RCTR) | 2.232720525 | 0.698970004 | 5.164411053 | 0.331154271 | ||
| Biosynthesis of protectins | 2.015767806 | 2.743509765 | 6.775045376 | 0.47752193 | 0.400234833 | 0.046905279 |
| Biosynthesis of specialized proresolving mediators (SPMs) | 0.444337382 | 1.369215857 | 0.152098448 | |||
| Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein | -0.587555066 | 4 | 0.587555066 | |||
| Biotin transport and metabolism | 2.775451622 | 2.387389826 | 7.938293071 | 0.464985215 | 0.438644321 | 0.057300129 |
| Blockage of phagosome acidification | 1.887664824 | 0.698970004 | 4.474299652 | 0.270445349 | ||
| Blood group systems biosynthesis | -0.325274725 | 1.035612126 | 0.084214612 | |||
| BMAL1:CLOCK,NPAS2 activates circadian gene expression | 2.347229284 | 2.324282455 | 7.018741023 | 0.452955287 | 0.768391413 | 0.090106632 |
| Branched-chain amino acid catabolism | 2.42533399 | 2.017033339 | 6.867701319 | -0.257613169 | 0.147729615 | 0.013428742 |
| Breakdown of the nuclear lamina | 2.265122298 | 0.954242509 | 5.484487106 | 0.600821918 | 0.373389358 | 0.043299048 |
| Budding | 2.943361751 | 1.51851394 | 7.405237442 | 0.528279191 | ||
| Budding and maturation of HIV virion | 3.213287838 | 2.255272505 | 8.68184818 | 0.629722405 | 1.028983427 | 0.165472927 |
| Butyrate Response Factor 1 (BRF1) binds and destabilizes mRNA | 3.018304931 | 2.975431809 | 9.012041671 | 0.787064949 | 1.536011348 | 0.283760374 |
| Butyrophilin (BTN) family interactions | 2.153509 | 1.505149978 | 5.812167978 | -0.43546177 | 0.210508824 | 0.020949494 |
| C-type lectin receptors (CLRs) | 0.489871149 | 4 | 0.489871149 | |||
| Ca-dependent events | 0.493369576 | 3.935963445 | 0.485471154 | |||
| Ca2+ activated K+ channels | 2.758691613 | 2.255272505 | 7.772655731 | 0.602303895 | 1.365903777 | 0.198955646 |
| Ca2+ pathway | 2.451294438 | 2.872156273 | 7.77474515 | 0.204794763 | 0.078560635 | 0.007541579 |
| Calcineurin activates NFAT | 2.770051578 | 2.133538908 | 7.673642065 | 0.421862871 | 0.389451898 | 0.047445552 |
| Calcitonin-like ligand receptors | 3.036959419 | 2.113943352 | 8.187862189 | 0.500815326 | 1.075250952 | 0.14873686 |
| Calmodulin induced events | 2.586639083 | 2.985426474 | 8.15870464 | 0.484031809 | 3.478698207 | 0.472593621 |
| Calnexin/calreticulin cycle | 2.753984481 | 0.301029996 | 5.808998958 | 0.563245901 | 0.701264829 | 0.080961383 |
| Cam-PDE 1 activation | 2.407899328 | 2.064457989 | 6.880256646 | 0.516173246 | 0.543782042 | 0.067093324 |
| CaM pathway | 3.590444885 | 0.698970004 | 7.879859775 | 0.484031809 | 3.478698207 | 0.460083029 |
| CaMK IV-mediated phosphorylation of CREB | 2.556344414 | 2.613841822 | 7.72653065 | 0.534281398 | 1.501254934 | 0.20501274 |
| Cap-dependent Translation Initiation | 0.860219913 | 4 | 0.860219913 | |||
| Carboxyterminal post-translational modifications of tubulin | 2.755460067 | 3.288249226 | 8.799169359 | 0.630289298 | 4 | 0.649584119 |
| Cardiac conduction | 0.382027023 | 3.722810633 | 0.355553566 | |||
| Cargo concentration in the ER | 2.443195228 | 1.633468456 | 6.519858912 | 0.308172964 | 0.153247777 | 0.014211365 |
| Cargo recognition for clathrin-mediated endocytosis | 2.802450094 | 3.757927183 | 9.362827372 | 0.374479437 | 1.253073541 | 0.172535016 |
| Cargo trafficking to the periciliary membrane | 0.424597458 | 0.647817482 | 0.068765414 | |||
| Carnitine metabolism | 2.261673782 | 2.936513742 | 7.459861307 | 0.404092433 | 0.674306058 | 0.078791299 |
| Carnitine synthesis | 1.659617141 | 0 | 3.319234281 | -0.604657534 | 0.43091043 | 0.038142359 |
| CASP8 activity is inhibited | 2.670496925 | 1.681241237 | 7.022235087 | -0.325192097 | 0.433215769 | 0.043902625 |
| Caspase-mediated cleavage of cytoskeletal proteins | 2.134224396 | 2.833784375 | 7.102233167 | 0.344866029 | 0.208594603 | 0.02186744 |
| Caspase activation via Death Receptors in the presence of ligand | 3.187369628 | 2.071882007 | 8.446621264 | 0.340592946 | 0.332244188 | 0.04041321 |
| Caspase activation via Dependence Receptors in the absence of ligand | 2.39047218 | 2.276461804 | 7.057406165 | 0.47162051 | 0.465162453 | 0.055865269 |
| Caspase activation via extrinsic apoptotic signalling pathway | 0.316062385 | 0.344865711 | 0.02724977 | |||
| Catecholamine biosynthesis | 1.605437701 | 0.301029996 | 3.511905397 | -0.548519737 | 0.752137818 | 0.063167136 |
| Cation-coupled Chloride cotransporters | 2.337474549 | 0 | 4.674949098 | -0.541917808 | 0.288732548 | 0.028341505 |
| CD163 mediating an anti-inflammatory response | 2.811282093 | 2.72427587 | 8.346840055 | 0.58536314 | 1.214636101 | 0.183344993 |
| CD209 (DC-SIGN) signaling | 2.762756056 | 3.126780577 | 8.652292689 | 0.517580406 | 2.451087515 | 0.358871339 |
| CD22 mediated BCR regulation | 3.219568438 | 2.328379603 | 8.767516479 | 0.474388678 | 0.241460515 | 0.034408176 |
| CD28 co-stimulation | 2.825156536 | 3.169968174 | 8.820281245 | 0.545472664 | 4 | 0.608264946 |
| CD28 dependent PI3K/Akt signaling | 2.76577557 | 3.704750904 | 9.236302045 | 0.686942086 | 4 | 0.700461821 |
| CD28 dependent Vav1 pathway | 2.76699052 | 2.843232778 | 8.377213818 | 0.359870009 | 0.386120101 | 0.047551283 |
| Cdc20:Phospho-APC/C mediated degradation of Cyclin A | 3.244585419 | 3.054613055 | 9.543783892 | 0.783946722 | 4 | 0.764826868 |
| CDC6 association with the ORC:origin complex | 2.70224945 | 0 | 5.4044989 | -0.750136986 | 0.86719543 | 0.115852923 |
| CDK-mediated phosphorylation and removal of Cdc6 | 3.158288959 | 3.165244326 | 9.481822244 | 0.774145951 | 4 | 0.756729934 |
| CDT1 association with the CDC6:ORC:origin complex | 3.351342375 | 3.118595365 | 9.821280115 | 0.81513038 | 4 | 0.794734494 |
| Cell-Cell communication | 0.380715426 | 1.294906911 | 0.123247759 | |||
| Cell-cell junction organization | 0.411976954 | 0.34817501 | 0.03586002 | |||
| Cell-extracellular matrix interactions | 2.305855543 | 0.602059991 | 5.213771077 | 0.451320836 | 0.402371623 | 0.037735639 |
| Cell Cycle | 0.422686548 | 4 | 0.422686548 | |||
| Cell Cycle Checkpoints | 0.516541592 | 4 | 0.516541592 | |||
| Cell Cycle, Mitotic | 0.450389625 | 4 | 0.450389625 | |||
| Cell death signalling via NRAGE, NRIF and NADE | 0.447775966 | 1.847018667 | 0.206762642 | |||
| Cell junction organization | 0.250645725 | 0.105820165 | 0.006630843 | |||
| Cell recruitment (pro-inflammatory response) | 0.36445318 | 0.62268746 | 0.056735106 | |||
| Cell redox homeostasis | 2.376341605 | 0.954242509 | 5.70692572 | 0.378879147 | ||
| Cell surface interactions at the vascular wall | 3.168571984 | 3.152899596 | 9.490043564 | 0.331683866 | 1.888465225 | 0.253017998 |
| Cellular hexose transport | 2.405045016 | 2.311753861 | 7.121843892 | 0.494232157 | 0.952731918 | 0.117906229 |
| Cellular response to heat stress | 0.380969188 | 1.728635643 | 0.164639229 | |||
| Cellular response to hypoxia | 3.008628714 | 2.440909082 | 8.458166511 | 0.637200187 | 4 | 0.635447513 |
| Cellular responses to external stimuli | 0.531746175 | 4 | 0.531746175 | |||
| Cellular responses to stress | 0.531746175 | 4 | 0.531746175 | |||
| Cellular Senescence | 0.381406499 | 3.137791061 | 0.299193476 | |||
| Centrosome maturation | 0.433052919 | 1.568652939 | 0.169827434 | |||
| Ceramide signalling | 1.81200832 | 0.301029996 | 3.925046637 | -0.585365854 | 0.611819829 | 0.057460666 |
| cGMP effects | 2.244351386 | 2.956648579 | 7.445351351 | 0.461647373 | 1.106348938 | 0.137027076 |
| Chaperone Mediated Autophagy | 2.785142006 | 2.82672252 | 8.397006533 | 0.450349306 | 0.851502953 | 0.114711685 |
| Chaperonin-mediated protein folding | 0.496216125 | 4 | 0.496216125 | |||
| Chemokine receptors bind chemokines | 2.522059816 | 3.1430148 | 8.187134433 | 0.251425372 | 0.242927836 | 0.026028369 |
| Chk1/Chk2(Cds1) mediated inactivation of Cyclin B:Cdk1 complex | 2.962985952 | 2.725094521 | 8.651066424 | 0.444824578 | 1.061891286 | 0.145800661 |
| CHL1 interactions | 2.349938813 | 1.301029996 | 6.000907621 | -0.311848601 | 0.19072431 | 0.016497834 |
| Cholesterol biosynthesis | 2.637321007 | 2.564666064 | 7.839308079 | 0.299908117 | 0.290386125 | 0.031570423 |
| Cholesterol biosynthesis via desmosterol | 4.005918674 | 2.017033339 | 10.02887069 | 0.546875 | 0.637203727 | 0.106941301 |
| Cholesterol biosynthesis via lathosterol | 4.005918674 | 2.017033339 | 10.02887069 | 0.546875 | 0.637203727 | 0.106941301 |
| Choline catabolism | 2.144242787 | 0 | 4.288485575 | 0.254099332 | ||
| Chondroitin sulfate biosynthesis | 1.747886255 | 0 | 3.49577251 | 0.184364568 | ||
| Chondroitin sulfate/dermatan sulfate metabolism | -0.287164015 | 0.102836792 | 0.007382757 | |||
| Chorismate via Shikimate Pathway | 1.585480873 | 0.301029996 | 3.471991741 | 0.18227258 | ||
| ChREBP activates metabolic gene expression | 2.55760479 | 2.720159303 | 7.835368883 | 0.453315786 | 0.614131793 | 0.07851302 |
| Chromatin modifying enzymes | 1.927262632 | 1.113943352 | 4.968468617 | 0.172378404 | 0.041392685 | 0.002307705 |
| Chromatin organization | 0.172378404 | 0.041392685 | 0.001783801 | |||
| Chromosome Maintenance | 0.254863118 | 0.224382824 | 0.014296727 | |||
| Chylomicron assembly | 2.349557229 | 1.414973348 | 6.114087805 | 0.563561862 | 0.707055507 | 0.084439981 |
| Chylomicron clearance | 2.917544346 | 1.505149978 | 7.340238671 | 0.513839408 | 0.345595043 | 0.044589896 |
| Chylomicron remodeling | 2.084532142 | 0.84509804 | 5.014162325 | 0.506576464 | 0.326411727 | 0.03202608 |
| Cilium Assembly | 2.797482852 | 3.137986733 | 8.732952437 | 0.43319345 | 3.200727706 | 0.438195722 |
| Circadian Clock | 2.500119249 | 2.977266212 | 7.97750471 | 0.323652834 | 0.757466652 | 0.085949154 |
| Citric acid cycle (TCA cycle) | 2.593595773 | 1.838849091 | 7.026040637 | -0.191548285 | 0.180890142 | 0.015318654 |
| Class A/1 (Rhodopsin-like receptors) | 2.547422647 | 3.451939869 | 8.546785163 | 0.366878075 | 4 | 0.504858219 |
| Class B/2 (Secretin family receptors) | 2.985509353 | 3.194791758 | 9.165810464 | 0.393459791 | 1.867541532 | 0.256826212 |
| Class C/3 (Metabotropic glutamate/pheromone receptors) | 2.265945878 | 2.900367129 | 7.432258884 | 0.216939547 | 0.098251119 | 0.00914697 |
| Class I MHC mediated antigen processing & presentation | 0.456595245 | 4 | 0.456595245 | |||
| Class I peroxisomal membrane protein import | 1.335048002 | 0 | 2.670096003 | 0.111730018 | ||
| Classical antibody-mediated complement activation | 2.542233756 | 0.477121255 | 5.561588767 | 0.413150685 | 0.043502622 | 0.004067395 |
| Classical Kir channels | 1.961564334 | 0 | 3.923128668 | -0.654246575 | 0.562009475 | 0.057607646 |
| Clathrin-mediated endocytosis | 2.784005229 | 3.760196229 | 9.328206687 | 0.331277086 | 1.118751896 | 0.147499216 |
| Cleavage of the damaged purine | 3.937025917 | 1.716003344 | 9.590055178 | 0.484716486 | 0.521777871 | 0.080562353 |
| CLEC7A (Dectin-1) induces NFAT activation | 2.554593424 | 1.51851394 | 6.627700787 | -0.33287596 | 0.467977875 | 0.045493675 |
| CLEC7A (Dectin-1) signaling | 3.021649264 | 3.420450859 | 9.463749388 | 0.575112885 | 4 | 0.656281037 |
| CLEC7A/inflammasome pathway | 2.157049477 | 1.968482949 | 6.282581902 | 0.382198727 | 0.213479179 | 0.021118972 |
| Cobalamin (Cbl, vitamin B12) transport and metabolism | 2.38904545 | 2.878521796 | 7.656612695 | 0.505213289 | 0.746115018 | 0.098506316 |
| Coenzyme A biosynthesis | 2.008542065 | 0 | 4.017084131 | -0.820997807 | 2.703167381 | 0.336702892 |
| Cohesin Loading onto Chromatin | 2.784985662 | 0 | 5.569971324 | 0.673788003 | 0.221282525 | 0.027922638 |
| Collagen biosynthesis and modifying enzymes | 2.538384511 | 1.579783597 | 6.656552618 | 0.246483437 | 0.04514143 | 0.00391766 |
| Collagen chain trimerization | -0.712328767 | 0.750689861 | 0.133684496 | |||
| Collagen degradation | 2.457177419 | 3.277150614 | 8.191505451 | 0.440291799 | 2.007154617 | 0.262554139 |
| Collagen formation | 0.268823441 | 0.267127096 | 0.017952506 | |||
| Common Pathway of Fibrin Clot Formation | 2.735604707 | 3.188928484 | 8.660137897 | 0.262252983 | 0.113130111 | 0.012964525 |
| Complement cascade | 0.263276327 | 0.147827082 | 0.009729843 | |||
| Complex I biogenesis | 1.950385677 | 1 | 4.900771354 | -0.80315651 | 1.69227485 | 0.226300576 |
| Condensation of Prometaphase Chromosomes | 2.345564402 | 2.981818607 | 7.672947412 | 0.664746228 | 2.140820627 | 0.325785663 |
| Condensation of Prophase Chromosomes | 2.787023776 | 2.741151599 | 8.31519915 | 0.499531168 | 0.995483899 | 0.139177966 |
| Constitutive Signaling by Aberrant PI3K in Cancer | 2.718133601 | 3.60906055 | 9.045327753 | 0.504622652 | 4 | 0.599449901 |
| Constitutive Signaling by AKT1 E17K in Cancer | 2.458588894 | 3.16613397 | 8.083311758 | 0.413337806 | 1.290261812 | 0.162630367 |
| Constitutive Signaling by EGFRvIII | 3.135163505 | 3.228400359 | 9.498727369 | 0.508948849 | 1.652090981 | 0.258140663 |
| Constitutive Signaling by Ligand-Responsive EGFR Cancer Variants | 3.255098206 | 3.230959556 | 9.741155968 | 0.531143323 | 2.212638209 | 0.358783395 |
| Constitutive Signaling by NOTCH1 HD Domain Mutants | 3.16047486 | 1.755874856 | 8.076824576 | 0.504950302 | 0.617772617 | 0.084889596 |
| Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants | 2.670981929 | 3.455758203 | 8.797722061 | 0.554377404 | 4 | 0.611553507 |
| Constitutive Signaling by NOTCH1 PEST Domain Mutants | 2.670981929 | 3.455758203 | 8.797722061 | 0.554377404 | 4 | 0.611553507 |
| Constitutive Signaling by NOTCH1 t(7;9)(NOTCH1:M1580_K2555) Translocation Mutant | 2.72915141 | 1.672097858 | 7.130400678 | 0.40154604 | 0.183885387 | 0.020646749 |
| Constitutive Signaling by Overexpressed ERBB2 | 3.22420874 | 2.840106094 | 9.288523573 | 0.467859478 | 0.97068936 | 0.14405384 |
| Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase | 2.243461093 | 0.301029996 | 4.787952182 | -0.596491228 | 0.89424458 | 0.095181105 |
| Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding | 2.469301787 | 2.899820502 | 7.838424076 | 0.371773751 | 0.435195366 | 0.051218393 |
| Cooperation of Prefoldin and TriC/CCT in actin and tubulin folding | 0.546025276 | 2.868919078 | 0.391625583 | |||
| COPI-dependent Golgi-to-ER retrograde traffic | 2.885033577 | 3.053846427 | 8.823913582 | 0.404597025 | 2.405876212 | 0.323599082 |
| COPI-independent Golgi-to-ER retrograde traffic | 2.74174189 | 3.041787319 | 8.525271099 | 0.556861632 | 4 | 0.598740105 |
| COPI-mediated anterograde transport | 2.78457754 | 3.02201574 | 8.591170819 | 0.413709464 | 1.786956475 | 0.237023574 |
| COPII-mediated vesicle transport | 2.466390255 | 2.428134794 | 7.360915305 | -0.197636064 | 0.230448921 | 0.0206862 |
| Costimulation by the CD28 family | 3.036548849 | 3.324693914 | 9.397791612 | 0.446686153 | 3.030316067 | 0.445960226 |
| COX reactions | 2.312956995 | 2.344392274 | 6.970306264 | 0.71240756 | 0.275451609 | 0.041062518 |
| Creatine metabolism | 2.786012759 | 1.892094603 | 7.464120121 | 0.336436782 | 0.158030145 | 0.017137729 |
| Creation of C4 and C2 activators | -0.371334612 | 0.093476837 | 0.008677796 | |||
| CREB phosphorylation | 2.472072879 | 2.785329835 | 7.729475593 | 0.461884876 | 0.642728594 | 0.081979599 |
| CREB1 phosphorylation through NMDA receptor-mediated activation of RAS signaling | 2.392905384 | 2.644438589 | 7.430249357 | 0.551034982 | 3.363099333 | 0.45345974 |
| CREB1 phosphorylation through the activation of Adenylate Cyclase | 2.794959848 | 2.540329475 | 8.130249171 | 0.516512532 | 1.325163675 | 0.184922187 |
| CREB1 phosphorylation through the activation of CaMKII/CaMKK/CaMKIV cascasde | 2.593647817 | 2.499687083 | 7.686982716 | 0.511331555 | 1.083744803 | 0.144335429 |
| CREB3 factors activate genes | 2.722263059 | 0 | 5.444526117 | 0.554368666 | 0.052322698 | 0.005736668 |
| Cristae formation | 3.144812386 | 1.806179974 | 8.095804746 | -0.478177326 | 1.639798029 | 0.220242152 |
| CRMPs in Sema3A signaling | 2.169299613 | 3.032215703 | 7.370814929 | 0.514582171 | 0.910924736 | 0.117974535 |
| cRNA Synthesis | 2.641954315 | 1.322219295 | 6.606127925 | 0.457981733 | ||
| Cross-presentation of particulate exogenous antigens (phagosomes) | 2.073855291 | 1.740362689 | 5.888073272 | -0.358925144 | 0.27733533 | 0.025218174 |
| Cross-presentation of soluble exogenous antigens (endosomes) | 3.276915779 | 2.944482672 | 9.498314231 | 0.79473929 | 4 | 0.767877411 |
| Crosslinking of collagen fibrils | 2.182792993 | 1.079181246 | 5.444767233 | -0.273450357 | 0.064772051 | 0.004827362 |
| CS/DS degradation | 2.771464701 | 0.77815125 | 6.321080652 | -0.544682018 | 0.742153597 | 0.088861384 |
| CTLA4 inhibitory signaling | 2.724335181 | 3.467163966 | 8.915834328 | 0.555049276 | 1.855797718 | 0.286712747 |
| Cyclin A/B1/B2 associated events during G2/M transition | 2.433733605 | 3.1775365 | 8.04500371 | 0.366839203 | 0.83123878 | 0.099530931 |
| Cyclin A:Cdk2-associated events at S phase entry | 2.467746928 | 3.238798563 | 8.174292419 | 0.611324225 | 4 | 0.607864707 |
| Cyclin D associated events in G1 | 2.687348493 | 3.432809005 | 8.807505991 | 0.415198605 | 1.719872614 | 0.233244331 |
| CYP2E1 reactions | 2.326014333 | 2.88536122 | 7.537389887 | 0.400473134 | 0.462246702 | 0.054265671 |
| Cysteine formation from homocysteine | 2.915388108 | 0.477121255 | 6.30789747 | 0.563407286 | 0.064968224 | 0.007919959 |
| Cysteine synthesis from O-acetylserine | 1.981857061 | 0 | 3.963714121 | 0.225529271 | ||
| Cytochrome c-mediated apoptotic response | 0.472008782 | 0.81442922 | 0.096104436 | |||
| Cytochrome P450 - arranged by substrate type | 1.6726434 | 0 | 3.345286801 | 0.25976811 | 0.376316198 | 0.01721292 |
| Cytokine Signaling in Immune system | 0.353861012 | 4 | 0.353861012 | |||
| Cytosolic iron-sulfur cluster assembly | 2.398638076 | 0.698970004 | 5.496246156 | 0.353088972 | 0.005649439 | 0.000481034 |
| Cytosolic sulfonation of small molecules | 2.43671464 | 0.698970004 | 5.572399284 | -0.310751509 | 0.186684333 | 0.015091042 |
| Cytosolic tRNA aminoacylation | 2.476482385 | 1.230448921 | 6.183413691 | -0.405153509 | 0.281802553 | 0.0283262 |
| DAG and IP3 signaling | 2.218593286 | 2.830588669 | 7.267775241 | 0.438989973 | 2.942626999 | 0.349386285 |
| DAP12 interactions | 4.50092904 | 0.301029996 | 9.302888075 | 0.487370692 | 2.219372176 | 0.33518692 |
| DAP12 signaling | 2.675795563 | 3.606273853 | 8.957864979 | 0.527640405 | 2.633216227 | 0.399226287 |
| DARPP-32 events | 2.497728885 | 2.996511672 | 7.991969442 | 0.478682113 | 1.875672801 | 0.249529081 |
| Dasatinib-resistant KIT mutants | 2.617771423 | 2.158362492 | 7.393905339 | 0.918104629 | 0.749379508 | 0.135074794 |
| DCC mediated attractive signaling | 2.664092417 | 3.317227349 | 8.645412183 | 0.313928514 | 0.137010539 | 0.016560173 |
| DDX58/IFIH1-mediated induction of interferon-alpha/beta | 3.146361343 | 1.041392685 | 7.334115371 | 0.312923937 | 0.606505214 | 0.062973518 |
| De novo synthesis of IMP | 1.551630587 | 0 | 3.103261174 | 0.149835447 | ||
| Deactivation of the beta-catenin transactivating complex | 2.759240738 | 3.369215857 | 8.887697334 | 0.600223716 | 3.786033731 | 0.604930966 |
| Deadenylation-dependent mRNA decay | -0.246628131 | 0.853958305 | 0.052652535 | |||
| Deadenylation of mRNA | 2.530498419 | 1.763427994 | 6.824424831 | -0.314030938 | 1.498878144 | 0.145983061 |
| Death Receptor Signalling | 0.328989983 | 1.508203794 | 0.124045985 | |||
| Dectin-1 mediated noncanonical NF-kB signaling | 3.237236748 | 2.983175072 | 9.457648569 | 0.740665893 | 4 | 0.738742805 |
| Dectin-2 family | 2.820774434 | 2.994317153 | 8.63586602 | 0.35385557 | 0.236233 | 0.029702908 |
| Defective ABCA1 causes TGD | 0.837634982 | 1.260211732 | 0.263899358 | |||
| Defective ABCB11 causes PFIC2 and BRIC2 | 0.815119145 | 0.427387533 | 0.08709294 | |||
| Defective ABCC2 causes DJS | 0.809641194 | 0.416164829 | 0.084236047 | |||
| Defective ABCC8 can cause hypo- and hyper-glycemias | -0.437808219 | 0.096060398 | 0.010514008 | |||
| Defective ABCC9 causes CMD10, ATFB12 and Cantu syndrome | 0.476986301 | 0.114083696 | 0.01360409 | |||
| Defective AHCY causes Hypermethioninemia with S-adenosylhomocysteine hydrolase deficiency (HMAHCHD) | 2.092640797 | 0.477121255 | 4.662402848 | 0.286992738 | ||
| Defective AVP does not bind AVPR1A,B and causes neurohypophyseal diabetes insipidus (NDI) | 2.776910261 | 2.245512668 | 7.799333191 | 0.842191781 | 1.314569394 | 0.231349311 |
| Defective AVP does not bind AVPR2 and causes neurohypophyseal diabetes insipidus (NDI) | 3.002099096 | 2.029383778 | 8.03358197 | 0.927143248 | 0.794607048 | 0.150680358 |
| Defective B3GALT6 causes EDSP2 and SEMDJL1 | 0.713229252 | 0.275451609 | 0.049115036 | |||
| Defective B3GALTL causes Peters-plus syndrome (PpS) | 1.691740574 | 0.477121255 | 3.860602403 | -0.519736842 | 0.661330404 | 0.056135605 |
| Defective B3GAT3 causes JDSSDHD | 0.713229252 | 0.275451609 | 0.049115036 | |||
| Defective B4GALT1 causes B4GALT1-CDG (CDG-2d) | 2.796313514 | 0 | 5.592627027 | 0.36882432 | ||
| Defective B4GALT7 causes EDS, progeroid type | 0.713229252 | 0.275451609 | 0.049115036 | |||
| Defective binding of RB1 mutants to E2F1,(E2F2, E2F3) | 2.484285335 | 2.706717782 | 7.675288452 | 0.531181191 | 1.857708808 | 0.251742762 |
| Defective CFTR causes cystic fibrosis | 3.338270742 | 2.827369273 | 9.503910757 | 0.802873991 | 4 | 0.772233482 |
| Defective cofactor function of FVIIIa variant | 3.514569506 | 2.456366033 | 9.485505045 | 0.774458756 | 1.442380478 | 0.272998029 |
| Defective CP causes aceruloplasminemia (ACERULOP) | 2.746803831 | 1.568201724 | 7.061809387 | 0.868255273 | 0.571700333 | 0.097037294 |
| Defective CYP11A1 causes Adrenal insufficiency, congenital, with 46,XY sex reversal (AICSR) | 0.593536018 | 0.103115854 | 0.015300743 | |||
| Defective CYP11B1 causes Adrenal hyperplasia 4 (AH4) | 1.834697134 | 1.361727836 | 5.031122105 | 0.653245686 | 0.1952033 | 0.022773975 |
| Defective CYP11B2 causes Corticosterone methyloxidase 1 deficiency (CMO-1 deficiency) | 1.765967143 | 1.079181246 | 4.611115531 | 0.661188715 | 0.200565028 | 0.022512204 |
| Defective CYP17A1 causes Adrenal hyperplasia 5 (AH5) | 2.45201941 | 1.62324929 | 6.527288111 | 0.826622843 | 0.455567869 | 0.071814175 |
| Defective CYP19A1 causes Aromatase excess syndrome (AEXS) | 2.060552187 | 1.73239376 | 5.853498133 | 0.733497672 | 0.298487777 | 0.040984133 |
| Defective CYP1B1 causes Glaucoma | 2.008562513 | 0.77815125 | 4.795276276 | 0.612161052 | 0.133233055 | 0.014454538 |
| Defective CYP21A2 causes Adrenal hyperplasia 3 (AH3) | 2.953778363 | 1.322219295 | 7.22977602 | 0.703916735 | 0.258171792 | 0.039076505 |
| Defective CYP24A1 causes Hypercalcemia, infantile (HCAI) | 3.623582979 | 1.041392685 | 8.288558643 | 0.870720351 | 0.578087132 | 0.107445828 |
| Defective CYP26B1 causes Radiohumeral fusions with other skeletal and craniofacial anomalies (RHFCA) | 2.74167968 | 0.602059991 | 6.085419351 | 0.645576554 | 0.185976667 | 0.024048069 |
| Defective CYP27A1 causes Cerebrotendinous xanthomatosis (CTX) | 3.129526989 | 0 | 6.259053977 | 0.427449727 | ||
| Defective CYP27B1 causes Rickets vitamin D-dependent 1A (VDDR1A) | 3.414576441 | 0.698970004 | 7.528122886 | 0.766091482 | 0.337353038 | 0.054981207 |
| Defective EXT1 causes exostoses 1, TRPS2 and CHDS | 0.713229252 | 0.275451609 | 0.049115036 | |||
| Defective EXT2 causes exostoses 2 | 0.713229252 | 0.275451609 | 0.049115036 | |||
| Defective F8 accelerates dissociation of the A2 domain | 5.036920984 | 0.602059991 | 10.67590196 | 0.774034511 | 0.350486053 | 0.071698562 |
| Defective F8 binding to the cell membrane | 5.036920984 | 0.602059991 | 10.67590196 | 0.774034511 | 0.350486053 | 0.071698562 |
| Defective F8 binding to von Willebrand factor | 5.342825153 | 1 | 11.68565031 | 0.774246575 | 0.9404323 | 0.204655439 |
| Defective F8 cleavage by thrombin | 4.395585317 | 2.305351369 | 11.096522 | 0.774458756 | 1.442380478 | 0.302967454 |
| Defective F8 secretion | 5.036920984 | 0.602059991 | 10.67590196 | 0.774034511 | 0.350486053 | 0.071698562 |
| Defective F8 sulfation at Y1699 | 3.409969638 | 0.602059991 | 7.421999268 | 0.420652745 | 0.047397666 | 0.005613291 |
| Defective F9 activation | 2.903955315 | 2.178976947 | 7.986887577 | 0.922465753 | 2.025037055 | 0.381601735 |
| Defective F9 secretion | 3.007810544 | 1.919078092 | 7.93469918 | 0.98767461 | 1.455779172 | 0.285216068 |
| Defective F9 variant does not activate FX | 3.514569506 | 2.456366033 | 9.485505045 | 0.774458756 | 1.442380478 | 0.272998029 |
| Defective factor IX causes hemophilia B | 0.700277149 | 1.794716648 | 0.314199764 | |||
| Defective factor IX causes thrombophilia | 3.514569506 | 2.456366033 | 9.485505045 | 0.774458756 | 1.442380478 | 0.272998029 |
| Defective factor VIII causes hemophilia A | 0.687841821 | 1.937691887 | 0.333206379 | |||
| Defective factor XII causes hereditary angioedema | 2.601519071 | 2.51054501 | 7.713583152 | 0.851192107 | 2.116982979 | 0.372605434 |
| Defective GALE can cause Epimerase-deficiency galactosemia (EDG) | 2.591596578 | 0 | 5.183193156 | 0.332806528 | ||
| Defective GALK1 can cause Galactosemia II (GALCT2) | 2.518483803 | 0.954242509 | 5.991210116 | 0.56066831 | 0.057142886 | 0.006713052 |
| Defective gamma-carboxylation of F9 | 2.793195982 | 1.919078092 | 7.505470057 | 0.763529904 | 0.877876797 | 0.142537248 |
| Defective GCK causes maturity-onset diabetes of the young 2 (MODY2) | 2.163374529 | 1.041392685 | 5.368141743 | 0.57189811 | 0.069931643 | 0.007751655 |
| Defective GCLC causes Hemolytic anemia due to gamma-glutamylcysteine synthetase deficiency (HAGGSD) | 2.807283625 | 0.301029996 | 5.915597246 | 0.397235927 | ||
| Defective GFPT1 causes CMSTA1 | 2.352157702 | 0 | 4.704315405 | 0.290679775 | ||
| Defective GGT1 causes Glutathionuria (GLUTH) | 3.177084184 | 0.698970004 | 7.053138372 | 0.497305127 | ||
| Defective HEXA causes GM2G1 | 2.879894388 | 0.698970004 | 6.45875878 | 0.445017707 | ||
| Defective HEXB causes GM2G2 | 2.663035014 | 0.301029996 | 5.627100024 | 0.371856901 | ||
| Defective HK1 causes hexokinase deficiency (HK deficiency) | 1.614620906 | 0 | 3.229241813 | 0.160917931 | ||
| Defective HLCS causes multiple carboxylase deficiency | 2.096658586 | 1.278753601 | 5.472070773 | 0.494019284 | 0.290414074 | 0.029753422 |
| Defective LARGE causes MDDGA6 and MDDGB6 | 2.011074196 | 0 | 4.022148392 | 0.230669719 | ||
| Defective LFNG causes SCDO3 | 3.401746715 | 1.414973348 | 8.218466778 | 0.646268112 | 1.116813782 | 0.175232429 |
| Defective MAN1B1 causes MRT15 | 1.714952492 | 0 | 3.429904984 | 0.178570219 | ||
| Defective MAOA causes Brunner syndrome (BRUNS) | 2.006543645 | 1.698970004 | 5.712057294 | 0.561490003 | 0.058113377 | 0.006623806 |
| Defective MGAT2 causes MGAT2-CDG (CDG-2a) | 2.491451644 | 0.84509804 | 5.828001329 | 0.771843331 | 0.348585418 | 0.049419048 |
| Defective MPI causes MPI-CDG (CDG-1b) | 1.945112611 | 0.301029996 | 4.191255217 | 0.245546003 | ||
| Defective MTR causes methylmalonic aciduria and homocystinuria type cblG | 1.551341237 | 0 | 3.102682474 | 0.149784539 | ||
| Defective MTRR causes methylmalonic aciduria and homocystinuria type cblE | 1.551341237 | 0 | 3.102682474 | 0.149784539 | ||
| Defective NEU1 causes sialidosis | 2.013218208 | 0.77815125 | 4.804587666 | -0.597423952 | 0.647052205 | 0.069084848 |
| Defective PAPSS2 causes SEMD-PA | 3.942700933 | 0 | 7.885401865 | 0.570519132 | ||
| Defective PMM2 causes PMM2-CDG (CDG-1a) | 1.889856015 | 0 | 3.77971203 | 0.209342654 | ||
| Defective POMGNT1 causes MDDGA3, MDDGB3 and MDDGC3 | 3.765928541 | 0 | 7.531857083 | -0.702739726 | 0.713967054 | 0.110741602 |
| Defective POMT1 causes MDDGA1, MDDGB1 and MDDGC1 | 3.765928541 | 0 | 7.531857083 | 0.539417889 | ||
| Defective POMT2 causes MDDGA2, MDDGB2 and MDDGC2 | 3.765928541 | 0 | 7.531857083 | 0.539417889 | ||
| Defective RIPK1-mediated regulated necrosis | 0.590136986 | 0.336845789 | 0.04969629 | |||
| Defective SERPING1 causes hereditary angioedema | 2.651725785 | 2.120573931 | 7.4240255 | 0.850958904 | 1.39375064 | 0.240065327 |
| Defective SLC11A2 causes hypochromic microcytic anemia, with iron overload 1 (AHMIO1) | 1.727851824 | 0 | 3.455703648 | 0.180839721 | ||
| Defective SLC16A1 causes symptomatic deficiency in lactate transport (SDLT) | 2.458141228 | 0.84509804 | 5.761380495 | 0.552725281 | 0.052322698 | 0.00593974 |
| Defective SLC1A1 is implicated in schizophrenia 18 (SCZD18) and dicarboxylic aminoaciduria (DCBXA) | 1.793527138 | 0 | 3.587054276 | 0.192394601 | ||
| Defective SLC1A3 causes episodic ataxia 6 (EA6) | 2.011514426 | 0.301029996 | 4.324058847 | 0.257228704 | ||
| Defective SLC22A12 causes renal hypouricemia 1 (RHUC1) | 2.300463722 | 0.602059991 | 5.202987434 | 0.521227061 | 0.019112041 | 0.001956734 |
| Defective SLC22A5 causes systemic primary carnitine deficiency (CDSP) | 2.032746471 | 0.477121255 | 4.542614197 | 0.276454962 | ||
| Defective SLC27A4 causes ichthyosis prematurity syndrome (IPS) | 2.179580005 | 0 | 4.35916001 | 0.260316544 | ||
| Defective SLC2A1 causes GLUT1 deficiency syndrome 1 (GLUT1DS1) | 2.258514469 | 1.716003344 | 6.233032282 | 0.684196111 | 0.234204636 | 0.03186077 |
| Defective SLC2A2 causes Fanconi-Bickel syndrome (FBS) | 2.581879218 | 0.84509804 | 6.008856476 | 0.731854286 | 0.298487777 | 0.041520897 |
| Defective SLC2A9 causes hypouricemia renal 2 (RHUC2) | 3.026144768 | 0 | 6.052289536 | 0.409260712 | ||
| Defective SLC33A1 causes spastic paraplegia 42 (SPG42) | 2.648559329 | 0.698970004 | 5.996088663 | 0.862229526 | 0.559201787 | 0.08680848 |
| Defective SLC34A1 causes hypophosphatemic nephrolithiasis/osteoporosis 1 (NPHLOP1) | 2.194461218 | 0 | 4.388922436 | 0.262934737 | ||
| Defective SLC40A1 causes hemochromatosis 4 (HFE4) (duodenum) | 2.746803831 | 1.568201724 | 7.061809387 | 0.868255273 | 0.571700333 | 0.097037294 |
| Defective SLC40A1 causes hemochromatosis 4 (HFE4) (macrophages) | 2.746803831 | 1.568201724 | 7.061809387 | 0.868255273 | 0.571700333 | 0.097037294 |
| Defective SLC5A1 causes congenital glucose/galactose malabsorption (GGM) | 2.414633851 | 1.204119983 | 6.033387685 | 0.821144892 | 0.441249172 | 0.066444135 |
| Defective SLC5A2 causes renal glucosuria (GLYS1) | 2.254349347 | 1.301029996 | 5.80972869 | 0.881676253 | 0.618515789 | 0.09603306 |
| Defective SLC5A7 causes distal hereditary motor neuronopathy 7A (HMN7A) | 2.323637597 | 1.230448921 | 5.877724115 | 0.574637086 | 0.072937153 | 0.008589136 |
| Defective SLC6A2 causes orthostatic intolerance (OI) | 2.896930759 | 2.505149978 | 8.299011497 | 0.984935634 | 1.407475511 | 0.281883764 |
| Defective SLC6A3 causes Parkinsonism-dystonia infantile (PKDYS) | 2.900018898 | 3.17868924 | 8.978727036 | 0.991783073 | 1.591460158 | 0.334045194 |
| Defective SLC6A5 causes hyperekplexia 3 (HKPX3) | 3.129348763 | 1.361727836 | 7.620425363 | 0.812927965 | 0.42061917 | 0.071515043 |
| Defective SLCO1B1 causes hyperbilirubinemia, Rotor type (HBLRR) | 2.634460029 | 2.328379603 | 7.597299662 | 0.96740619 | 1.095637043 | 0.207113507 |
| Defective SLCO1B3 causes hyperbilirubinemia, Rotor type (HBLRR) | 2.592582015 | 2.298853076 | 7.484017107 | 0.969049575 | 1.128637304 | 0.21193457 |
| Defective SLCO2A1 causes primary, autosomal recessive hypertrophic osteoarthropathy 2 (PHOAR2) | 2.204060833 | 0.602059991 | 5.010181658 | 0.789098877 | 0.384138445 | 0.051236215 |
| Defective ST3GAL3 causes MCT12 and EIEE15 | 2.812631269 | 0 | 5.625262539 | 0.371695257 | ||
| Defective TBXAS1 causes Ghosal hematodiaphyseal dysplasia (GHDD) | 2.417314052 | 1.568201724 | 6.402829828 | 0.615173925 | 0.139046991 | 0.018020535 |
| Defective TPMT causes Thiopurine S-methyltransferase deficiency (TPMT deficiency) | 1.536717874 | 0 | 3.073435749 | 0.147211712 | ||
| Defective TPR may confer susceptibility towards thyroid papillary carcinoma (TPC) | 2.27683677 | 1.079181246 | 5.632854787 | 0.572211565 | 0.535926741 | 0.061256125 |
| Defective translocation of RB1 mutants to the nucleus | 2.534533405 | 0.301029996 | 5.370096807 | 0.349248394 | ||
| Defective UGT1A1 causes hyperbilirubinemia | 3.44995963 | 0.84509804 | 7.7450173 | 0.540125993 | 0.04190292 | 0.005762905 |
| Defective UGT1A4 causes hyperbilirubinemia | 3.050398871 | 0.301029996 | 6.401827738 | 0.440009498 | ||
| Defects in biotin (Btn) metabolism | 0.494019284 | 0.290414074 | 0.035867538 | |||
| Defects in vitamin and cofactor metabolism | 0.400058094 | 0.18214123 | 0.018216768 | |||
| Defects of contact activation system (CAS) and kallikrein/kinin system (KKS) | 0.684179007 | 3.282841468 | 0.561512804 | |||
| Defensins | 4.911698236 | 0.77815125 | 10.60154772 | 0.55113283 | 1.426791101 | 0.250754637 |
| Degradation of AXIN | 3.268735533 | 2.924795996 | 9.462267061 | 0.756078226 | 4 | 0.746687236 |
| Degradation of beta-catenin by the destruction complex | 3.169230053 | 3.152288344 | 9.490748451 | 0.716563104 | 4 | 0.728399006 |
| Degradation of cysteine and homocysteine | 3.322659137 | 0 | 6.645318273 | -0.480547945 | 0.15506398 | 0.017971841 |
| Degradation of DVL | 3.407189921 | 2.903632516 | 9.718012358 | 0.789374255 | 4 | 0.776528935 |
| Degradation of GABA | 2.393494393 | 0.602059991 | 5.389048777 | -0.589589041 | 0.390074176 | 0.044206008 |
| Degradation of GLI1 by the proteasome | 3.170698162 | 2.968015714 | 9.309412039 | 0.725114689 | 4 | 0.723319805 |
| Degradation of GLI2 by the proteasome | 3.166504394 | 2.948412966 | 9.281421755 | 0.692965981 | 4 | 0.705801456 |
| Degradation of the extracellular matrix | 2.444935792 | 3.380753771 | 8.270625355 | 0.32548894 | 1.308499078 | 0.153721425 |
| Depolymerisation of the Nuclear Lamina | 2.613844632 | 3.000867722 | 8.228556985 | 0.840416781 | 3.395433968 | 0.615601047 |
| Deposition of new CENPA-containing nucleosomes at the centromere | 2.678132762 | 0.77815125 | 6.134416774 | -0.280504524 | 0.079993771 | 0.006743863 |
| Depurination | 0.484716486 | 0.521777871 | 0.063228584 | |||
| Depyrimidination | 0.549246385 | 0.843447774 | 0.11581516 | |||
| Deregulated CDK5 triggers multiple neurodegenerative pathways in Alzheimer's disease models | 2.590812698 | 2.673941999 | 7.855567395 | 0.293306767 | 0.272305844 | 0.029437162 |
| Detoxification of Reactive Oxygen Species | 2.605945067 | 2.571708832 | 7.783598966 | -0.215549623 | 0.409646482 | 0.039922288 |
| Deubiquitination | 0.436091126 | 4 | 0.436091126 | |||
| Developmental Biology | 0.498146943 | 4 | 0.498146943 | |||
| Digestion | 2.354429827 | 1 | 5.708859655 | -0.247113799 | 0.482260894 | 0.035997146 |
| Digestion and absorption | 0.268634622 | 0.198198729 | 0.01331076 | |||
| Digestion of dietary carbohydrate | 2.329296584 | 1.556302501 | 6.214895669 | 0.360374613 | 0.3096907 | 0.029521751 |
| Digestion of dietary lipid | 2.207777291 | 1.146128036 | 5.561682617 | -0.504795834 | 0.397940009 | 0.041765611 |
| Dimerization of procaspase-8 | 2.670496925 | 1.681241237 | 7.022235087 | -0.325192097 | 0.433215769 | 0.043902625 |
| Dimycocersyl phthiocerol biosynthesis | 2.395468129 | 0.477121255 | 5.268057512 | 0.340272023 | ||
| Disassembly of the destruction complex and recruitment of AXIN to the membrane | 2.845144856 | 2.959518377 | 8.649808089 | 0.326898819 | 0.395370902 | 0.048451069 |
| Diseases associated with glycosaminoglycan metabolism | -0.295013475 | 0.119737543 | 0.008831047 | |||
| Diseases associated with glycosylation precursor biosynthesis | -0.440296459 | 1.308780157 | 0.144062817 | |||
| Diseases associated with N-glycosylation of proteins | -0.228344298 | 0.005395032 | 0.000307981 | |||
| Diseases associated with O-glycosylation of proteins | 0.265037686 | 0.117501193 | 0.007785561 | |||
| Diseases associated with the TLR signaling cascade | 0.326564311 | 0.482147338 | 0.039363028 | |||
| Diseases associated with visual transduction | -0.284813596 | 0.037301411 | 0.002655987 | |||
| Diseases of carbohydrate metabolism | -0.175034432 | 0.116505569 | 0.005098122 | |||
| Diseases of cellular response to stress | 0.830136986 | 1.189630658 | 0.246889102 | |||
| Diseases of Cellular Senescence | 0.830136986 | 1.189630658 | 0.246889102 | |||
| Diseases of glycosylation | -0.223087659 | 1.279257141 | 0.07134662 | |||
| Diseases of hemostasis | 0.684179007 | 3.282841468 | 0.561512804 | |||
| Diseases of Immune System | 0.326564311 | 0.482147338 | 0.039363028 | |||
| Diseases of metabolism | 0.124585771 | 0.004798883 | 0.000149468 | |||
| Diseases of mitotic cell cycle | 0.435967188 | 1.281079769 | 0.139627186 | |||
| Diseases of programmed cell death | 0.320885048 | 0.44389945 | 0.035610174 | |||
| Diseases of signal transduction by growth factor receptors and second messengers | 0.483027973 | 4 | 0.483027973 | |||
| Diseases of the neuronal system | -0.284813596 | 0.037301411 | 0.002655987 | |||
| Disinhibition of SNARE formation | 2.441101507 | 2.926342447 | 7.80854546 | 0.811729241 | 1.719995994 | 0.296354724 |
| Disorders of Developmental Biology | 0.548637099 | 1.301534696 | 0.178517555 | |||
| Disorders of Nervous System Development | 0.548637099 | 1.301534696 | 0.178517555 | |||
| Disorders of transmembrane transporters | 0.599937046 | 4 | 0.599937046 | |||
| Displacement of DNA glycosylase by APEX1 | 2.37882144 | 2.250420002 | 7.008062882 | 0.310757113 | 0.037632536 | 0.003738946 |
| Dissolution of Fibrin Clot | 2.320686376 | 2.414973348 | 7.0563461 | 0.339643347 | 0.193787675 | 0.020073998 |
| DNA Damage Bypass | 0.425240846 | 1.16089976 | 0.123415499 | |||
| DNA Damage Recognition in GG-NER | 3.131463417 | 1.949390007 | 8.212316841 | 0.379025218 | 0.4366268 | 0.053888136 |
| DNA Damage Reversal | -0.543859649 | 0.742153597 | 0.100906849 | |||
| DNA Damage/Telomere Stress Induced Senescence | 2.635637609 | 3.036628895 | 8.307904114 | 0.467224801 | 1.74885734 | 0.237279678 |
| DNA Double-Strand Break Repair | 0.244227878 | 0.330806386 | 0.020198035 | |||
| DNA Double Strand Break Response | 0.228705491 | 0.130780344 | 0.007477546 | |||
| DNA methylation | 2.144966421 | 0.77815125 | 5.068084092 | -0.298868188 | 0.137322718 | 0.010003617 |
| DNA Repair | 0.203789419 | 0.149787842 | 0.007631294 | |||
| DNA Replication | 0.64204053 | 4 | 0.64204053 | |||
| DNA replication initiation | 2.598842939 | 0.84509804 | 6.042783919 | -0.420273973 | 0.069981174 | 0.007039701 |
| DNA Replication Pre-Initiation | 0.727620172 | 4 | 0.727620172 | |||
| DNA strand elongation | 0.17093846 | 0.003930294 | 0.00016796 | |||
| Dopamine clearance from the synaptic cleft | 3.045670364 | 3.354684554 | 9.446025281 | 0.473451114 | 0.23816623 | 0.035995002 |
| Dopamine Neurotransmitter Release Cycle | 2.849506152 | 1.913813852 | 7.612826157 | 0.25879625 | 0.0220444 | 0.002218963 |
| Dopamine receptors | 2.953572564 | 3.803730171 | 9.710875298 | 0.909788868 | 4 | 0.836368046 |
| Downregulation of ERBB2 signaling | 2.910112915 | 3.395850376 | 9.216076206 | 0.564223726 | 3.043383454 | 0.485464708 |
| Downregulation of ERBB2:ERBB3 signaling | 2.730757395 | 3.23350376 | 8.69501855 | 0.715781978 | 2.731460368 | 0.469099232 |
| Downregulation of ERBB4 signaling | 3.145216622 | 3.122870923 | 9.413304167 | 0.800107479 | 2.354979312 | 0.451082103 |
| Downregulation of SMAD2/3:SMAD4 transcriptional activity | 2.509659095 | 3.056904851 | 8.076223042 | 0.331356401 | 0.336431005 | 0.038926874 |
| Downregulation of TGF-beta receptor signaling | 2.656804307 | 2.691965103 | 8.005573716 | 0.337398794 | 0.322948589 | 0.037316543 |
| Downstream signal transduction | 2.931443305 | 3.668665415 | 9.531552025 | 0.400058339 | 1.387834572 | 0.198547654 |
| Downstream signaling events of B Cell Receptor (BCR) | 0.639183299 | 4 | 0.639183299 | |||
| Downstream signaling of activated FGFR1 | 2.936295611 | 2.36361198 | 8.236203203 | 0.487120609 | 1.69773137 | 0.232995305 |
| Downstream signaling of activated FGFR2 | 0.470694236 | 1.569139725 | 0.184646256 | |||
| Downstream signaling of activated FGFR3 | 0.519289485 | 2.075604958 | 0.269459957 | |||
| Downstream signaling of activated FGFR4 | 0.519085311 | 2.075604958 | 0.269354011 | |||
| Downstream TCR signaling | 3.086096119 | 3.615423953 | 9.787616191 | 0.632982876 | 4 | 0.701924049 |
| Drug-mediated inhibition of ERBB2 signaling | 3.346562468 | 2.812244697 | 9.505369633 | 0.708169711 | 1.096594048 | 0.198745774 |
| Drug resistance in ERBB2 KD mutants | 0.708169711 | 1.096594048 | 0.194143672 | |||
| Drug resistance in ERBB2 TMD/JMD mutants | 3.346562468 | 2.812244697 | 9.505369633 | 0.708169711 | 1.096594048 | 0.198745774 |
| Drug resistance of KIT mutants | 0.918104629 | 0.749379508 | 0.172002199 | |||
| Drug resistance of PDGFR mutants | 0.882497946 | 0.618515789 | 0.136459728 | |||
| DSCAM interactions | 2.878762868 | 3.345961542 | 9.103487279 | 0.592488272 | 1.446640706 | 0.23377103 |
| Dual Incision in GG-NER | 2.562048971 | 2.292256071 | 7.416354014 | 0.275488455 | 0.167184746 | 0.01675381 |
| Dual incision in TC-NER | 2.824418928 | 2.492760389 | 8.141598245 | 0.252760945 | 0.156806043 | 0.016734984 |
| E2F-enabled inhibition of pre-replication complex formation | 2.276871539 | 2.222716471 | 6.776459549 | 0.839726027 | 1.260211732 | 0.204769468 |
| E2F mediated regulation of DNA replication | 0.224666963 | 0.033325063 | 0.00187176 | |||
| E3 ubiquitin ligases ubiquitinate target proteins | 2.477776523 | 2.220108088 | 7.175661134 | 0.213059792 | 0.04062724 | 0.003628154 |
| Early Phase of HIV Life Cycle | 2.899236821 | 2.298853076 | 8.097326718 | 0.436405799 | 0.701444623 | 0.090562621 |
| ECM proteoglycans | 2.296956639 | 2.603144373 | 7.19705765 | 0.281411183 | 0.263620169 | 0.025867298 |
| Effects of PIP2 hydrolysis | 2.348018685 | 3.134177108 | 7.830214477 | -0.136952328 | 0 | 0 |
| EGFR downregulation | 2.952586511 | 3.078819183 | 8.983992205 | 0.301676497 | 0.193300699 | 0.023911904 |
| EGFR interacts with phospholipase C-gamma | 2.336615834 | 3.067070856 | 7.740302525 | 0.39808705 | 0.255052107 | 0.030533368 |
| EGFR Transactivation by Gastrin | 2.663972906 | 3.215901813 | 8.543847625 | 0.573332289 | 1.656798956 | 0.251806013 |
| EGR2 and SOX10-mediated initiation of Schwann cell myelination | 2.907578475 | 2.619093331 | 8.43425028 | 0.36163811 | 0.482397122 | 0.059869421 |
| Eicosanoid ligand-binding receptors | 2.092534737 | 0 | 4.185069473 | 0.411163177 | 0.951893696 | 0.071864054 |
| Eicosanoids | 2.418490379 | 1.86923172 | 6.706212477 | -0.385353812 | 0.427716399 | 0.044818529 |
| Elastic fibre formation | 2.171008578 | 2.372912003 | 6.71492916 | 0.251428655 | 0.150275176 | 0.013247869 |
| Elevation of cytosolic Ca2+ levels | 2.355377001 | 2.666517981 | 7.377271983 | 0.290619713 | 0.263788356 | 0.026800556 |
| EML4 and NUDC in mitotic spindle formation | 2.889195997 | 3.235023159 | 9.013415153 | 0.523770089 | 4 | 0.607377275 |
| Endogenous sterols | 2.716201055 | 2.77524626 | 8.207648369 | 0.324248244 | 0.381619005 | 0.044463148 |
| Endosomal Sorting Complex Required For Transport (ESCRT) | 3.29064576 | 1.278753601 | 7.860045121 | 0.460851164 | 0.21061637 | 0.027191438 |
| Endosomal/Vacuolar pathway | 2.414859124 | 1.973127854 | 6.802846102 | 0.450781464 | 0.40098188 | 0.045796297 |
| Energy dependent regulation of mTOR by LKB1-AMPK | 2.579534227 | 3.32858345 | 8.487651904 | 0.575125567 | 2.684305194 | 0.406626154 |
| eNOS activation | 2.724600358 | 3.178976947 | 8.628177663 | 0.369361023 | 0.342826394 | 0.043735963 |
| Entry of Influenza Virion into Host Cell via Endocytosis | 2.813254352 | 1.51851394 | 7.145022643 | 0.505388163 | ||
| Enzymatic degradation of dopamine by COMT | 1.811295796 | 2.301029996 | 5.923621588 | -0.438630137 | 0.096060398 | 0.0097359 |
| Enzymatic degradation of Dopamine by monoamine oxidase | 1.811295796 | 2.301029996 | 5.923621588 | -0.438630137 | 0.096060398 | 0.0097359 |
| EPH-ephrin mediated repulsion of cells | 2.750091305 | 3.62797999 | 9.1281626 | 0.534245992 | 4 | 0.61853495 |
| EPH-Ephrin signaling | 2.642117627 | 3.192846115 | 8.477081369 | 0.452751516 | 4 | 0.54419898 |
| EPHA-mediated growth cone collapse | 2.633593457 | 3.426185825 | 8.693372738 | 0.499584327 | 2.356622775 | 0.340987806 |
| EPHB-mediated forward signaling | 2.776895193 | 3.690639012 | 9.244429398 | 0.45411208 | 2.37603163 | 0.347177485 |
| Ephrin signaling | 2.687533619 | 3.266231697 | 8.641298935 | 0.574156342 | 2.127731224 | 0.326273348 |
| Epigenetic regulation of gene expression | 0.255008553 | 0.304121073 | 0.019388369 | |||
| ER-Phagosome pathway | 3.286083336 | 3.027757205 | 9.599923877 | 0.663921668 | 4 | 0.707710556 |
| ER Quality Control Compartment (ERQC) | 3.084698851 | 1.041392685 | 7.210790388 | 0.735399814 | 0.796055454 | 0.12342765 |
| ER to Golgi Anterograde Transport | 0.347708415 | 1.386346685 | 0.120511102 | |||
| ERBB2 Activates PTK6 Signaling | 2.593881722 | 3.266701967 | 8.45446541 | 0.636187077 | 1.882520946 | 0.298732553 |
| ERBB2 Regulates Cell Motility | 2.753754626 | 3.255754787 | 8.763264039 | 0.656150179 | 2.100155666 | 0.346873384 |
| ERCC6 (CSB) and EHMT2 (G9a) positively regulate rRNA expression | 2.796691618 | 2.79518459 | 8.388567827 | 0.520291629 | 1.142611462 | 0.163794123 |
| ERK/MAPK targets | 2.483760929 | 3.504470862 | 8.47199272 | 0.395429347 | 0.877174623 | 0.112996621 |
| ERKs are inactivated | 2.180844441 | 3.035029282 | 7.396718163 | 0.265280798 | 0.092002679 | 0.009079022 |
| Erythrocytes take up carbon dioxide and release oxygen | 2.276321811 | 2.536558443 | 7.089202064 | 0.508681617 | 0.511025918 | 0.063950359 |
| Erythrocytes take up oxygen and release carbon dioxide | 2.276321811 | 2.536558443 | 7.089202064 | 0.508681617 | 0.511025918 | 0.063950359 |
| Erythropoietin activates Phosphoinositide-3-kinase (PI3K) | 3.022832332 | 3.736157375 | 9.78182204 | 0.690307039 | 3.378148934 | 0.616754744 |
| Erythropoietin activates Phospholipase C gamma (PLCG) | 2.431774098 | 2.474216264 | 7.337764461 | 0.422377196 | 0.39072736 | 0.045933461 |
| Erythropoietin activates RAS | 3.231527597 | 3.004751156 | 9.467806349 | 0.514548736 | 1.474216264 | 0.2307916 |
| Erythropoietin activates STAT5 | 2.584213872 | 2.472756449 | 7.641184193 | 0.412211359 | 0.34817501 | 0.04185113 |
| ESR-mediated signaling | 2.790910793 | 3.589726256 | 9.171547842 | 0.34767508 | 2.868701039 | 0.37830113 |
| Essential fructosuria | 1.92125311 | 0.301029996 | 4.143536216 | 0.241348174 | ||
| Establishment of Sister Chromatid Cohesion | 2.784985662 | 0 | 5.569971324 | 0.673788003 | 0.221282525 | 0.027922638 |
| Estrogen-dependent gene expression | 2.518806201 | 3.695831773 | 8.733444175 | 0.260231217 | 0.375881251 | 0.04333576 |
| Estrogen-dependent nuclear events downstream of ESR-membrane signaling | 2.466239717 | 3.644143051 | 8.576622484 | 0.475691362 | 1.89023932 | 0.265013511 |
| Estrogen-stimulated signaling through PRKCZ | 2.546536633 | 3.114944416 | 8.208017682 | 0.701671571 | 2.0354419 | 0.333190765 |
| Estrogen biosynthesis | 2.052728816 | 2.222716471 | 6.328174102 | 0.411184211 | 0.206299777 | 0.021277502 |
| Ethanol oxidation | 1.870119631 | 2.627365857 | 6.367605118 | -0.346573218 | 0.980523303 | 0.093709525 |
| Eukaryotic Translation Elongation | 2.54570103 | 0 | 5.09140206 | 0.903381997 | 4 | 0.594849872 |
| Eukaryotic Translation Initiation | 0.860219913 | 4 | 0.860219913 | |||
| Eukaryotic Translation Termination | 4.387380501 | 2.894869657 | 11.66963066 | 0.904343252 | 4 | 0.934695781 |
| Evasion of Oncogene Induced Senescence Due to Defective p16INK4A binding to CDK4 | 2.309790471 | 1.886490725 | 6.506071667 | 0.835113667 | 0.480534196 | 0.076128308 |
| Evasion of Oncogene Induced Senescence Due to Defective p16INK4A binding to CDK4 and CDK6 | 2.339514958 | 2.120573931 | 6.799603846 | 0.830136986 | 1.189630658 | 0.192230057 |
| Evasion of Oncogene Induced Senescence Due to p16INK4A Defects | 0.830136986 | 1.189630658 | 0.246889102 | |||
| Evasion of Oxidative Stress Induced Senescence Due to Defective p16INK4A binding to CDK4 | 2.309790471 | 1.886490725 | 6.506071667 | 0.835113667 | 0.480534196 | 0.076128308 |
| Evasion of Oxidative Stress Induced Senescence Due to Defective p16INK4A binding to CDK4 and CDK6 | 2.339514958 | 2.120573931 | 6.799603846 | 0.830136986 | 1.189630658 | 0.192230057 |
| Evasion of Oxidative Stress Induced Senescence Due to p16INK4A Defects | 0.830136986 | 1.189630658 | 0.246889102 | |||
| Events associated with phagocytolytic activity of PMN cells | 2.148067882 | 1.113943352 | 5.410079116 | -0.559726027 | 0.30673697 | 0.033699823 |
| Export of Viral Ribonucleoproteins from Nucleus | 0.288942665 | 0.044046224 | 0.003181708 | |||
| Expression and Processing of Neurotrophins | 0.731719671 | 1.753466155 | 0.320761419 | |||
| Extension of Telomeres | 0.330161242 | 0.541125198 | 0.044664642 | |||
| Extra-nuclear estrogen signaling | 2.777308753 | 3.981274833 | 9.535892339 | 0.459982656 | 4 | 0.602437717 |
| Extracellular matrix organization | 4.341773087 | 0.477121255 | 9.160667428 | 0.274705383 | 1.161584987 | 0.142422383 |
| Extrinsic Pathway of Fibrin Clot Formation | 3.041693404 | 3.164055292 | 9.2474421 | 0.838497395 | 3.342549925 | 0.649135046 |
| Factors involved in megakaryocyte development and platelet production | 2.526428137 | 3.38219721 | 8.435053484 | 0.368213924 | 2.588530315 | 0.323412272 |
| Fanconi Anemia Pathway | 2.83096022 | 2.198657087 | 7.860577527 | 0.313571167 | 0.328243025 | 0.036336822 |
| FasL/ CD95L signaling | 2.520128591 | 0.77815125 | 5.818408432 | 0.317384494 | 0.043285039 | 0.003672268 |
| Fatty acid metabolism | 0.218038129 | 0.233278211 | 0.012715886 | |||
| Fatty acids | 2.621986709 | 2.511883361 | 7.755856779 | 0.222120586 | 0.022781095 | 0.002230703 |
| Fatty Acids bound to GPR40 (FFAR1) regulate insulin secretion | 2.137001822 | 1.740362689 | 6.014366334 | -0.360197368 | 0.155157352 | 0.014385899 |
| Fatty acyl-CoA biosynthesis | 2.499489461 | 2.663700925 | 7.662679847 | 0.292896029 | 0.245399634 | 0.025905425 |
| FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | 3.347652175 | 3.067814511 | 9.763118862 | 0.808014392 | 4 | 0.78817601 |
| FBXW7 Mutants and NOTCH1 in Cancer | 0.616837173 | 0.733153016 | 0.113059008 | |||
| Fc epsilon receptor (FCERI) signaling | 2.874730613 | 2.887054378 | 8.636515604 | 0.52306278 | 4 | 0.587579689 |
| FCERI mediated Ca+2 mobilization | 2.613097341 | 3.245018871 | 8.471213553 | 0.30549984 | 0.438434463 | 0.05154569 |
| FCERI mediated MAPK activation | 2.634172119 | 3.429106008 | 8.697450247 | 0.413039385 | 1.709884827 | 0.229001267 |
| FCERI mediated NF-kB activation | 3.106556657 | 3.335457901 | 9.548571215 | 0.640495057 | 4 | 0.69334801 |
| Fcgamma receptor (FCGR) dependent phagocytosis | 0.389561853 | 2.423615684 | 0.236037054 | |||
| FCGR activation | 3.17779751 | 3.258876629 | 9.61447165 | 0.614045825 | 2.057718295 | 0.351624518 |
| FCGR3A-mediated IL10 synthesis | 2.784592262 | 2.972665592 | 8.541850117 | 0.48879536 | 3.629709316 | 0.513206657 |
| FCGR3A-mediated phagocytosis | 2.799016059 | 3.146438135 | 8.744470254 | 0.471666562 | 2.487978692 | 0.352951418 |
| Fertilization | -0.439298438 | 0.226468947 | 0.024871864 | |||
| FGFR1 ligand binding and activation | 0.532959416 | 0.775952286 | 0.103387769 | |||
| FGFR1 mutant receptor activation | 0.388135766 | 0.631138774 | 0.061241883 | |||
| FGFR1b ligand binding and activation | 2.936295611 | 2.36361198 | 8.236203203 | 0.6007181 | 0.880241776 | 0.13330284 |
| FGFR1c and Klotho ligand binding and activation | 3.235601506 | 2.330413773 | 8.801616785 | 0.915365653 | 0.735293047 | 0.145633714 |
| FGFR1c ligand binding and activation | 3.272875843 | 2.36361198 | 8.909363665 | 0.659497479 | 1.217483944 | 0.203889918 |
| FGFR2 alternative splicing | 2.972842343 | 0.301029996 | 6.246714683 | -0.459429825 | 0.403335041 | 0.04360812 |
| FGFR2 ligand binding and activation | 0.477612545 | 0.632171666 | 0.07548328 | |||
| FGFR2 mutant receptor activation | 0.424635738 | 0.489193057 | 0.051932214 | |||
| FGFR2b ligand binding and activation | 2.949791452 | 2.170261715 | 8.069844618 | 0.535017803 | 0.782582865 | 0.110407401 |
| FGFR2c ligand binding and activation | 3.290870296 | 2.170261715 | 8.752002308 | 0.660507539 | 1.227029262 | 0.203153079 |
| FGFR3 ligand binding and activation | 0.659637114 | 1.217483944 | 0.200774399 | |||
| FGFR3 mutant receptor activation | 3.033432602 | 2.209515015 | 8.276380219 | 0.659637114 | 1.217483944 | 0.193971909 |
| FGFR3b ligand binding and activation | 3.320144162 | 2.187520721 | 8.827809045 | 0.830684932 | 1.211907052 | 0.227614151 |
| FGFR3c ligand binding and activation | 3.279532596 | 2.209515015 | 8.768580206 | 0.659637114 | 1.217483944 | 0.201700552 |
| FGFR4 ligand binding and activation | 3.119116736 | 2.29666519 | 8.534898663 | 0.659336585 | 1.217483944 | 0.197985492 |
| FGFR4 mutant receptor activation | 2.637869158 | 1.959041392 | 7.234779708 | 0.911804985 | 0.726149667 | 0.128825562 |
| FGFRL1 modulation of FGFR1 signaling | 2.56243219 | 0.954242509 | 6.079106889 | 0.276745621 | 0.004447824 | 0.00036971 |
| Fibronectin matrix formation | 2.823209217 | 1.204119983 | 6.850538416 | 0.497944642 | 0.301662617 | 0.03641698 |
| FLT3 Signaling | 2.65547774 | 2.940516485 | 8.251471964 | 0.474887719 | 4 | 0.543628081 |
| FMO oxidises nucleophiles | -0.867360921 | 2.201338775 | 0.477338807 | |||
| Folding of actin by CCT/TriC | 2.642322673 | 0.477121255 | 5.761766602 | 0.512876712 | 0.166378198 | 0.018059554 |
| Formation of a pool of free 40S subunits | 4.372108713 | 2.901458321 | 11.64567575 | 0.914536557 | 4 | 0.938556619 |
| Formation of annular gap junctions | 2.668988332 | 1.361727836 | 6.699704501 | 0.418303805 | 0.227995435 | 0.024810563 |
| Formation of apoptosome | 2.619967998 | 1.278753601 | 6.518689596 | 0.471749863 | 0.61901022 | 0.070051277 |
| Formation of ATP by chemiosmotic coupling | 3.592613844 | 1.806179974 | 8.991407662 | 0.479572251 | 0.496852496 | 0.072558228 |
| Formation of Fibrin Clot (Clotting Cascade) | 0.562944818 | 3.952206723 | 0.556218573 | |||
| Formation of HIV-1 elongation complex containing HIV-1 Tat | 2.775378265 | 2.826074803 | 8.376831333 | 0.559254361 | 1.514537726 | 0.224257066 |
| Formation of HIV elongation complex in the absence of HIV Tat | 2.968865603 | 2.603144373 | 8.540875579 | 0.516537675 | 1.325163675 | 0.191944376 |
| Formation of Incision Complex in GG-NER | 2.656764985 | 2.499687083 | 7.813217053 | 0.314837608 | 0.406426625 | 0.044807908 |
| Formation of Senescence-Associated Heterochromatin Foci (SAHF) | 2.305953824 | 2.69019608 | 7.302103727 | 0.394678356 | 0.1726716 | 0.019621809 |
| Formation of TC-NER Pre-Incision Complex | 3.22269461 | 2.307496038 | 8.752885257 | 0.376242742 | 0.570352293 | 0.074170478 |
| Formation of the beta-catenin:TCF transactivating complex | 2.568067264 | 3.250420002 | 8.38655453 | 0.236121149 | 0.121319595 | 0.013078665 |
| Formation of the cornified envelope | 2.643917483 | 2.51851394 | 7.806348907 | 0.360598491 | 0.597863877 | 0.069280458 |
| Formation of the Early Elongation Complex | 2.737273932 | 2.225309282 | 7.699857146 | 0.468337064 | 0.447819898 | 0.057309237 |
| Formation of the Editosome | 2.162473389 | 0.301029996 | 4.625976774 | 0.283788346 | ||
| Formation of the HIV-1 Early Elongation Complex | 2.605185205 | 2.025305865 | 7.235676276 | 0.468337064 | 0.447819898 | 0.054628285 |
| Formation of the ternary complex, and subsequently, the 43S complex | 4.17886706 | 2.534026106 | 10.89176023 | 0.869182633 | 4 | 0.876985789 |
| Formation of tubulin folding intermediates by CCT/TriC | 2.81460137 | 2.920645001 | 8.549847741 | 0.634289025 | 4 | 0.63872169 |
| Formation of xylulose-5-phosphate | 1.832474659 | 0.477121255 | 4.142070574 | -0.644012058 | 0.817069316 | 0.085013956 |
| Formyl peptide receptors bind formyl peptides and many other ligands | 2.745267069 | 2.5289167 | 8.019450839 | 0.522519199 | 0.572745738 | 0.079536348 |
| FOXO-mediated transcription | 0.25747287 | 0.344610567 | 0.022181968 | |||
| FOXO-mediated transcription of cell cycle genes | 2.901476664 | 2.687528961 | 8.49048229 | 0.242191064 | 0.041796398 | 0.00459353 |
| FOXO-mediated transcription of cell death genes | 2.761034731 | 1.949390007 | 7.471459468 | 0.361196567 | 0.431509525 | 0.048171816 |
| FOXO-mediated transcription of oxidative stress, metabolic and neuronal genes | 2.633155825 | 2.904715545 | 8.171027195 | 0.28877951 | 0.256946089 | 0.028676719 |
| Free fatty acid receptors | 2.161999627 | 1.812913357 | 6.136912611 | -0.360197368 | 0.155157352 | 0.014631128 |
| Free fatty acids regulate insulin secretion | -0.360296134 | 0.281195751 | 0.025328435 | |||
| FRS-mediated FGFR1 signaling | 2.896378926 | 2.509202522 | 8.301960375 | 0.43197692 | 0.77815125 | 0.102089035 |
| FRS-mediated FGFR2 signaling | 2.901777262 | 2.380211242 | 8.183765767 | 0.408898272 | 0.707187285 | 0.08966082 |
| FRS-mediated FGFR3 signaling | 3.006356067 | 2.404833717 | 8.417545851 | 0.489324916 | 1.170261715 | 0.163665282 |
| FRS-mediated FGFR4 signaling | 2.969507376 | 2.462397998 | 8.401412751 | 0.489223288 | 1.170261715 | 0.163406916 |
| Frs2-mediated activation | 2.790755098 | 3.368286885 | 8.94979708 | 0.713383302 | 3.3795803 | 0.590498755 |
| Fructose biosynthesis | 1.697968744 | 1.86332286 | 5.259260349 | -0.491780822 | 0.178365057 | 0.017734345 |
| Fructose catabolism | 1.955069038 | 2.624282096 | 6.534420172 | 0.453486956 | 0.084972867 | 0.009439348 |
| Fructose metabolism | 0.328399123 | 0.058728247 | 0.004821576 | |||
| Fusion of the Influenza Virion to the Host Cell Endosome | 2.943361751 | 1.51851394 | 7.405237442 | 0.528279191 | ||
| G-protein activation | 2.682721891 | 3.279438788 | 8.644882571 | 0.328940545 | 0.214017407 | 0.026267973 |
| G-protein beta:gamma signalling | 0.399238074 | 0.8162413 | 0.081468651 | |||
| G-protein mediated events | 0.401091794 | 2.474303024 | 0.24810566 | |||
| G alpha (12/13) signalling events | 2.752038722 | 3.634779458 | 9.138856901 | 0.313950412 | 0.376466907 | 0.047899661 |
| G alpha (i) signalling events | 2.567120395 | 4.387193994 | 9.521434783 | 0.29083569 | 2.642942502 | 0.341678537 |
| G alpha (q) signalling events | 2.667611726 | 4.416790238 | 9.75201369 | 0.347427132 | 4 | 0.557309474 |
| G alpha (s) signalling events | 2.675680617 | 3.941859127 | 9.293220361 | 0.353249169 | 3.2709999 | 0.438765164 |
| G alpha (z) signalling events | 2.989892517 | 3.408070286 | 9.38785532 | 0.575049324 | 4 | 0.652333946 |
| G beta:gamma signalling through BTK | 2.456764967 | 2.62838893 | 7.541918865 | 0.483963985 | 0.261768892 | 0.033477687 |
| G beta:gamma signalling through CDC42 | 3.234383867 | 1.633468456 | 8.102236189 | 0.371162281 | 0.126658717 | 0.015327808 |
| G beta:gamma signalling through PI3Kgamma | 2.507889372 | 3.164947373 | 8.180726116 | 0.529389752 | 1.364699076 | 0.193524352 |
| G beta:gamma signalling through PLC beta | 2.276920174 | 0.477121255 | 5.030961603 | -0.629385965 | 0.987666265 | 0.112281304 |
| G protein gated Potassium channels | 0.439770008 | 0.827485878 | 0.090975868 | |||
| G0 and Early G1 | 2.223436846 | 3.13001195 | 7.576885641 | 0.374287204 | 0.738279738 | 0.084630203 |
| G1 Phase | 0.415198605 | 1.719872614 | 0.178522177 | |||
| G1/S-Specific Transcription | 2.5149718 | 2.699837726 | 7.729781326 | 0.265249924 | 0.154498403 | 0.015909303 |
| G1/S DNA Damage Checkpoints | 0.730338528 | 4 | 0.730338528 | |||
| G1/S Transition | 0.533296295 | 4 | 0.533296295 | |||
| G2 Phase | 2.219853321 | 2.657055853 | 7.096762495 | 0.719276452 | 1.673762021 | 0.253680375 |
| G2/M Checkpoints | 3.3093952 | 3.108226656 | 9.727017057 | 0.525579325 | 4 | 0.645096015 |
| G2/M DNA damage checkpoint | 2.41530344 | 2.945468585 | 7.776075465 | 0.302160985 | 0.573810196 | 0.062077571 |
| G2/M DNA replication checkpoint | 2.716195894 | 2.58546073 | 8.017852518 | 0.627639155 | 1.340223717 | 0.203697778 |
| G2/M Transition | 0.55138528 | 4 | 0.55138528 | |||
| GAB1 signalosome | 2.692826774 | 3.632153484 | 9.017807031 | 0.523493613 | 1.670439654 | 0.253683661 |
| GABA B receptor activation | 3.191924719 | 1.968482949 | 8.352332386 | 0.440581747 | 1.765694331 | 0.234695384 |
| GABA receptor activation | 2.268063253 | 3.72664572 | 8.262772226 | 0.491410107 | 4 | 0.552472247 |
| GABA synthesis | 3.310132572 | 0 | 6.620265145 | -0.446575342 | 0.104632278 | 0.011648693 |
| GABA synthesis, release, reuptake and degradation | 2.952837149 | 0.301029996 | 6.206704293 | -0.314936848 | 0.641931503 | 0.057479317 |
| Galactose catabolism | 2.55504019 | 0.954242509 | 6.06432289 | 0.449315068 | 0.079040947 | 0.008259948 |
| Gamma-carboxylation of protein precursors | 2.605162015 | 3.07371835 | 8.28404238 | 0.666245917 | 1.776442271 | 0.284669305 |
| Gamma-carboxylation, transport, and amino-terminal cleavage of proteins | 0.705130763 | 2.566270386 | 0.452389049 | |||
| Gamma carboxylation, hypusine formation and arylsulfatase activation | 3.100079906 | 0.954242509 | 7.154402322 | 0.342374442 | 0.522569529 | 0.054970989 |
| Gap-filling DNA repair synthesis and ligation in GG-NER | 2.717702589 | 2.075546961 | 7.51095214 | 0.315997542 | 0.116705419 | 0.012428549 |
| Gap-filling DNA repair synthesis and ligation in TC-NER | 3.000326417 | 2.494154594 | 8.494807428 | 0.295993576 | 0.252923692 | 0.029512052 |
| Gap junction assembly | 2.79062862 | 2.950851459 | 8.532108699 | 0.687122022 | 4 | 0.664223046 |
| Gap junction degradation | 2.668988332 | 1.361727836 | 6.699704501 | 0.418303805 | 0.227995435 | 0.024810563 |
| Gap junction trafficking | 0.544067625 | 3.188877124 | 0.433741201 | |||
| Gap junction trafficking and regulation | 0.558802764 | 3.564849271 | 0.498011906 | |||
| Gastrin-CREB signalling pathway via PKC and MAPK | 2.506035292 | 3.321805484 | 8.333876067 | 0.545064092 | 2.827883373 | 0.412140973 |
| GDP-fucose biosynthesis | 3.080015461 | 0 | 6.160030923 | 0.418738695 | ||
| Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation | 2.71491574 | 2.315970345 | 7.745801824 | 0.314722657 | 0.371392777 | 0.040617227 |
| Gene expression (Transcription) | 0.329930477 | 4 | ||||
| 2, YAF9, YAP-1, YAP1, YEATS4, YES1, YGK3, YNG1, YNG2, YPK1, YWHAB, YWHABB, YWHABL, YWHAE, YWHAE1, YWHAE2, YWHAG, YWHAG2, YWHAH, YWHAQ, YWHAQA, YWHAQB, YWHAZ, YY1, ZBTB12.2, ZBTB17, ZBTB39, ZBTB41, ZBTB47A, ZBTB49, ZC3H11A, ZC3H11B, ZC3H6, ZC3H8, ZFH-2, ZFH2, ZFHX3, ZFP1, ZFP101, ZFP108, ZFP109, ZFP11, ZFP111, ZFP112, ZFP113, ZFP114, ZFP120, ZFP13, ZFP133, ZFP136, ZFP14, ZFP157, ZFP160, ZFP169, ZFP180, ZFP184, ZFP189, ZFP2, ZFP202, ZFP212, ZFP213, ZFP229, ZFP235, ZFP248, ZFP263, ZFP267, ZFP27, ZFP28, ZFP282, ZFP286, ZFP286A, ZFP30, ZFP317, ZFP324, ZFP334, ZFP345, ZFP37, ZFP383, ZFP385A, ZFP386, ZFP39, ZFP398, ZFP40, ZFP418, ZFP420, ZFP429, ZFP433, ZFP442, ZFP445, ZFP454, ZFP455, ZFP456, ZFP457, ZFP458, ZFP459, ZFP46, ZFP472, ZFP473, ZFP496, ZFP51, ZFP52, ZFP53, ZFP54, ZFP551, ZFP558, ZFP560, ZFP563, ZFP566, ZFP57, ZFP583, ZFP595, ZFP60, ZFP600, ZFP606, ZFP61, ZFP612, ZFP617, ZFP626, ZFP641, ZFP647, ZFP65, ZFP655, ZFP658, ZFP668, ZFP677, ZFP688, ZFP69, ZFP69B, ZFP703, ZFP704, ZFP706, ZFP707, ZFP709, ZFP710, ZFP729A, ZFP735, ZFP738, ZFP74, ZFP746, ZFP748, ZFP759, ZFP760, ZFP763, ZFP764, ZFP770, ZFP772, ZFP775, ZFP78, ZFP780B, ZFP780B-PS1, ZFP786, ZFP790, ZFP791, ZFP799, ZFP804B, ZFP81, ZFP810, ZFP811, ZFP819, ZFP82, ZFP839, ZFP84, ZFP866, ZFP867, ZFP87, ZFP870, ZFP871, ZFP872, ZFP873, ZFP874A, ZFP874B, ZFP882, ZFP9, ZFP90, ZFP934, ZFP937, ZFP938, ZFP94, ZFP942, ZFP944, ZFP945, ZFP948, ZFP950, ZFP952, ZFP954, ZFP955A, ZFP955B, ZFP960, ZFP961, ZFP963, ZFP964, ZFP97, ZFP970, ZFP971, ZFP974, ZFP975, ZFP976, ZFP997, ZFPM1, ZG16, ZGC:112234;, ZGC:113050, ZGC:113102, ZGC:113377, ZGC:113452, ZGC:113984;, ZGC:158291, ZGC:158398, ZGC:162472, ZGC:173486, ZGC:173585, ZGC:173729, ZGC:174153, ZGC:174574, ZGC:56493, ZGC:77262, ZGC:86599, ZGC:91910, ZIK1, ZIM2, ZIM3, ZK328.6, ZKSCAN1, ZKSCAN3, ZKSCAN4, ZKSCAN5, ZKSCAN7, ZKSCAN8, ZMP:0000001236, ZNF10, ZNF100, ZNF101, ZNF1018, ZNF1028, ZNF1035, ZNF1071, ZNF1109, ZNF1114, ZNF112, ZNF1138, ZNF114, ZNF1175, ZNF12, ZNF124, ZNF133, ZNF135, ZNF136, ZNF138, ZNF14, ZNF140, ZNF141, ZNF143, ZNF143A, ZNF146, ZNF154, ZNF155, ZNF157, ZNF16, ZNF160, ZNF169, ZNF17, ZNF175, ZNF177, ZNF18, ZNF180, ZNF181, ZNF184, ZNF189, ZNF19, ZNF192, ZNF195, ZNF197, ZNF2, ZNF20, ZNF200, ZNF202, ZNF205, ZNF208, ZNF211, ZNF212, ZNF213, ZNF214, ZNF215, ZNF221, ZNF222, ZNF223, ZNF224, ZNF225, ZNF226, ZNF227, ZNF23, ZNF230, ZNF233, ZNF234, ZNF235, ZNF248, ZNF25, ZNF250, ZNF253, ZNF254, ZNF256, ZNF257, ZNF26, ZNF263, ZNF264, ZNF266, ZNF267, ZNF268, ZNF271, ZNF273, ZNF274, ZNF28, ZNF282, ZNF283, ZNF285, ZNF286A, ZNF287, ZNF3, ZNF30, ZNF300, ZNF302, ZNF304, ZNF311, ZNF317, ZNF320, ZNF324, ZNF324B, ZNF331, ZNF333, ZNF334, ZNF337, ZNF33A, ZNF33B, ZNF34, ZNF343, ZNF347, ZNF350, ZNF354A, ZNF354B, ZNF354C, ZNF37A, ZNF382, ZNF383, ZNF384, ZNF385A, ZNF391, ZNF394, ZNF398, ZNF41, ZNF415, ZNF416, ZNF417, ZNF418, ZNF419, ZNF420, ZNF425, ZNF426, ZNF429, ZNF43, ZNF430, ZNF431, ZNF432, ZNF433, ZNF436, ZNF439, ZNF440, ZNF441, ZNF442, ZNF443, ZNF445, ZNF446, ZNF45, ZNF454, ZNF460, ZNF461, ZNF467, ZNF468, ZNF470, ZNF471, ZNF473, ZNF479, ZNF480, ZNF483, ZNF484, ZNF485, ZNF486, ZNF490, ZNF492, ZNF493, ZNF496, ZNF500, ZNF506, ZNF510, ZNF514, ZNF517, ZNF519, ZNF521, ZNF528, ZNF529, ZNF530, ZNF540, ZNF543, ZNF544, ZNF546, ZNF547, ZNF548, ZNF549, ZNF550, ZNF551, ZNF552, ZNF554, ZNF555, ZNF556, ZNF557, ZNF558, ZNF559, ZNF560, ZNF561, ZNF562, ZNF563, ZNF564, ZNF565, ZNF566, ZNF567, ZNF568, ZNF569, ZNF570, ZNF571, ZNF572, ZNF573, ZNF574, ZNF575, ZNF577, ZNF582, ZNF583, ZNF584, ZNF585A, ZNF585B, ZNF586, ZNF587, ZNF589, ZNF595, ZNF596, ZNF597, ZNF599, ZNF600, ZNF605, ZNF606, ZNF607, ZNF610, ZNF611, ZNF613, ZNF614, ZNF615, ZNF616, ZNF619, ZNF620, ZNF621, ZNF624, ZNF625, ZNF626, ZNF627, ZNF641, ZNF648, ZNF649, ZNF655, ZNF658, ZNF658B, ZNF660, ZNF662, ZNF664, ZNF665, ZNF667, ZNF668, ZNF669, ZNF670, ZNF671, ZNF675, ZNF676, ZNF677, ZNF678, ZNF679, ZNF680, ZNF681, ZNF682, ZNF684, ZNF688, ZNF689, ZNF691, ZNF692, ZNF696, ZNF697, ZNF699, ZNF70, ZNF700, ZNF701, ZNF702P, ZNF703, ZNF704, ZNF705A, ZNF705D, ZNF705E, ZNF705F, ZNF705G, ZNF706, ZNF706L, ZNF707, ZNF708, ZNF709, ZNF71, ZNF710, ZNF710A, ZNF711, ZNF713, ZNF714, ZNF716, ZNF717, ZNF718, ZNF720, ZNF721, ZNF724, ZNF726, ZNF726P1, ZNF727, ZNF728, ZNF729, ZNF730, ZNF732, ZNF735, ZNF736, ZNF737, ZNF738, ZNF74, ZNF740, ZNF746, ZNF747, ZNF749, ZNF750, ZNF75A, ZNF75CP, ZNF75D, ZNF761, ZNF764, ZNF767P, ZNF77, ZNF770, ZNF771, ZNF772, ZNF773, ZNF774, ZNF775, ZNF776, ZNF777, ZNF778, ZNF782, ZNF784, ZNF785, ZNF786, ZNF79, ZNF790, ZNF791, ZNF792, ZNF793, ZNF799, ZNF804B, ZNF805, ZNF835, ZNF839, ZNF840P, ZNF852, ZNF860, ZNF875, ZNF879, ZNF92, ZNF985, ZNF988, ZNF99, ZNF996, ZSCAN25, ZSCAN32, ZTF-8, ZW, ZWF1 | 0.329930477 | |||||
| Gene Silencing by RNA | 0.477074022 | 0.719641978 | 0.085830623 | |||
| Generation of second messenger molecules | 2.533662335 | 2.814913181 | 7.882237851 | 0.371950768 | 0.582158094 | 0.068856368 |
| Generic Transcription Pathway | 3.047777312 | 1.785329835 | 7.880884459 | 0.348753663 | 4 | 0.461442756 |
| GLI proteins bind promoters of Hh responsive genes to promote transcription | 2.584330468 | 0.301029996 | 5.469690932 | -0.560547945 | 0.311357167 | 0.034478794 |
| GLI3 is processed to GLI3R by the proteasome | 3.11396962 | 3.173477643 | 9.401416883 | 0.705029702 | 4 | 0.718023764 |
| Global Genome Nucleotide Excision Repair (GG-NER) | 0.276223669 | 0.355181111 | 0.024527357 | |||
| Glucagon-like Peptide-1 (GLP1) regulates insulin secretion | 2.725989222 | 3.486146997 | 8.93812544 | 0.335853222 | 0.659733237 | 0.084039246 |
| Glucagon-type ligand receptors | 2.985342272 | 3.282848603 | 9.253533148 | 0.426459303 | 1.167739185 | 0.166726697 |
| Glucagon signaling in metabolic regulation | 3.113568113 | 2.459392488 | 8.686528713 | 0.531045283 | 2.820142949 | 0.418897742 |
| Glucocorticoid biosynthesis | 2.46629885 | 2.444044796 | 7.376642495 | 0.46630683 | 0.840678951 | 0.103867129 |
| Gluconeogenesis | 2.68179378 | 1.681241237 | 7.044828798 | -0.479504814 | 3.636714944 | 0.439757865 |
| Glucose metabolism | 0.183298269 | 0.053302909 | 0.002442583 | |||
| Glucuronidation | 2.486993547 | 1.342422681 | 6.316409775 | -0.46346642 | 3.226635021 | 0.35338849 |
| Glutamate and glutamine metabolism | 2.515399331 | 1.982271233 | 7.013069896 | -0.289163237 | 0.239059751 | 0.023121723 |
| Glutamate binding, activation of AMPA receptors and synaptic plasticity | 0.466836346 | 2.038346413 | 0.237893548 | |||
| Glutamate Neurotransmitter Release Cycle | 2.654821851 | 1.986771734 | 7.296415437 | -0.328115996 | 0.242384483 | 0.025509249 |
| Glutathione conjugation | 2.254742516 | 1.230448921 | 5.739933953 | -0.274326553 | 0.797817811 | 0.062584691 |
| Glutathione synthesis and recycling | 2.992183905 | 0.602059991 | 6.586427801 | -0.525753425 | 0.258916319 | 0.031274677 |
| Glycerophospholipid biosynthesis | 2.331599169 | 0 | 4.663198337 | 0.211291604 | 0.141362662 | 0.008012224 |
| Glycerophospholipid catabolism | 1.994047482 | 0 | 3.988094964 | -0.748698273 | 1.316892429 | 0.151636649 |
| Glycine degradation | 3.549082534 | 0.698970004 | 7.797135072 | 0.562754325 | ||
| Glycogen breakdown (glycogenolysis) | 2.21419288 | 2.465382851 | 6.893768611 | 0.293861678 | 0.188658955 | 0.018067504 |
| Glycogen metabolism | 0.268672841 | 0.144143442 | 0.009681857 | |||
| Glycogen storage disease type 0 (muscle GYS1) | 2.16618187 | 0 | 4.332363739 | 0.257959283 | ||
| Glycogen storage disease type Ia (G6PC) | 2.068166944 | 0 | 4.136333888 | 0.240714587 | ||
| Glycogen storage disease type Ib (SLC37A4) | 2.47379029 | 0 | 4.94758058 | 0.31207975 | ||
| Glycogen storage disease type II (GAA) | 2.346617785 | 1.672097858 | 6.365333427 | 0.688304574 | 0.24007357 | 0.033192104 |
| Glycogen storage disease type XV (GYG1) | 2.16618187 | 0 | 4.332363739 | 0.257959283 | ||
| Glycogen storage diseases | 0.226989711 | 0.020747956 | 0.001177393 | |||
| Glycogen synthesis | 3.014914482 | 1.041392685 | 7.07122165 | 0.416706525 | 0.123466194 | 0.01400259 |
| Glycolysis | 2.98156528 | 1.662757832 | 7.625888391 | 0.216108234 | 0.109144469 | 0.010422342 |
| Glycoprotein hormones | 2.302452787 | 1.939519253 | 6.544424827 | 0.313896998 | 0.038882305 | 0.003645873 |
| Glycosaminoglycan metabolism | -0.251729344 | 0.695948413 | 0.043797659 | |||
| Glycosphingolipid metabolism | 2.225213435 | 2.584331224 | 7.034758095 | -0.248207391 | 1.336612169 | 0.122807482 |
| Glyoxylate metabolism and glycine degradation | 2.401169683 | 1.322219295 | 6.124558661 | -0.310164835 | 0.902403287 | 0.079307932 |
| Golgi-to-ER retrograde transport | 0.36670871 | 2.103143144 | 0.192810227 | |||
| Golgi Associated Vesicle Biogenesis | 2.739094433 | 1.643452676 | 7.121641543 | 0.256527194 | 0.082716906 | 0.007778715 |
| Golgi Cisternae Pericentriolar Stack Reorganization | 2.428812518 | 3.201397124 | 8.059022161 | 0.599705315 | 1.492398197 | 0.222407834 |
| GP1b-IX-V activation signalling | 3.724373155 | 3.220892249 | 10.66963856 | 0.797653046 | 4 | 0.829761926 |
| GPCR downstream signalling | 0.322200366 | 4 | ||||
| .1, SI:DKEY-73N8.3, SI:DKEY-94E7.2, SI:DKEYP-118H3.5, SIF, SIFAR, SLC24A1, SMA-5, SOS, SOS-1, SOS1, SOS2, SPAC1786.02, SPAC1A6.03C, SPAC22H12.03, SPAC977.09C, SPBC1348.10C, SPBC800.10C, SPCC1450.09C, SPE-41, SPK1, SPN100A, SPN27A, SPN28B, SPN28DA, SPN28DB, SPN28DC, SPN28F, SPN31A, SPN38F, SPN42DA, SPN42DB, SPN42DD, SPN42DE, SPN43AA, SPN43AB, SPN43AD, SPN47C, SPN53F, SPN55B, SPN75F, SPN77BA, SPN77BB, SPN77BC, SPN85F, SPN88EA, SPN88EB, SPO1, SRC, SRO-1, SRP-1, SRP-2, SRP-3, SRP-6, SRP-7, SRP-8, SS1R, SS2R, SS3R, SST, SST.1, SST1.1, SST2, SSTR1, SSTR1A, SSTR1B, SSTR2, SSTR2B, SSTR3, SSTR4, SSTR5, STE6, STI, STPG4, STRA6, SUCNR1, T2R10, T2R10C, T2R14, T2R15, T2R17, T2R19, T2R20, T2R22, T2R24, T2R32, T2R34, T2R37, T2R38, T2R43, T2R45, T2R46, T2R48, T2R49, T2R52, T2R53, T2R65A, T2R9, TAAR1, TAAR13A, TAAR13B, TAAR13C, TAAR13D, TAAR13E, TAAR19T, TAAR1B, TAAR2, TAAR3, TAAR3P, TAAR5, TAAR6, TAAR8, TAAR8B, TAAR9, TAC1, TAC3, TAC3A, TACR1, TACR1A, TACR2, TACR3, TACR3L, TAG-89, TAPBP, TAS1R1, TAS1R2, TAS1R2.1, TAS1R3, TAS2R1, TAS2R10, TAS2R106, TAS2R119, TAS2R120, TAS2R13, TAS2R131, TAS2R135, TAS2R136, TAS2R14, TAS2R140, TAS2R145, TAS2R16, TAS2R19, TAS2R20, TAS2R200.1, TAS2R200.2, TAS2R203, TAS2R3, TAS2R30, TAS2R31, TAS2R38, TAS2R39, TAS2R4, TAS2R40, TAS2R41, TAS2R42, TAS2R43, TAS2R45, TAS2R46, TAS2R5, TAS2R50, TAS2R60, TAS2R7, TAS2R8, TAS2R9, TAX-2, TAX-4, TBXA2R, TCF3, TDA5, TIAM-1, TIAM1, TIAM1A, TIAM2, TIAM2A, TKR-1, TMEM26, TMIGD3, TOS3, TPA-1, TPK1, TPK2, TPK3, TRAP1, TRE1, TRH, TRHR, TRHR-1, TRHRA, TRIO, TRIOB, TRISSINR, TRP-1, TRPC3, TRPC6, TRPC6B, TRPC7, TRPC7A, TSHB, TSHBA, TSHR, TSPAN17, TTC19, TTR, TYRA-2, TYRA-3, TYRR, TYRRII, UBE2N, UIG-1, UNC-73, UTS2, UTS2B, UTS2D, UTS2R, VAV, VAV-1, VAV1, VAV2, VAV3, VAV3B, VIP, VIPR1, VIPR1B, VIPR2, VPAC2, XB5807236, XCL1, XCL2, XCR1, XCR1A.1, XENTR_V90028168MG, XENTR_V90028851MG, YDL114W, YPK1, ZC3H12B, ZC4H2, ZGC:101744, ZGC:162608, ZK697.8, ZNF750 | 0.322200366 | |||||
| GPCR ligand binding | 0.343431502 | 4 | 0.343431502 | |||
| GPVI-mediated activation cascade | 2.80813808 | 3.632760888 | 9.249037049 | 0.374737507 | 1.127459832 | 0.153621058 |
| GRB2 events in EGFR signaling | 2.814068824 | 3.08170727 | 8.709844919 | 0.410606212 | 0.577126751 | 0.077210095 |
| GRB2 events in ERBB2 signaling | 2.971993262 | 3.204933522 | 9.148920047 | 0.53227242 | 1.486014968 | 0.229819282 |
| GRB7 events in ERBB2 signaling | 3.067642286 | 2.806179974 | 8.941464546 | 0.664839682 | 0.897437484 | 0.151263133 |
| Growth hormone receptor signaling | 2.64960199 | 3.424554977 | 8.723758956 | 0.409312215 | 0.758121253 | 0.101437613 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | 4.173041351 | 2.92788341 | 11.27396611 | 0.870577636 | 4 | 0.897400971 |
| HATs acetylate histones | 2.499052102 | 2.29666519 | 7.294769395 | -0.245038589 | 1.26620697 | 0.120083282 |
| HCMV Early Events | 2.617278711 | 3.251151343 | 8.485708766 | 0.480758709 | 3.599689998 | 0.502739595 |
| HCMV Infection | 0.461327245 | 3.091351754 | 0.356531197 | |||
| HCMV Late Events | 2.336434301 | 1 | 5.672868602 | 0.486160592 | 0.267582827 | 0.027844438 |
| HCN channels | 2.161917907 | 0.602059991 | 4.925895806 | -0.610197368 | 0.923208276 | 0.101488108 |
| HDACs deacetylate histones | 2.78650158 | 3.147676324 | 8.720679484 | 0.558917094 | 2.564449916 | 0.39098167 |
| HDL assembly | 2.536838506 | 2.522444234 | 7.596121245 | 0.637367483 | 1.532926293 | 0.226512459 |
| HDL clearance | 1.439622222 | 0.77815125 | 3.657395695 | -0.423287671 | 0.078048796 | 0.005479485 |
| HDL remodeling | 2.308569498 | 2.562292864 | 7.17943186 | 0.323605815 | 0.127718231 | 0.013176737 |
| HDMs demethylate histones | 2.048754076 | 2.269512944 | 6.367021095 | 0.260109437 | 0.173576254 | 0.014711529 |
| HDR through Homologous Recombination (HRR) | 2.670181792 | 2.51851394 | 7.858877525 | 0.269449076 | 0.211080566 | 0.022198031 |
| HDR through Homologous Recombination (HRR) or Single Strand Annealing (SSA) | 0.265267132 | 0.355153758 | 0.023552655 | |||
| HDR through MMEJ (alt-NHEJ) | 2.22757855 | 2.10720997 | 6.56236707 | 0.302083333 | 0.029595541 | 0.002738227 |
| HDR through Single Strand Annealing (SSA) | 2.641664143 | 2.828015064 | 8.11134335 | 0.254520576 | 0.146499386 | 0.015610073 |
| Hedgehog 'off' state | 2.942087322 | 3.22685757 | 9.111032215 | 0.621961167 | 4 | 0.661508796 |
| Hedgehog 'on' state | 3.222593228 | 3.011147361 | 9.456333817 | 0.716756162 | 4 | 0.726720112 |
| Hedgehog ligand biogenesis | 3.321007494 | 2.961895474 | 9.603910461 | 0.753492463 | 4 | 0.752701618 |
| Heme biosynthesis | 2.404553576 | 2.459392488 | 7.268499639 | -0.34559429 | 0.628234823 | 0.067263634 |
| Heme degradation | 2.493782109 | 3.113274692 | 8.10083891 | 0.398272914 | 0.645853426 | 0.080336036 |
| Hemostasis | 0.289518273 | 3.629516327 | 0.262702825 | |||
| Heparan sulfate/heparin (HS-GAG) metabolism | 0.369392544 | 0.183035674 | 0.016903003 | |||
| Hh mutants abrogate ligand secretion | 0.803757214 | 4 | 0.803757214 | |||
| Highly calcium permeable nicotinic acetylcholine receptors | 1.921366557 | 3.07371835 | 6.916451464 | 0.377394509 | 0.424654678 | 0.045226667 |
| Highly calcium permeable postsynaptic nicotinic acetylcholine receptors | 2.029605095 | 3.13893394 | 7.198144131 | 0.405191274 | 0.652484442 | 0.074128686 |
| Highly sodium permeable postsynaptic acetylcholine nicotinic receptors | 2.005622502 | 3.003460532 | 7.014705536 | 0.440061586 | 0.554546103 | 0.064107093 |
| Histamine receptors | 2.113627851 | 3.211921084 | 7.439176786 | 0.577576754 | 0.783331763 | 0.108308732 |
| Histidine catabolism | 2.134176459 | 1.079181246 | 5.347534163 | 0.519405283 | 0.368609407 | 0.038342304 |
| HIV elongation arrest and recovery | 3.159791186 | 2.471291711 | 8.790874084 | 0.564968965 | 1.077108237 | 0.166008232 |
| HIV Infection | 0.524755778 | 4 | 0.524755778 | |||
| HIV Life Cycle | 0.415589789 | 2.145780263 | 0.222941092 | |||
| HIV Transcription Elongation | 0.559254361 | 1.514537726 | 0.211752957 | |||
| HIV Transcription Initiation | 2.549132026 | 2.029383778 | 7.127647831 | 0.242677495 | 0.042908549 | 0.003964169 |
| Homologous DNA Pairing and Strand Exchange | 2.659464602 | 2.238046103 | 7.556975308 | 0.29436143 | 0.234828079 | 0.02451232 |
| Homology Directed Repair | 0.278733311 | 0.46419487 | 0.032346643 | |||
| Hormone ligand-binding receptors | 2.991532083 | 3.07114529 | 9.054209457 | 0.648062277 | 1.621260889 | 0.272221017 |
| Host Interactions of HIV factors | 0.611173457 | 4 | 0.611173457 | |||
| HS-GAG biosynthesis | 4.843162618 | 0.84509804 | 10.53142328 | 0.447785936 | 0.077360889 | 0.012526623 |
| HS-GAG degradation | 2.657444777 | 1.826074803 | 7.140964357 | 0.442629492 | 0.286955709 | 0.033732276 |
| HSF1-dependent transactivation | 2.667586543 | 3.410777233 | 8.745950319 | 0.351341455 | 0.658238598 | 0.083491752 |
| HSF1 activation | 2.739402657 | 3.109578547 | 8.588383861 | 0.470546269 | 0.87629681 | 0.122427068 |
| HSP90 chaperone cycle for steroid hormone receptors (SHR) | 2.728454706 | 3.532117116 | 8.989026529 | 0.512427097 | 4 | 0.60044759 |
| HuR (ELAVL1) binds and stabilizes mRNA | 2.293948968 | 2.876794976 | 7.464692911 | 0.533424151 | 0.606083927 | 0.080655683 |
| Hyaluronan biosynthesis and export | 2.273369892 | 0.477121255 | 5.023861039 | 0.318790122 | ||
| Hyaluronan metabolism | -0.381448957 | 0.734245765 | 0.07001932 | |||
| Hyaluronan uptake and degradation | 2.485478083 | 1.431363764 | 6.40231993 | -0.381344307 | 0.578719149 | 0.058083173 |
| Hydrolysis of LPC | 2.854014531 | 2.041392685 | 7.749421746 | 0.392017858 | 0.167592074 | 0.019955725 |
| Hydrolysis of LPE | 3.678862617 | 0.301029996 | 7.65875523 | 0.67981375 | 0.226978272 | 0.034927038 |
| Hydroxycarboxylic acid-binding receptors | 2.052984659 | 1.176091259 | 5.282060577 | -0.537407509 | 0.485473329 | 0.051180849 |
| Hypusine synthesis from eIF5A-lysine | 2.704986251 | 0.698970004 | 6.108942506 | -0.605753425 | 0.433979661 | 0.054087931 |
| IGF1R signaling cascade | 0.47445583 | 2.898686136 | 0.343824634 | |||
| IkBA variant leads to EDA-ID | 2.329213213 | 2.392696953 | 7.05112338 | 0.356277641 | 0.240714923 | 0.02541938 |
| IKBKB deficiency causes SCID | 2.525399045 | 2.155336037 | 7.206134127 | 0.532760462 | 0.413710863 | 0.053641373 |
| IKBKG deficiency causes anhidrotic ectodermal dysplasia with immunodeficiency (EDA-ID) (via TLR) | 2.525399045 | 2.155336037 | 7.206134127 | 0.532760462 | 0.413710863 | 0.053641373 |
| IKK complex recruitment mediated by RIP1 | 3.074600713 | 2.788875116 | 8.938076541 | 0.509097632 | 2.245185267 | 0.334619264 |
| IL-6-type cytokine receptor ligand interactions | 2.774925262 | 2.902546779 | 8.452397303 | 0.392974242 | 0.284716548 | 0.036517525 |
| Imatinib-resistant KIT mutants | 2.617771423 | 2.158362492 | 7.393905339 | 0.918104629 | 0.749379508 | 0.135074794 |
| Imatinib-resistant PDGFR mutants | 2.318185123 | 2.017033339 | 6.653403585 | 0.882497946 | 0.618515789 | 0.102826733 |
| Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell | 2.942329077 | 2.646403726 | 8.53106188 | 0.241939534 | 0.184419525 | 0.020358892 |
| Inactivation of APC/C via direct inhibition of the APC/C complex | 2.0418342 | 0 | 4.0836684 | -0.841600438 | 1.906363685 | 0.244000712 |
| Inactivation of CDC42 and RAC1 | 2.657270552 | 0.84509804 | 6.159639144 | -0.470136986 | 0.129880434 | 0.014070494 |
| Inactivation, recovery and regulation of the phototransduction cascade | 2.69873907 | 3.376029182 | 8.773507321 | 0.337516117 | 0.513951981 | 0.064484739 |
| Incretin synthesis, secretion, and inactivation | 0.387458244 | 0.49662653 | 0.048105511 | |||
| Infection with Mycobacterium tuberculosis | 0.360168636 | 0.47096766 | 0.042406945 | |||
| Infectious disease | 0.480222965 | 4 | 0.480222965 | |||
| Inflammasomes | 0.463920023 | 1.227124617 | 0.14232192 | |||
| Influenza Infection | 2.943361751 | 1.51851394 | 7.405237442 | 0.826895197 | 4 | 0.675975293 |
| Influenza Viral RNA Transcription and Replication | 0.894835487 | 4 | 0.894835487 | |||
| Influenza Virus Induced Apoptosis | 3.049655892 | 0.954242509 | 7.053554293 | 0.672730771 | 0.612713702 | 0.088958377 |
| Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits | 2.599434789 | 2.530199698 | 7.729069276 | 0.439770008 | 0.827485878 | 0.103253462 |
| Inhibition of DNA recombination at telomere | 3.711017358 | 1.397940009 | 8.819974725 | 0.620401228 | 0.982614685 | 0.158621871 |
| Inhibition of nitric oxide production | 2.161195661 | 1.838849091 | 6.161240412 | 0.555890411 | 0.250111405 | 0.029781774 |
| Inhibition of PKR | 2.707677037 | 1.322219295 | 6.737573368 | 0.687482881 | 0.235664451 | 0.033689701 |
| Inhibition of replication initiation of damaged DNA by RB1/E2F1 | 2.487200621 | 1.079181246 | 6.053582488 | -0.42085048 | 0.782242658 | 0.0788544 |
| Inhibition of Signaling by Overexpressed EGFR | 2.566125945 | 2.765668555 | 7.897920445 | 0.510128905 | 0.514753791 | 0.069879017 |
| Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components | -0.841600438 | 1.906363685 | 0.401099128 | |||
| Inhibition of TSC complex formation by PKB | 2.667308687 | 2.356025857 | 7.690643231 | 0.909613804 | 0.712785706 | 0.130450198 |
| Initial triggering of complement | 2.731125299 | 0.477121255 | 5.939371852 | 0.327665126 | 0.136876442 | 0.012001925 |
| Initiation of Nuclear Envelope (NE) Reformation | 2.47541536 | 2.58546073 | 7.536291449 | 0.484865909 | 1.173186268 | 0.150086193 |
| InlA-mediated entry of Listeria monocytogenes into host cells | 3.276310398 | 2.658011397 | 9.210632193 | 0.795438242 | 2.280100671 | 0.42944876 |
| InlB-mediated entry of Listeria monocytogenes into host cell | 3.482208477 | 2.713490543 | 9.677907497 | 0.679177552 | 0.961895474 | 0.172987669 |
| Innate Immune System | 0.279853275 | 3.995843735 | ||||
| F3D7_1116700, PF3D7_1120100, PF3D7_1121900, PF3D7_1122800, PF3D7_1124100, PF3D7_1126100, PF3D7_1129200, PF3D7_1130400, PF3D7_1133300, PF3D7_1138400, PF3D7_1140100, PF3D7_1144900, PF3D7_1202400, PF3D7_1202900, PF3D7_1203200, PF3D7_1211800, PF3D7_1213600, PF3D7_1215000, PF3D7_1219600, PF3D7_1223500, PF3D7_1229400, PF3D7_1238900, PF3D7_1242000, PF3D7_1242800, PF3D7_1248500, PF3D7_1306600, PF3D7_1311400, PF3D7_1311500, PF3D7_1314500, PF3D7_1316000, PF3D7_1320500, PF3D7_1323200, PF3D7_1323500, PF3D7_1328100, PF3D7_1329400, PF3D7_1333300, PF3D7_1341900, PF3D7_1342400, PF3D7_1348100, PF3D7_1352600, PF3D7_1354400, PF3D7_1357000, PF3D7_1357300, PF3D7_1365900, PF3D7_1366500, PF3D7_1367500, PF3D7_1368100, PF3D7_1402300, PF3D7_1403900, PF3D7_1405200, PF3D7_1417600, PF3D7_1428300, PF3D7_1429800, PF3D7_1431900, PF3D7_1434200, PF3D7_1436000, PF3D7_1438900, PF3D7_1439100, PF3D7_1439800, PF3D7_1440300, PF3D7_1444800, PF3D7_1451100, PF3D7_1451700, PF3D7_1455100, PF3D7_1464700, PF3D7_1466300, PFK, PFK-1.1, PFK-1.2, PFK1, PFK2, PFKA, PFKL, PFKLB, PGAM1, PGAM1A, PGAM1B, PGES-2, PGI, PGI1, PGLYM78, PGLYM87, PGLYRP1, PGLYRP1-LIKE.1, PGLYRP2, PGLYRP3, PGLYRP3B, PGLYRP4, PGLYRP5, PGLYRP6, PGM1, PGM2, PGM2A, PGMA, PGMB, PGRMC1, PGRN-1, PGRP-LA, PGRP-LB, PGRP-LC, PGRP-LD, PGRP-LE, PGRP-LF, PGRP-SA, PGRP-SB1, PGRP-SB2, PGRP-SD, PHF5A, PHO-1, PHO-10, PHO-11, PHO-12, PHO-13, PHO-14, PHO-4, PHO-5, PHO-6, PHO-7, PHO-8, PHO-9, PHO2A, PI030, PI3, PI31, PI3K21B, PI3K92E, PIGE-108A11.3, PIGR, PIGRL4.2, PIK3C3, PIK3CA, PIK3CB, PIK3R1, PIK3R2, PIK3R4, PIN1, PINOPSIN100496420, PIRA, PIRA2, PKA-C1, PKA-C2, PKC-1, PKC1, PKC98E, PKCDELTA, PKH3, PKM, PKMA, PKP1, PKP1B, PKU70, PKU80, PLA2G10, PLA2G2A, PLA2G5, PLA2G6, PLAC8, PLAC8.1, PLACL2, PLAU, PLAUB, PLAUR, PLC-3, PLCG1, PLCG2, PLD, PLD-1, PLD1, PLD1A, PLD2, PLD3, PLD4, PLDA, PLDB, PLDC, PLDY, PLDZ, PLEKHO2, PLI, PLIN5, PLL, PLPP4, PLPP5, PMD1, PMK-1, PMK-2, PMO25, PMP2, PMT-2, PNP, PNP-1, PNP1, PNP5A, POC1A, POE, POLG, POLR1C, POLR1D, POLR2E, POLR2F, POLR2H, POLR2K, POLR2L, POLR3A, POLR3B, POLR3C, POLR3D, POLR3E, POLR3F, POLR3G, POLR3GL, POLR3H, POLR3K, POP5, PORB, POT1, POXA, PP2A-29B, PP2B-14D, PPA1, PPA2, PPB1, PPBP, PPIA, PPIAB, PPIE, PPP2CA, PPP2CAA, PPP2CB, PPP2R1A, PPP2R1B, PPP2R1BA, PPP2R1BB, PPP2R2C, PPP2R5D, PPP3CA, PPP3CB, PPP3CC, PPP3R1, PPP3R1B, PPPA, PPTR-2, PQN-84, PRCP, PRDX-6, PRDX4, PRDX6, PRE7, PRE8, PRG2, PRG3, PRGJ, PRICKLE2, PRKACA, PRKACB, PRKACG, PRKCD, PRKCDA, PRKCE, PRKCEB, PRKCQ, PRKCSH, PRKDC, PRLA, PRM15, PRORP, PROS1, PROSALPHA1, PROSALPHA2, PROSALPHA3, PROSALPHA4, PROSALPHA4T1, PROSALPHA4T2, PROSALPHA5, PROSALPHA6, PROSALPHA7, PROSBETA1, PROSBETA2, PROSBETA2R1, PROSBETA2R2, PROSBETA3, PROSBETA4, PROSBETA5, PROSBETA5R1, PROSBETA5R2, PROSBETA6, PROSBETA7, PRPS2, PRSS1, PRSS2, PRSS3, PRSS57, PRSS59.1, PRSS59.2, PRTN3, PRTN3-PROV, PRTP, PRX1, PRX6005, PSAA, PSAB, PSAP, PSEN1, PSENA, PSENB, PSG18, PSG22, PSG29, PSGB1, PSMA1, PSMA2, PSMA3, PSMA4, PSMA5, PSMA6, PSMA6A, PSMA6B, PSMA7, PSMA8, PSMB1, PSMB10, PSMB11, PSMB11A, PSMB2, PSMB3, PSMB4, PSMB5, PSMB6, PSMB7, PSMB7.2, PSMB8, PSMB8A, PSMB9, PSMB9A, PSMC1, PSMC1A, PSMC2, PSMC3, PSMC4, PSMC5, PSMC6, PSMD1, PSMD10, PSMD11, PSMD11B, PSMD12, PSMD13, PSMD14, PSMD2, PSMD3, PSMD4, PSMD4A, PSMD5, PSMD6, PSMD7, PSMD8, PSMD9, PSME1, PSME2, PSME3, PSME4, PSME4B, PSMF1, PSN, PSRA, PSTPIP1, PSTPIP1A, PTAFR, PTCD2, PTGDS, PTGDSA, PTGDSB.1, PTGDSB.2, PTGES2, PTGFR, PTK2, PTK2AB, PTP, PTP-1, PTP-2, PTP1, PTP2, PTP3, PTP52F, PTPA1-1; PTPA1-2, PTPB, PTPMEG, PTPN11, PTPN4, PTPN4A, PTPN6, PTPRB, PTPRC, PTPRJ, PTPRJB.1, PTPRN2, PTR1, PTX3, PTX3B, PUP1, PUP2, PXN, PXN-1, PXN-2, PYCARD, PYGB, PYGL, PYGM, PYK, PYK-1, PYK-2, PYK1, PYK2, PYKA, PYP1, PYP2, PYP3, Q19348, Q19831, Q21673, Q2Q423, Q6Q1Q8, Q6Q1Q9, Q8I3I8, Q8I3J6, Q8I4A8, Q8IDU4, Q8IDV2, Q8IDV5, Q8UWG6, Q8WNW4, Q90633, Q90WU1, Q9VIT8, QDPR, QPCT, QRICH1, QSOX1, R04B3.2, R07B1.3, R09B5.11, R09H10.3, R4GJ86, R4GMA6, RAB-10, RAB-14, RAB-18, RAB-3, RAB-5, RAB-6.2, RAB-7, RAB10, RAB14, RAB14L, RAB18, RAB18B, RAB1C, RAB24, RAB27, RAB27A, RAB3, RAB31, RAB32D, RAB37, RAB3A, RAB3AB, RAB3D, RAB3DA, RAB4, RAB41, RAB44, RAB4B, RAB5, RAB5A, RAB5B, RAB5C, RAB6, RAB6A, RAB7, RAB7A, RAB7B, RAB8A, RAB8B, RAB9, RAB9B, RABJ, RABK1, RABK2, RABK3, RABL, RABM, RABN1, RABN2, RABO, RABV, RABZ, RAC-2, RAC1, RAC1A, RAC1B, RAC1C, RAC2, RACB, RACC, RACD, RACE, RACF1, RACF2, RACG, RACH, RACJ, RACL, RACM, RACN, RACQ, RAD32, RAF, RAF1, RAF1A, RAL2, RALA, RAP-1, RAP-2, RAP-3, RAP1, RAP1A, RAP1AA, RAP1B, RAP2A, RAP2B, RAP2C, RAP2L, RAPA, RAPB, RAPC, RAS, RAS85D, RASGRP1, RASGRP2, RASGRP4, RBM12, RBSN, RCC1L, RCDBB, RCG_21066, RCG_21069, RCG_64228, RCJMB04_11A3, RCJMB04_11H6, RCJMB04_15C17, RCJMB04_17B10, RCJMB04_18I8, RCJMB04_19D10, RCJMB04_21A21, RCJMB04_25F21, RCJMB04_29D24, RCJMB04_29G15, RCJMB04_3O20, RCJMB04_8F10, REDB, REG, REG3A, REG3B, REG3D, REG3G, REL, RELA, RELB, RELL1, RETN, RGA3, RGA4, RGA5, RGAA, RGD1308751, RGD1564614, RGD1564657, RGD1564827, RHA-1, RHAU, RHO-1, RHO1, RHO2, RHO3, RHO4, RHO5, RHOA, RHOAB, RHOF, RHOG, RHOGAP16F, RHOGAP18B, RHOGB, RIC-4, RIP-1, RIPK1, RIPK2, RIPK3, RL, RME-8, RNASE17, RNASE2, RNASE2A, RNASE2B, RNASE3, RNASE6, RNASE7, RNASE8, RNASEL2, RNASEL3, RNASEL4, RNASET2, RNASET2A, RNASET2B, RNASEX25, RNF125, RNF135, RNF216, RNF38, RNG2, RNST-2, RNY1, ROCK1, ROK, ROL-3, RPL40, RPN-1, RPN-11, RPN-2, RPN-3, RPN-5, RPN-6.1, RPN-7, RPN-8, RPN-9, RPN1, RPN10, RPN11, RPN12, RPN2, RPN3, RPN5, RPN501, RPN502, RPN6, RPN7, RPN8, RPN9, RPS27A, RPS3A, RPS6KA, RPS6KA1, RPS6KA2, RPS6KA3, RPS6KA3A, RPS6KA5, RPT-1, RPT-5, RPT1, RPT2, RPT3, RPT4, RPT5, RPT6, RSKN-1, RSKN-2, RSR1, RT1-A1, RT1-A2, RT1-CE1, RT1-CE10, RT1-CE3, RT1-CE4, RT1-CE5, RT1-CE7, RT1-M2, RT1-M3-1, RT1-M4, RT1-M5, RT1-M6-1, RT1-M6-2, RT1-N2, RT1-N3, RT1-S3, RTN4RL2A, RTN4RL2B, RYH1, S100A1, S100A10A, S100A10B, S100A11, S100A12, S100A7, S100A7A, S100A8, S100A9, S100B, S100P, S100Z, S6KII, SAA1, SAC7, SACY-1, SANTA-MARIA, SAP-R, SARM1, SARNP, SC:D0175, SC:D0202, SCAMP, SCAMP1, SCAR, SCAV-2, SCAV-3, SCAV-4, SCAV-6, SCB, SCL-1, SCL-10, SCL-11, SCL-12, SCL-13, SCL-14, SCL-15, SCL-17, SCL-18, SCL-19, SCL-2, SCL-20, SCL-23, SCL-27, SCL-3, SCL-5, SCL-6, SCL-7, SCL-8, SCL-9, SCM-1, SCPR-A, SCPR-C, SCRA, SCS2, SDCBP, SDHAF4, SEC26, SEC4, SEC9, SEK-1, SEK-4, SEK-5, SEL-12, SELL, SEM1, SEMA4G, SEMG1, SERPINA1, SERPINA10A, SERPINA1C, SERPINA1L, SERPINA3, SERPINA3-8, SERPINA3G, SERPINA3K-PROV, SERPINA7, SERPINB1, SERPINB10, SERPINB10A, SERPINB12, SERPINB14, SERPINB1A, SERPINB1L1, SERPINB1L3, SERPINB1L4, SERPINB2, SERPINB3, SERPINB3A, SERPINB3B, SERPINB3C, SERPINB3D, SERPINB6, SERPINB6A, SERPING1, SERPINH2, SFT-4, SFTPA, SFTPA1, SFTPA2, SFTPBA, SFTPD, SGT1, SHC, SHC1, SHK1, SHOT, SHR5, SI, SI:BUSM1-169I8.11, SI:CABZ01036022.1, SI:CH1073-205C8.3, SI:CH1073-280E3.1, SI:CH1073-416D2.3, SI:CH1073-66L23.1, SI:CH211-114C12.5, SI:CH211-114L13.12, SI:CH211-120G10.1, SI:CH211-125E6.11, SI:CH211-125E6.14, SI:CH211-125E6.5, SI:CH211-12P12.2, SI:CH211-135N15.2, SI:CH211-137J23.6, SI:CH211-137J23.7, SI:CH211-137J23.8, SI:CH211-14A17.10, SI:CH211-15B10.6, SI:CH211-160J14.2, SI:CH211-163C2.2, SI:CH211-165F21.2, SI:CH211-165F21.7, SI:CH211-173A9.6, SI:CH211-173N18.3, SI:CH211-173N18.4, SI:CH211-175L6.2, SI:CH211-175M2.4, SI:CH211-186E20.2, SI:CH211-186E20.7, SI:CH211-194P6.10, SI:CH211-194P6.12, SI:CH211-196F2.3, SI:CH211-212K18.7, SI:CH211-214K5.3, SI:CH211-215E19.4, SI:CH211-215E19.6, SI:CH211-215E19.8, SI:CH211-220F21.1, SI:CH211-220F21.2, SI:CH211-220F21.3, SI:CH211-232P21.6, SI:CH211-243A20.4, SI:CH211-264F5.2, SI:CH211-264F5.6, SI:CH211-286B5.6, SI:CH211-66E2.3, SI:CH211-66E2.5, SI:CH211-74M13.1, SI:CH211-8C17.3, SI:CH211-93I7.4, SI:CH211-9D9.7, SI:CH211-9D9.8, SI:CH73-111E15.1, SI:CH73-122G19.1, SI:CH73-144L3.1, SI:CH73-147O17.1, SI:CH73-173P19.2, SI:CH73-177H5.2, SI:CH73-208G10.1, SI:CH73-343L4.3, SI:CH73-343L4.5, SI:CH73-343L4.6, SI:CH73-343L4.8, SI:CH73-364H19.2, SI:CH73-368J24.17, SI:CH73-380L3.2, SI:CH73-54F23.4, SI:CH73-74H11.1, SI:DKEY-10H3.2, SI:DKEY-10H3.3, SI:DKEY-10H3.7, SI:DKEY-117A8.1, SI:DKEY-117A8.2, SI:DKEY-117A8.3, SI:DKEY-117A8.4, SI:DKEY-11F4.16, SI:DKEY-11O1.2, SI:DKEY-11O1.3, SI:DKEY-11O1.7, SI:DKEY-11P23.7, SI:DKEY-12F6.5, SI:DKEY-174K12.3, SI:DKEY-181C13.1, SI:DKEY-183C6.9, SI:DKEY-187I8.2, SI:DKEY-18P12.4, SI:DKEY-1C7.1, SI:DKEY-1C7.2, SI:DKEY-211E20.10, SI:DKEY-219E21.2, SI:DKEY-219E21.4, SI:DKEY-222F2.1, SI:DKEY-222N6.2, SI:DKEY-225F5.5, SI:DKEY-234I14.12, SI:DKEY-234I14.13, SI:DKEY-234I14.14, SI:DKEY-234I14.15, SI:DKEY-237I9.8, SI:DKEY-238D18.5, SI:DKEY-238D18.7, SI:DKEY-238D18.9, SI:DKEY-238J22.1, SI:DKEY-239J18.2, SI:DKEY-239J18.3, SI:DKEY-23A13.11, SI:DKEY-24P1.7, SI:DKEY-250K15.10, SI:DKEY-250K15.7, SI:DKEY-269I1.4, SI:DKEY-30C15.13, SI:DKEY-33M11.8, SI:DKEY-37M8.11, SI:DKEY-42L23.3, SI:DKEY-42L23.4, SI:DKEY-42L23.5, SI:DKEY-42L23.6, SI:DKEY-42L23.7, SI:DKEY-42L23.9, SI:DKEY-52P2.5, SI:DKEY-61F9.1, SI:DKEY-77F5.10, SI:DKEY-90M5.4, SI:DKEY-93H22.7, SI:DKEYP-104B3.1, SI:DKEYP-104B3.18, SI:DKEYP-104B3.2, SI:DKEYP-104B3.21, SI:RP71-45K5.4, SIGIRR, SIGLEC10, SIGLEC11, SIGLEC12, SIGLEC14, SIGLEC15, SIGLEC16, SIGLEC5, SIGLEC8, SIGLEC9, SIGLECL1, SIGLECL2, SIKE1, SIRPA, SIRPB1, SIRPB1A, SIRPB1B, SIRPB1C, SIRPD, SKH1, SKI7, SKO1, SKP1, SKPA, SL, SLA-6, SLA-8, SLC1, SLC11A1, SLC15A4, SLC19A3.2, SLC27A2, SLC27A2A, SLC27A3, SLC2A14, SLC2A3, SLC2A3B, SLC2A5, SLC44A2, SLC52A1, SLCF-2, SLCO4C1, SLIK, SLPI, SLT2, SMA-5, SMF-1, SMF-2, SMF-3, SMK1, SMYD2, SNAP-29, SNAP23, SNAP23.1, SNAP24, SNAP25, SNAP25A, SNAP29, SNB-1, SNB-2, SNB-5, SNB-6, SNB-7, SNC1, SNC2, SNG-1, SNMP1, SNMP2, SOCS1, SOS, SOS1, SP-A, SP108, SPAC1039.11C, SPAC1486.06, SPAC17A5.08, SPAC1805.16C, SPAC1851.02, SPAC26A3.11, SPAC26H5.04, SPAC30D11.01C, SPAC6F6.11C, SPAC6G10.10C, SPAC926.06C, SPAG11, SPBC119.09C, SPBC1703.07, SPBC1778.05C, SPBC17D1.07C, SPBC215.01, SPBC32F12.10, SPBC4.06, SPBC530.09C, SPBC725.05C, SPBC887.12, SPC-1, SPCC1840.05C, SPCC285.04, SPCC965.06, SPCC970.06, SPE-4, SPE-5, SPE-8, SPIA, SPIA1, SPIN1, SPK1, SPM1, SPN100A, SPN27A, SPN28B, SPN28DA, SPN28DB, SPN28DC, SPN28F, SPN31A, SPN38F, SPN42DA, SPN42DB, SPN42DC, SPN42DD, SPN42DE, SPN43AA, SPN43AB, SPN43AD, SPN47C, SPN53F, SPN55B, SPN75F, SPN77BA, SPN77BB, SPN77BC, SPN85F, SPN88EA, SPN88EB, SPNS1, SPO14, SPO20, SPP-10, SPTAN1, SPV-1, SRA-1, SRC, SRC-1, SRC-2, SRC42A, SRC64B, SRP-1, SRP-2, SRP-3, SRP-6, SRP-7, SRP-8, SRP14, SRP14-1; SRP14-2, SRPK1, SRV2, SSA1, SSA2, SSA3, SSA4, ST6GAL2, STA-1, STA-2, STAR, STAT5A, STAT5B, STAT6, STAT92E, STATH, STBD1, STE20, STE7, STING1, STK10, STK11IP, STO-2, STOM, STOM_TV1, STPG4, STRAP, STV1, STY1, SU(P), SUGT1, SUP-17, SUPERDEATH, SURF1-1; SURF1-2, SURF4, SURF4.2, SURO-1, SUT1, SUT2, SUT3, SVEP1L, SVIP, SVS3B, SWC3, SWO1, SXA1, SYB, SYB1, SYBA, SYBB, SYK, SYMA, SYNGR, SYNGR1, SYNGR1A, T19B4.3, T19C3.5, T22D1.3, T2R15, TAB1, TAB2, TAB3, TAG-120, TAG-225, TAG-325, TANK, TAP, TARM1, TAT-1, TAT-2, TAX-6, TAX1BP1, TAX1BP1B, TBB-6, TBC-10, TBC1D10B, TBC1D10C, TBK1, TBP1, TCIRG1, TCIRG1B, TCN1, TCN2, TCNBB, TEC, TEF1; TEF2, TEF101, TEF102, TEF103, TEP-1, TEP1, TEP2, TEP3, TEP4, TER94, TEST2, TESTIN, TF, TFA, TICAM1, TICAM2, TIFA, TIMP, TIMP-1, TIMP2, TIMP2A, TIRA, TIRAP, TKFC, TLI-1, TLR1, TLR10, TLR15, TLR1A, TLR1LB, TLR2, TLR2-1, TLR2B, TLR3, TLR4, TLR4AL, TLR4BA, TLR5, TLR6, TLR7, TLR8, TLR8A, TLR9, TMA108, TMBIM1, TMBIM1A, TMC6, TMC6B, TMED-3, TMED7, TMEM159, TMEM176, TMEM176L.1, TMEM176L.2, TMEM176L.3A, TMEM176L.3B, TMEM176L.4, TMEM179B, TMEM229B, TMEM268, TMEM30A, TMEM30AA, TMEM63, TMEM63A, TMEM87A, TMPRSS4, TNFAIP3, TNFAIP6, TNFIP6, TNFRSF1B, TNIP2, TOB1, TOLL-4, TOLL-9, TOLLIP, TOM1, TOR2A, TPA-1, TPPNR1L, TPX1, TRADV30.0.6, TRADV8.0, TRAF2, TRAF3, TRAF6, TRAPPC1, TRAPPC1-1; TRAPPC1-2, TREM1, TREM2, TREX1, TRIM 35-44, TRIM107, TRIM108, TRIM109, TRIM21, TRIM25, TRIM27.1, TRIM27.2, TRIM32, TRIM35-1, TRIM35-12, TRIM35-13, TRIM35-14, TRIM35-19, TRIM35-20, TRIM35-21, TRIM35-22, TRIM35-23, TRIM35-24, TRIM35-27, TRIM35-28, TRIM35-29, TRIM35-3, TRIM35-31, TRIM35-33, TRIM35-34, TRIM35-39, TRIM35-40, TRIM35-7, TRIM35-9, TRIM39.2, TRIM47, TRIM56, TRIM58, TRIM69, TRPM2, TRPP-1, TRX-1, TRX-2, TRX2, TRXC, TRY, TRY-1, TRY10, TRY1P1L1, TRY2, TRY4, TRY5, TSA1, TSA2, TSF2, TSP-14, TSP39D, TSP3A, TSP42EF, TSP42EK, TSP5D, TSPA, TSPAN14, TSPAN17, TSPB, TSPC, TSPD, TSPE, TTM-5, TTR, TUB2, TUBB, TUBB4B, TUBB4BL, TUBB5, TUBGCP5, TXK, TXN, TXNDC5, TXNIP, TXNIPA, TYROBP, UBA1, UBA3, UBA52, UBA7, UBB, UBC, UBCB, UBE2D1, UBE2D2, UBE2D3, UBE2K, UBE2L6, UBE2M, UBE2N, UBE2NA, UBE2NB, UBE2V1, UBI-P5E, UBI-P63E, UBQ-1, UBQB, UBQC, UBQD, UBQF, UBQG, UBQH, UBQI, UBQJ, UBQO, UBR-4, UBR4, UBX3, UCP10, UEV1A, UNC-122, UNC-13-4A, UNC-32, UNC-71, UNC-87, UNC-89, UNC13D, UNC93B1, UPK2, V1-11, V1-13, V1-16, V1-20, V1-3, V1-5, V1-7, V1-9, V2-11, V2-15, V2-17, V2-19, V2-8, V3-2, V3-3, V3-4, V4-1, V4-2, V4-6, V5-1, V5-4, V5-6, V9GZ90, VAB-10, VAMP8, VAP-2, VAPA, VAPAL, VAT1, VATA, VATB, VATC, VATD-1; VATD-2, VATE, VATF, VATH, VATM, VATP, VAV, VAV1, VAV2, VAV3, VAV3B, VCL, VCLA, VCP, VDUP1, VEM-1, VH1, VHA-1, VHA-10, VHA-11, VHA-12, VHA-13, VHA-14, VHA-15, VHA-16, VHA-17, VHA-4, VHA-5, VHA-6, VHA-7, VHA-8, VHA-9, VHA100-1, VHA100-2, VHA100-3, VHA100-4, VHA100-5, VHA13, VHA14-1, VHA16-1, VHA26, VHA36-1, VHA44, VHA55, VHA68-2, VHAAC39-1, VHAAC39-2, VHAM9.7-A, VHAM9.7-B, VHAM9.7-C, VHAM9.7-D, VHAPPA1-1, VHASFD, VID28, VINC, VMA1, VMA10, VMA13, VMA16, VMA2, VMA3, VMA4, VMA5, VMA6, VMA7, VMA8, VMA9, VNN1, VPH1, VPS35L, VPS73, VRK3, VSIG10L, VTN, VTNB, W03G11.3, WAS, WASB, WASF1, WASF2, WASF3, WASF3B, WASL, WASLA, WBP1, WDR76, WFDC2, WIPF1, WIPF1B, WIPF2, WIPF3, WIPI2, WIS1, WKD, WRD, WRM-1, WUN-LIKE, WVE-1, X5LQ17, XB5745823, XB5820455 [PROVISIONAL:MR1], XB5894692 [PROVISIONAL:AZGP1], XB5909790, XB5946283 [PROVISIONAL:FCRL5], XB5960289 [PROVISIONAL:FCRL4], XB986811 [PROVISIONAL:CTSK], XB994119, XB994846 [PROVISIONAL:PYCARD], XB995368 [PROVISIONAL:C1RA], XBX-6, XRCC5, XRCC6, YBR241C, YBR284W, YES1, YF5, YFR018C, YIPPEE, YJL070C, YKU70, YKU80, YLR446W, YNK1, YPEL5, YPT10, YPT2, YPT4, YPT5, YPT52, YPT6, YPT7, YPT71, YRK, ZBP1, ZFAND4, ZGC:101540, ZGC:136767, ZGC:136930, ZGC:153151, ZGC:153219, ZGC:153704, ZGC:154125, ZGC:158482, ZGC:158846, ZGC:171509, ZGC:172053, ZGC:173729, ZGC:174153, ZGC:174259, ZGC:174904, ZGC:194252, ZGC:194990, ZGC:56493, ZGC:63882, ZGC:64051, ZGC:77929, ZGC:92027, ZGC:92162, ZGC:92590, ZGC:92594, ZK652.6, ZK686.3, ZK697.8, ZMAT5, ZMP:0000000650, ZMP:0000000729, ZMP:0000000924, ZMP:0000000937, ZMP:0000001139, ZMP:0000001316 | 0.279562489 | |||||
| Inositol phosphate metabolism | -0.259350935 | 0.644887496 | 0.041813044 | |||
| Inositol transporters | 3.842439337 | 0 | 7.684878673 | 0.640920296 | 0.1795042 | 0.026809594 |
| Insertion of tail-anchored proteins into the endoplasmic reticulum membrane | 2.53793389 | 2.568201724 | 7.644069504 | 0.366968642 | 0.119819292 | 0.013729289 |
| Insulin effects increased synthesis of Xylulose-5-Phosphate | 1.913742534 | 0 | 3.827485067 | 0.213545236 | ||
| Insulin processing | 3.17358852 | 1.079181246 | 7.426358286 | -0.232429311 | 0.141877159 | 0.01347237 |
| Insulin receptor recycling | 2.517631441 | 2.71432976 | 7.749592641 | 0.230983281 | 0.021839149 | 0.002160898 |
| Insulin receptor signalling cascade | 3.297228 | 2.777426822 | 9.371882823 | 0.471471188 | 3.329185447 | 0.499145498 |
| Integration of energy metabolism | 3.604467738 | 0.602059991 | 7.810995467 | 0.373948361 | 2.97531841 | 0.349923174 |
| Integration of provirus | 3.114054267 | 2.133538908 | 8.361647441 | 0.548340428 | 0.637203727 | 0.093356447 |
| Integration of viral DNA into host genomic DNA | 2.884935624 | 1.77815125 | 7.548022499 | 0.540839956 | ||
| Integrin cell surface interactions | 2.618351586 | 3.188365926 | 8.425069097 | 0.243890346 | 0.219510685 | 0.023986214 |
| Integrin signaling | 2.755484408 | 3.544316142 | 9.055284958 | 0.54195746 | 3.16999211 | 0.490263838 |
| Interaction between L1 and Ankyrins | 2.974839119 | 2.788168371 | 8.737846609 | 0.469846849 | 1.296760545 | 0.18355623 |
| Interaction between PHLDA1 and AURKA | 2.742470911 | 2.571708832 | 8.056650655 | 0.898657902 | 0.667028215 | 0.124311131 |
| Interaction With Cumulus Cells And The Zona Pellucida | 2.796313514 | 0 | 5.592627027 | 0.36882432 | ||
| Interactions of Rev with host cellular proteins | 0.412182908 | 0.288000184 | 0.029677188 | |||
| Interactions of Tat with host cellular proteins | 2.498870478 | 2.42975228 | 7.427493236 | 0.876712329 | 1.623302227 | 0.284902534 |
| Interactions of Vpr with host cellular proteins | 0.552711692 | 0.660133688 | 0.091215902 | |||
| Interconversion of nucleotide di- and triphosphates | 2.742172102 | 2.627365857 | 8.11171006 | -0.245871377 | 0.659613691 | 0.069574355 |
| Interconversion of polyamines | 1.936149634 | 0.477121255 | 4.349420524 | -0.602082762 | 0.672235751 | 0.068218829 |
| Interferon alpha/beta signaling | 2.567071054 | 2.672097858 | 7.806239967 | 0.314863207 | 0.313034477 | 0.034484401 |
| Interferon gamma signaling | 2.533160621 | 3.248463718 | 8.314784959 | 0.32693068 | 0.691929615 | 0.081806116 |
| Interferon Signaling | 0.319329028 | 1.117445746 | 0.089208216 | |||
| Interleukin-1 family signaling | 0.486798067 | 4 | 0.486798067 | |||
| Interleukin-1 processing | 2.187729054 | 2.198657087 | 6.574115195 | 0.468867316 | 0.599986983 | 0.068111191 |
| Interleukin-1 signaling | 3.042012843 | 3.22685757 | 9.310883256 | 0.559449192 | 4 | 0.640562956 |
| Interleukin-10 signaling | 2.55040399 | 3.106190897 | 8.206998877 | 0.382709888 | 1.182378983 | 0.146391772 |
| Interleukin-12 family signaling | 0.357701869 | 0.941564669 | 0.084199861 | |||
| Interleukin-12 signaling | 2.681401763 | 3.08170727 | 8.444510797 | 0.346943982 | 0.722526037 | 0.088439883 |
| Interleukin-15 signaling | 2.812699268 | 2.977723605 | 8.603122142 | 0.321424024 | 0.26202866 | 0.031773436 |
| Interleukin-17 signaling | 3.317886266 | 0.301029996 | 6.936802528 | 0.425431036 | 3.324172263 | 0.374864454 |
| Interleukin-18 signaling | 2.39823379 | 2.008600172 | 6.805067753 | 0.82470556 | 0.448349512 | 0.072175106 |
| Interleukin-2 family signaling | 2.30859952 | 0.301029996 | 4.918229036 | 0.346965592 | 0.944658892 | 0.072669732 |
| Interleukin-2 signaling | 2.628519169 | 3.158663981 | 8.415702318 | 0.467346226 | 1.048247532 | 0.143696351 |
| Interleukin-20 family signaling | 2.507265364 | 3.072249898 | 8.086780626 | 0.380193766 | 0.446172118 | 0.054408996 |
| Interleukin-21 signaling | 2.569382096 | 2.767155866 | 7.905920058 | 0.348999351 | 0.182633933 | 0.021133356 |
| Interleukin-23 signaling | 2.706050554 | 2.953276337 | 8.365377444 | 0.573622155 | 0.998899126 | 0.149553076 |
| Interleukin-27 signaling | 2.572067659 | 3.095169351 | 8.239304669 | 0.423319616 | 0.489193057 | 0.063254659 |
| Interleukin-3, Interleukin-5 and GM-CSF signaling | 2.881575415 | 3.594060901 | 9.357211731 | 0.468499095 | 3.280429804 | 0.489996122 |
| Interleukin-35 Signalling | 2.579524097 | 2.927370363 | 8.086418557 | 0.423319616 | 0.489193057 | 0.06229006 |
| Interleukin-37 signaling | 2.432900208 | 2.776701184 | 7.642501601 | 0.335343028 | 0.425654822 | 0.047081623 |
| Interleukin-38 signaling | 2.347791248 | 2.499687083 | 7.195269579 | 0.818679814 | 0.434263051 | 0.071765827 |
| Interleukin-4 and Interleukin-13 signaling | 2.605137288 | 3.858055718 | 9.068330294 | 0.359785492 | 2.346892759 | 0.309917751 |
| Interleukin-6 family signaling | 0.405767219 | 0.654672329 | 0.066411143 | |||
| Interleukin-6 signaling | 2.691643592 | 3.113609151 | 8.496896334 | 0.423552018 | 0.643452676 | 0.085357451 |
| Interleukin-7 signaling | 2.797786046 | 3.172894698 | 8.768466789 | 0.346676818 | 0.409290024 | 0.051795034 |
| Interleukin-9 signaling | 2.569382096 | 2.767155866 | 7.905920058 | 0.348999351 | 0.182633933 | 0.021133356 |
| Interleukin receptor SHC signaling | 2.914380716 | 3.595275712 | 9.424037143 | 0.455152891 | 1.758039383 | 0.261179745 |
| Intestinal absorption | 0.449453566 | 0.303900426 | 0.034147282 | |||
| Intestinal hexose absorption | 2.283193111 | 1.568201724 | 6.134587947 | 0.592906021 | 0.614240266 | 0.075770985 |
| Intestinal infectious diseases | 1.525535475 | 0 | 3.051070949 | 0.145244286 | ||
| Intestinal lipid absorption | 2.837794868 | 0.954242509 | 6.629832245 | 0.552177486 | 0.051365047 | 0.006402832 |
| Intestinal saccharidase deficiencies | 2.372313261 | 0.954242509 | 5.698869032 | -0.483287671 | 0.158317137 | 0.01647058 |
| Intra-Golgi and retrograde Golgi-to-ER traffic | 0.296224631 | 0.986728264 | 0.073073304 | |||
| Intra-Golgi traffic | 2.485195216 | 1.176091259 | 6.146481691 | -0.452322067 | 2.497159481 | 0.264543193 |
| Intracellular metabolism of fatty acids regulates insulin secretion | 1.93006036 | 0.602059991 | 4.462180711 | 0.269379249 | ||
| Intracellular oxygen transport | 1.933155975 | 0 | 3.86631195 | 0.216960827 | ||
| Intracellular signaling by second messengers | 0.472986424 | 4 | 0.472986424 | |||
| Intraflagellar transport | 2.768781737 | 2.968482949 | 8.506046423 | 0.702682253 | 4 | 0.670658631 |
| Intrinsic Pathway for Apoptosis | 0.391674026 | 1.822421057 | 0.178448748 | |||
| Intrinsic Pathway of Fibrin Clot Formation | 3.205235821 | 3.251881455 | 9.662353096 | 0.656088896 | 3.235660945 | 0.571915077 |
| Invadopodia formation | 1.866061772 | 0 | 3.732123543 | -0.848493151 | 1.227128996 | 0.15255711 |
| Inwardly rectifying K+ channels | 0.262593573 | 0.155504728 | 0.010208636 | |||
| Ion channel transport | 2.639220494 | 0.301029996 | 5.579470984 | 0.300374294 | 1.124504225 | 0.089545701 |
| Ion homeostasis | 2.667650124 | 3.007320953 | 8.342621202 | 0.392096781 | 1.756963975 | 0.2226666 |
| Ion transport by P-type ATPases | 3.007815527 | 3.037824751 | 9.053455804 | 0.562068636 | 3.165884197 | 0.497512537 |
| Ionotropic activity of kainate receptors | 0.379526273 | 0.315235927 | 0.029910079 | |||
| IRAK1 recruits IKK complex | 3.119575156 | 2.589949601 | 8.829099914 | 0.553554186 | 1.355219555 | 0.207606306 |
| IRAK1 recruits IKK complex upon TLR7/8 or 9 stimulation | 3.119575156 | 2.589949601 | 8.829099914 | 0.553554186 | 1.355219555 | 0.207606306 |
| IRAK2 mediated activation of TAK1 complex | 2.687136953 | 1.86923172 | 7.243505626 | 0.53461204 | 0.782582865 | 0.102027305 |
| IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation | 3.216216745 | 2.158362492 | 8.590795983 | 0.565483973 | 1.457881897 | 0.221026431 |
| IRAK4 deficiency (TLR2/4) | 3.247383201 | 2.59439255 | 9.089158953 | 0.499313955 | 1.224137096 | 0.183331903 |
| IRAK4 deficiency (TLR5) | 2.052645423 | 1.963787827 | 6.069078673 | 0.435169038 | 0.160263127 | 0.016474284 |
| IRE1alpha activates chaperones | 2.059967989 | 1.792391689 | 5.912327668 | 0.302607071 | 0.284395408 | 0.023947037 |
| IRF3-mediated induction of type I IFN | 2.171867835 | 2.354108439 | 6.697844109 | 0.369393796 | 0.227601079 | 0.023370691 |
| IRF3 mediated activation of type 1 IFN | 2.266477215 | 2.281033367 | 6.813987798 | 0.604738218 | 0.385150469 | 0.051455595 |
| Iron uptake and transport | 2.364283076 | 2.294466226 | 7.023032378 | 0.273289066 | 0.184413609 | 0.017494148 |
| IRS-mediated signalling | 3.314364956 | 3.378034322 | 10.00676423 | 0.474240714 | 2.587362904 | 0.410005577 |
| IRS-related events triggered by IGF1R | 2.903475158 | 2.877946952 | 8.684897267 | 0.468473064 | 2.62938059 | 0.369941229 |
| IRS activation | 3.292948876 | 2.71432976 | 9.300227511 | 0.323208251 | 0.050938003 | 0.006646044 |
| ISG15 antiviral mechanism | 2.615688379 | 2.852479994 | 8.083856752 | 0.353914458 | 0.758317005 | 0.089954279 |
| JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 | 2.726819804 | 3.049218023 | 8.502857631 | 0.568547084 | 3.167014274 | 0.477765116 |
| Josephin domain DUBs | 3.61294742 | 1.968482949 | 9.194377788 | 0.872054795 | 1.593405129 | 0.315038238 |
| Keratan sulfate biosynthesis | 2.884300876 | 0.477121255 | 6.245723006 | -0.58513847 | 1.218444921 | 0.15086741 |
| Keratan sulfate degradation | 2.344467065 | 1.204119983 | 5.893054113 | -0.544532749 | 0.49398918 | 0.056411317 |
| Keratan sulfate/keratin metabolism | -0.545279912 | 2.035584005 | 0.277490767 | |||
| Keratinization | 0.360598491 | 0.597863877 | 0.053897203 | |||
| Killing mechanisms | 0.300481666 | 0.130863719 | 0.009830537 | |||
| Kinesins | 2.803994778 | 3.056523724 | 8.66451328 | 0.459442049 | 3.393642829 | 0.47274607 |
| KIT mutants bind TKIs | 2.617771423 | 2.158362492 | 7.393905339 | 0.918104629 | 0.749379508 | 0.135074794 |
| KSRP (KHSRP) binds and destabilizes mRNA | 2.78225886 | 3.108565024 | 8.673082743 | 0.801383429 | 3.357303906 | 0.611555134 |
| L13a-mediated translational silencing of Ceruloplasmin expression | 4.153603643 | 2.92788341 | 11.2350907 | 0.874662111 | 4 | 0.897437658 |
| L1CAM interactions | 2.608946916 | 1.672097858 | 6.889991689 | 0.419728545 | 4 | 0.445810872 |
| Lactose synthesis | 2.540929361 | 2.012837225 | 7.094695947 | 0.378356164 | 0.014989465 | 0.001632672 |
| Lagging Strand Synthesis | -0.331961591 | 0.386112802 | 0.032043655 | |||
| Laminin interactions | 2.066223369 | 1.643452676 | 5.775899414 | -0.311848601 | 0.19072431 | 0.015944353 |
| Late endosomal microautophagy | 2.652838518 | 2.307496038 | 7.613173073 | 0.322269385 | 0.191493387 | 0.020795684 |
| Late Phase of HIV Life Cycle | 2.786216774 | 1.113943352 | 6.686376899 | 0.431181352 | 1.593536582 | 0.175700539 |
| LDL clearance | 2.723499222 | 2.853698212 | 8.300696655 | 0.448424856 | 0.844609245 | 0.112530691 |
| LDL remodeling | 2.91694808 | 2.439332694 | 8.273228854 | 0.915068493 | 1.944475814 | 0.371803652 |
| Leading Strand Synthesis | -0.420504386 | 0.313724266 | 0.032980607 | |||
| Lectin pathway of complement activation | 3.108908384 | 0 | 6.217816768 | 0.4238221 | ||
| Leishmania infection | 0.345070974 | 4 | 0.345070974 | |||
| Leishmania parasite growth and survival | 0.388842908 | 4 | 0.388842908 | |||
| Leishmania phagocytosis | 0.471666562 | 2.487978692 | 0.293374089 | |||
| Leukotriene receptors | 2.87715131 | 2.758911892 | 8.513214512 | 0.786826117 | 2.674433079 | 0.476784797 |
| Lewis blood group biosynthesis | 2.727767564 | 0.301029996 | 5.756565123 | -0.325185389 | 0.766424204 | 0.065158851 |
| LGI-ADAM interactions | 3.317992829 | 1.73239376 | 8.368379417 | 0.850196607 | 3.624135237 | 0.668031124 |
| Ligand-receptor interactions | 2.713784612 | 1.602059991 | 7.029629216 | 0.832100794 | 0.47542479 | 0.078350098 |
| Linoleic acid (LA) metabolism | 2.436612028 | 0 | 4.873224056 | -0.367054501 | 0.387599175 | 0.030565151 |
| Lipophagy | 2.521357574 | 2.459392488 | 7.502107636 | 0.698201861 | 2.460277692 | 0.379267845 |
| Listeria monocytogenes entry into host cells | 0.710958406 | 2.13396867 | 0.379290741 | |||
| Localization of the PINCH-ILK-PARVIN complex to focal adhesions | 1.582619125 | 1.113943352 | 4.279181603 | 0.387129936 | 0.079560708 | 0.005864063 |
| Long-term potentiation | 2.440412462 | 3.185542155 | 8.066367079 | 0.512969231 | 1.954086167 | 0.270210758 |
| Loss of Function of FBXW7 in Cancer and NOTCH1 Signaling | 3.658339258 | 1.301029996 | 8.617708511 | 0.616837173 | 0.733153016 | 0.116112494 |
| Loss of function of MECP2 in Rett syndrome | 0.548637099 | 1.301534696 | 0.178517555 | |||
| Loss of Function of SMAD2/3 in Cancer | 0.421723816 | 0.320184867 | 0.033757396 | |||
| Loss of Function of SMAD4 in Cancer | 0.502465753 | 0.145286923 | 0.018250426 | |||
| Loss of Function of TGFBR1 in Cancer | 0.468478216 | 0.599986983 | 0.070270208 | |||
| Loss of Function of TGFBR2 in Cancer | 0.536659304 | 0.423656973 | 0.056839864 | |||
| Loss of MECP2 binding ability to 5mC-DNA | 2.360224661 | 2.267171728 | 6.98762105 | 0.671240423 | 0.606999218 | 0.087499457 |
| Loss of MECP2 binding ability to the NCoR/SMRT complex | 2.382133695 | 2.096910013 | 6.861177403 | 0.48703853 | 0.42845534 | 0.051198202 |
| Loss of Nlp from mitotic centrosomes | 2.525662198 | 3.309630167 | 8.360954562 | 0.464013389 | 1.889050959 | 0.256834911 |
| Loss of phosphorylation of MECP2 at T308 | 2.800157973 | 2.071882007 | 7.672197954 | 0.548761578 | 0.637203727 | 0.087723957 |
| Loss of proteins required for interphase microtubule organization from the centrosome | 2.525662198 | 3.309630167 | 8.360954562 | 0.464013389 | 1.889050959 | 0.256834911 |
| LRR FLII-interacting protein 1 (LRRFIP1) activates type I IFN production | 2.428849944 | 2.426511261 | 7.28421115 | 0.481096644 | 0.408950065 | 0.050794908 |
| LTC4-CYSLTR mediated IL4 production | 2.796211733 | 2.217483944 | 7.80990741 | 0.512585091 | 0.520406294 | 0.070215406 |
| Lysine catabolism | 2.16358815 | 0.77815125 | 5.10532755 | -0.482995063 | 0.815129193 | 0.078532564 |
| Lysosomal oligosaccharide catabolism | 2.422128342 | 0.698970004 | 5.543226688 | -0.637883772 | 1.36246319 | 0.165338318 |
| Lysosome Vesicle Biogenesis | 2.845338208 | 2.330413773 | 8.021090189 | -0.266023984 | 0.260285028 | 0.027805665 |
| Lysosphingolipid and LPA receptors | 2.391388843 | 2.025305865 | 6.808083551 | 0.429041998 | 0.668020868 | 0.074524747 |
| M Phase | 0.535821602 | 4 | 0.535821602 | |||
| Macroautophagy | 2.71089605 | 3.323870607 | 8.745662707 | 0.523873174 | 4 | 0.593615694 |
| Major pathway of rRNA processing in the nucleolus and cytosol | 4.261424785 | 3.035429738 | 11.55827931 | 0.860727302 | 4 | 0.907143283 |
| Manipulation of host energy metabolism | 1.937949765 | 0.477121255 | 4.353020785 | -0.616986301 | 0.459347133 | 0.047491849 |
| MAP kinase activation | 0.439113085 | 3.846440315 | 0.422255568 | |||
| MAP2K and MAPK activation | 2.763864817 | 3.538196578 | 9.065926212 | 0.495804176 | 2.83945208 | 0.423151703 |
| MAP3K8 (TPL2)-dependent MAPK1/3 activation | 2.780219704 | 2.785329835 | 8.345769244 | 0.499387388 | 1.224137096 | 0.171606465 |
| MAPK family signaling cascades | 0.436488381 | 4 | 0.436488381 | |||
| MAPK targets/ Nuclear events mediated by MAP kinases | 0.362108746 | 0.939340934 | 0.085035892 | |||
| MAPK1 (ERK2) activation | 2.617441236 | 3.27485032 | 8.509732792 | 0.511920282 | 1.212088912 | 0.174379542 |
| MAPK1/MAPK3 signaling | 0.479583309 | 4 | 0.479583309 | |||
| MAPK3 (ERK1) activation | 2.642403337 | 3.212720154 | 8.497526829 | 0.520767709 | 1.368629369 | 0.198198692 |
| MAPK6/MAPK4 signaling | 3.084682174 | 3.200029267 | 9.369393615 | 0.563140902 | 4 | 0.645427311 |
| Masitinib-resistant KIT mutants | 2.617771423 | 2.158362492 | 7.393905339 | 0.918104629 | 0.749379508 | 0.135074794 |
| MASTL Facilitates Mitotic Progression | 2.688222019 | 2.235528447 | 7.611972485 | 0.406661581 | 0.321233382 | 0.038268836 |
| Maturation of nucleoprotein | 2.252159785 | 2.08278537 | 6.587104941 | -0.353976201 | 0.736080646 | 0.073112871 |
| Maturation of protein 3a | 2.926976194 | 0 | 5.853952388 | -0.58513847 | 1.218444921 | 0.144710877 |
| Maturation of protein E | 4.454445211 | 1.041392685 | 9.950283107 | 0.937277458 | 0.851088404 | 0.183508105 |
| Maturation of replicase proteins | 5.18567279 | 2.396199347 | 12.76754493 | 1 | ||
| Maturation of spike protein | 2.314956872 | 0.301029996 | 4.930943739 | 0.058360184 | 0.149868357 | 0.00614689 |
| MECP2 regulates neuronal receptors and channels | 2.421416851 | 3.326949994 | 8.169783696 | 0.472043275 | 1.777146031 | 0.239022423 |
| MECP2 regulates transcription factors | 2.150148186 | 1.86332286 | 6.163619233 | 0.329221404 | 0.026724219 | 0.002425786 |
| MECP2 regulates transcription of genes involved in GABA signaling | 2.157384329 | 0 | 4.314768658 | -0.446697725 | 0.233492731 | 0.019055451 |
| MECP2 regulates transcription of neuronal ligands | 2.140612639 | 2.267171728 | 6.548397005 | 0.329320571 | 0.06069784 | 0.00581158 |
| Meiosis | 0.386200198 | 0.872546258 | 0.084244384 | |||
| Meiotic recombination | 2.533984585 | 2.603144373 | 7.671113543 | 0.314131582 | 0.242067558 | 0.026222562 |
| Meiotic synapsis | 3.139864096 | 1.991226076 | 8.270954267 | 0.546458013 | 1.529509324 | 0.221938802 |
| Melanin biosynthesis | 2.294264463 | 1.278753601 | 5.867282527 | 0.463287671 | 0.096208973 | 0.009977586 |
| Membrane binding and targetting of GAG proteins | 3.242992985 | 1.397940009 | 7.883925979 | 0.486160592 | 0.267582827 | 0.035475003 |
| Membrane Trafficking | 0.30220807 | 2.30582734 | 0.174209908 | |||
| MET activates PI3K/AKT signaling | 3.287490716 | 3.298197867 | 9.873179299 | 0.654579825 | 1.465162453 | 0.262680424 |
| MET activates PTK2 signaling | 2.673389575 | 3.316599302 | 8.663378452 | 0.48015301 | 1.091080469 | 0.154799943 |
| MET activates PTPN11 | 2.852578863 | 2.750508395 | 8.45566612 | 0.565789474 | 0.724905738 | 0.108665756 |
| MET activates RAP1 and RAC1 | 3.416312501 | 2.719331287 | 9.551956288 | 0.530718596 | 0.85032427 | 0.135761585 |
| MET activates RAS signaling | 3.029150425 | 2.73239376 | 8.790694609 | 0.520817734 | 1.142611462 | 0.169795243 |
| MET activates STAT3 | 2.768541832 | 2.751279104 | 8.288362768 | 0.603727049 | 0.672603881 | 0.102563838 |
| MET interacts with TNS proteins | 2.763154191 | 2.706717782 | 8.233026165 | 0.662921348 | 0.882714228 | 0.140504557 |
| MET promotes cell motility | 0.406665731 | 0.851779625 | 0.086597396 | |||
| MET Receptor Activation | 2.763023154 | 2.705007959 | 8.231054268 | 0.569627193 | 0.743521209 | 0.109659059 |
| MET receptor recycling | 3.624451911 | 2.712649702 | 9.961553523 | 0.565944612 | 0.952043545 | 0.161223385 |
| Metabolic disorders of biological oxidation enzymes | 0.280931467 | 0.311956219 | 0.02190958 | |||
| Metabolism of amine-derived hormones | -0.322958411 | 1.080792593 | 0.087262765 | |||
| Metabolism of amino acids and derivatives | 0.582340848 | 4 | 0.582340848 | |||
| Metabolism of Angiotensinogen to Angiotensins | 2.513367474 | 2.911157609 | 7.937892557 | 0.349464078 | 0.494908905 | 0.057500863 |
| Metabolism of carbohydrates | -0.152033427 | 1.058091895 | 0.040216334 | |||
| Metabolism of cofactors | 0.495846214 | 0.961875298 | 0.119235556 | |||
| Metabolism of fat-soluble vitamins | 0.188355927 | 0.035265053 | 0.001660595 | |||
| Metabolism of folate and pterines | 1.901825709 | 2.149219113 | 5.95287053 | -0.339724866 | 0.762539682 | 0.068145072 |
| Metabolism of ingested H2SeO4 and H2SeO3 into H2Se | 3.62823435 | 2.193124598 | 9.449593298 | 0.372312327 | 0.060255553 | 0.008347661 |
| Metabolism of ingested MeSeO2H into MeSeH | 2.466074526 | 2.506505032 | 7.438654084 | 0.806080526 | 0.409567824 | 0.068325322 |
| Metabolism of ingested SeMet, Sec, MeSec into H2Se | 2.604297895 | 1.204119983 | 6.412715772 | 0.273923773 | 0.030830158 | 0.00268443 |
| Metabolism of lipids | 0.226201535 | 0.747581046 | 0.042275995 | |||
| Metabolism of nitric oxide: NOS3 activation and regulation | 0.295242765 | 0.170261715 | 0.012567135 | |||
| Metabolism of non-coding RNA | 0.624391248 | 1.507232929 | 0.235275762 | |||
| Metabolism of nucleotides | -0.314441023 | 2.741676452 | 0.215523887 | |||
| Metabolism of polyamines | 2.13174166 | 1.322219295 | 5.585702614 | 0.732570057 | 4 | 0.534944452 |
| Metabolism of porphyrins | 0.27828356 | 0.238518815 | 0.016593966 | |||
| Metabolism of proteins | 0.377728112 | 4 | ||||
| DB1, MDC1, MDCDS_12730, MDGA1, MDGA2, MDM2, MDM4, MDV1, MEC-15, MEC-7, MED, MED-1, MED-2, MED6, MEFG1, MEGF11, MEI-P26, MEI4, MELTF, MEMB-1, MEMBRIN, MEN1, MEPE, MESH, METTL20, METTL21A, METTL22, MFB-1, MFGE8, MFGE8B, MFN1, MFS3, MGAT1, MGAT1B, MGAT2, MGAT3, MGAT3B, MGAT4A, MGAT4B, MGAT4C, MGAT5, MHC1A, MHCIA6, MIA2, MIA3, MICA, MIG-6, MIRO, MIRO-1, MIS16, MITF, MITFA, MKL1, MLEC, MLP1, MLT-4, MME, MMP1, MMP1B, MMP2, MMS21, MMY, MN1, MNL2, MNN1, MNP-1, MOE1, MOGS, MPDU-1, MPDU1, MPDU1B, MPI, MR1-LIKE, MRF1, MRPL-10, MRPL-11, MRPL-12, MRPL-13, MRPL-15, MRPL-16, MRPL-17, MRPL-18, MRPL-19, MRPL-2, MRPL-21, MRPL-22, MRPL-23, MRPL-24, MRPL-28, MRPL-30, MRPL-32, MRPL-34, MRPL-35, MRPL-36, MRPL-37, MRPL-38, MRPL-39, MRPL-4, MRPL-40, MRPL-41, MRPL-44, MRPL-45, MRPL-46, MRPL-47, MRPL-48, MRPL-49, MRPL-50, MRPL-51, MRPL-53, MRPL-54, MRPL-55, MRPL-9, MRPL1, MRPL10, MRPL11, MRPL12, MRPL13, MRPL14, MRPL15, MRPL16, MRPL17, MRPL18, MRPL19, MRPL2, MRPL20, MRPL21, MRPL22, MRPL23, MRPL24, MRPL27, MRPL28, MRPL3, MRPL30, MRPL32, MRPL33, MRPL34, MRPL35, MRPL36, MRPL37, MRPL38, MRPL39, MRPL4, MRPL40, MRPL41, MRPL42, MRPL43, MRPL44, MRPL45, MRPL46, MRPL47, MRPL48, MRPL49, MRPL50, MRPL51, MRPL52, MRPL53, MRPL54, MRPL55, MRPL57, MRPL58, MRPL9, MRPS-10, MRPS-11, MRPS-14, MRPS-15, MRPS-16, MRPS-17, MRPS-18.C, MRPS-18A, MRPS-18B, MRPS-18C, MRPS-2, MRPS-21, MRPS-22, MRPS-23, MRPS-24, MRPS-25, MRPS-26, MRPS-27, MRPS-28, MRPS-30, MRPS-31, MRPS-33, MRPS-34, MRPS-35, MRPS-5, MRPS-6, MRPS-7, MRPS-9, MRPS10, MRPS11, MRPS12, MRPS14, MRPS15, MRPS16, MRPS17, MRPS18A, MRPS18B, MRPS18C, MRPS2, MRPS21, MRPS22, MRPS23, MRPS24, MRPS25, MRPS26, MRPS27, MRPS28, MRPS29, MRPS30, MRPS31, MRPS33, MRPS34, MRPS35, MRPS36, MRPS5, MRPS6, MRPS7, MRPS9, MRRF, MRRF-1, MRRF1, MRRF2, MRRFP1, MRS6, MRTFA, MRTFAB, MSC2, MSH4, MSLN, MSPO, MSRA, MSRA.1, MSRB1, MSRB1B, MSRB2, MSRB3, MTA1, MTFMT, MTIF2, MTIF3, MTOR, MTQ-2, MTQ2, MTRF1L, MTS2, MUC1, MUC12, MUC13, MUC15, MUC16, MUC17, MUC19, MUC2, MUC20, MUC21, MUC3A, MUC3B, MUC4, MUC5.1, MUC5.2, MUC5AC, MUC5B, MUC6, MUC7, MUCL1, MUG137, MUG71, MUG84, MUG87, MUL1, MUL1B, MVD, MVD1, MXRA8, MXRA8A, MYC, MYCA, MYDGF, MYH4, MYL2, MYO5A, MYRIP, MYSM1, N, N6AMT1, NAB3, NAC, NAE1, NAGK, NANP, NANS, NANSA, NAPA, NAPAB, NAPB, NAPBB, NAPG, NAPGB, NAPSA, NARS1, NARS2, NAS2, NAT10, NAT8, NAT8B, NBET-1, NCOA1, NCOA2, NCOR2, NCSTN, NDC1, NDNL2, NDUFAF2, NEC, NED-8, NEDD8, NEGR1, NEK9, NEP-1, NEP-10, NEP-11, NEP-13, NEP-16, NEP-17, NEP-18, NEP-2, NEP-20, NEP-21, NEP-22, NEP-23, NEP-26, NEP-3, NEP-4, NEP-6, NEP-9, NEP1, NEP2, NEP3, NEP4, NEP5, NEP6, NEP7, NEPL10, NEPL11, NEPL12, NEPL13, NEPL14, NEPL15, NEPL16, NEPL17, NEPL18, NEPL19, NEPL20, NEPL21, NEPL4, NEPL5, NEPL6, NEPL7, NEPL8, NEPL9, NERO, NEU1, NEU2, NEU3, NEU3.1, NEU4, NEURL2, NEWGENE_1582994, NFKB2, NFKBIA, NFKBIAB, NFRKB, NFYA, NFYB, NFYC, NGFG, NGLY1, NGRN, NHL-2, NHR-2, NHR-23, NHR-25, NHR-258, NHR-277, NHR-3, NHR-38, NHR-48, NHR-7, NHR-8, NHR-85, NIC96, NICN1, NID1, NIP1, NIPPED-A, NIPSNAP2, NKX2-6, NLP, NLRB5, NLRP3, NOCT, NOD1, NOD2, NOL-58, NOL4L, NOMPC, NOP5, NOP56, NOP58, NOT, NOTUM, NOTUM1A, NPH, NPL, NPM1, NPM1A, NPP-1, NPP-10, NPP-11, NPP-12, NPP-13, NPP-14, NPP-15, NPP-16, NPP-18, NPP-19, NPP-2, NPP-20, NPP-21, NPP-22, NPP-23, NPP-3, NPP-5, NPP-6, NPP-7, NPP-8, NPP-9, NPPA, NR1H2, NR1H3, NR1H4, NR1I2, NR2C1, NR3C1, NR3C2, NR4A2, NR5A1, NR5A1A, NR5A1B, NR5A2, NR5A5, NRIP1, NRIP1A, NRN1, NRN1L, NRT, NSE1, NSE2, NSE3, NSE4, NSF, NSF-1, NSF2, NSFA, NSMCE1, NSMCE2, NSMCE3, NSMCE4A, NSP1, NST, NSTP-10, NSTP-3, NSTP-5, NSTP-6, NSTP-7, NSTP-8, NTM, NTNG1, NTNG2, NUB1, NUCB-1, NUCB1, NUDT14, NUDT3, NUP1, NUP107, NUP120, NUP124, NUP132, NUP133, NUP145, NUP153, NUP154, NUP155, NUP160, NUP170, NUP184, NUP186, NUP188, NUP189, NUP192, NUP2, NUP205, NUP210, NUP211, NUP214, NUP35, NUP37, NUP40, NUP42, NUP43, NUP44, NUP44A, NUP45, NUP50, NUP54, NUP57, NUP58, NUP62, NUP62L, NUP75, NUP82, NUP84, NUP85, NUP88, NUP93, NUP93-2, NUP98, NUP98-96, NUPL1, NUPL2, NURF-38, NUS1, NW, O-FUT2, O16651, O16792, O61211, OBSL1, OC116, OCM4.3, OCTBETA1R, OCTBETA2R, OCTBETA3R, ODAM, OGT, OGT.1, OPCML, OS9, OSBPL3, OST-1, OTOA, OTOP3, OTU1, OTUB-1, OTUB-2, OTUB1, OTUB1A, OTUB1B, OTUB2, OTUD3, OTUD5, OTUD7A, OTUD7B, OTULIN, OTULINA, OXA-1, OXA1L, P115, P24-1, P24-2, P2RY2, P3H3, P4HB, P53, P58IPK, PAB-1, PAB1, PABP, PABPC1, PABPC1A, PAC1, PAD-2, PAF1, PAFAH1B1, PAFAH1B1A, PAFAH1B1B, PAM1, PANX2, PAPLA1, PAPPA, PAPPA1, PAPPA2, PAPPAA, PAR-2, PARK, PARK2, PARK7, PARN, PARP, PARP-1, PARP1, PARS2, PAS-1, PAS-2, PAS-3, PAS-4, PAS-5, PAS-6, PAS-7, PAX6, PBS-1, PBS-2, PBS-3, PBS-4, PBS-5, PBS-6, PBS-7, PC, PCAF-1, PCGF2, PCM-1, PCM1, PCM2, PCMA, PCMT, PCMT1, PCN-1, PCN1, PCNA, PCNP, PCSK1, PCSK2, PCSK9, PCU4, PCYOX1, PDCL, PDE1A, PDF1, PDI, PDI-2, PDI-3, PDI-6, PDI-P5, PDI1, PDI2, PDIA3, PDIA5, PDIA6, PDLIM2, PDR-1, PEK, PEK-1, PENK, PENKA, PEX10, PEX12, PEX13, PEX14, PEX2, PEX26, PEX3, PEX5, PF10_0084, PF10_0264, PF11_0174, PF14_0358, PF3D7_0103900, PF3D7_0106800, PF3D7_0107600, PF3D7_0108000, PF3D7_0109850, PF3D7_0205900, PF3D7_0211200, PF3D7_0211600, PF3D7_0212300, PF3D7_0214000, PF3D7_0214100, PF3D7_0216800, PF3D7_0217800, PF3D7_0303000, PF3D7_0306800, PF3D7_0306900, PF3D7_0307200, PF3D7_0310600.1, PF3D7_0310700, PF3D7_0312300, PF3D7_0312800, PF3D7_0315200, PF3D7_0316300.2, PF3D7_0316800, PF3D7_0317000, PF3D7_0317600, PF3D7_0317800, PF3D7_0319300, PF3D7_0319600, PF3D7_0319700, PF3D7_0320100, PF3D7_0320300, PF3D7_0321200, PF3D7_0321500, PF3D7_0321600, PF3D7_0323100, PF3D7_0405100, PF3D7_0413600, PF3D7_0413900, PF3D7_0415900, PF3D7_0416800, PF3D7_0418200, PF3D7_0418500, PF3D7_0422300, PF3D7_0422400, PF3D7_0502800, PF3D7_0503800, PF3D7_0505300, PF3D7_0505800, PF3D7_0507100, PF3D7_0509000, PF3D7_0510500, PF3D7_0513800, PF3D7_0514000, PF3D7_0516900, PF3D7_0517000, PF3D7_0517500, PF3D7_0517700, PF3D7_0519400, PF3D7_0520000, PF3D7_0527200, PF3D7_0528200, PF3D7_0530200, PF3D7_0602500, PF3D7_0607000, PF3D7_0608500, PF3D7_0608700, PF3D7_0611700, PF3D7_0612100, PF3D7_0614500, PF3D7_0615300, PF3D7_0616700, PF3D7_0617800, PF3D7_0618300, PF3D7_0618700, PF3D7_0619400, PF3D7_0621900, PF3D7_0624700, PF3D7_0705600, PF3D7_0705700, PF3D7_0706400, PF3D7_0714000, PF3D7_0715900, PF3D7_0716800, PF3D7_0719600, PF3D7_0719700, PF3D7_0719900, PF3D7_0721600, PF3D7_0727400, PF3D7_0728000, PF3D7_0729000, PF3D7_0801800, PF3D7_0803800, PF3D7_0806400, PF3D7_0807300, PF3D7_0807500, PF3D7_0807600, PF3D7_0807800, PF3D7_0812600, PF3D7_0813800, PF3D7_0813900, PF3D7_0814000, PF3D7_0814400, PF3D7_0816700, PF3D7_0821700, PF3D7_0828500, PF3D7_0903200, PF3D7_0903700, PF3D7_0903900, PF3D7_0904600, PF3D7_0907700, PF3D7_0909200, PF3D7_0912900, PF3D7_0913200, PF3D7_0915100, PF3D7_0916600, PF3D7_0918300, PF3D7_0918600, PF3D7_0921000.1, PF3D7_0925700, PF3D7_0931000, PF3D7_0931800, PF3D7_1003500, PF3D7_1004000, PF3D7_1007900, PF3D7_1008400, PF3D7_1008700, PF3D7_1008800, PF3D7_1009000, PF3D7_1009700, PF3D7_1010100, PF3D7_1010500, PF3D7_1010600, PF3D7_1011400, PF3D7_1011700, PF3D7_1013900, PF3D7_1014000, PF3D7_1017400, PF3D7_1017900, PF3D7_1019400, PF3D7_1025100, PF3D7_1026800, PF3D7_1027800, PF3D7_1030500, PF3D7_1033900, PF3D7_1034000, PF3D7_1036900, PF3D7_1038100, PF3D7_1103100, PF3D7_1105000, PF3D7_1105100, PF3D7_1105400, PF3D7_1107300, PF3D7_1109900, PF3D7_1111300, PF3D7_1111900, PF3D7_1113100, PF3D7_1116400, PF3D7_1116700, PF3D7_1123400, PF3D7_1124900, PF3D7_1126200, PF3D7_1129200, PF3D7_1130000, PF3D7_1130100, PF3D7_1130200, PF3D7_1130400, PF3D7_1132200, PF3D7_1136400, PF3D7_1142500, PF3D7_1142600, PF3D7_1143400, PF3D7_1144000, PF3D7_1144500, PF3D7_1144900, PF3D7_1147200, PF3D7_1203200, PF3D7_1203900, PF3D7_1204300, PF3D7_1206200, PF3D7_1206700, PF3D7_1210100, PF3D7_1211800, PF3D7_1212700, PF3D7_1213900, PF3D7_1216300, PF3D7_1217800, PF3D7_1224300, PF3D7_1225800, PF3D7_1226800, PF3D7_1229500, PF3D7_1230700, PF3D7_1231100, PF3D7_1233900, PF3D7_1236800, PF3D7_1237000, PF3D7_1242700, PF3D7_1243700, PF3D7_1247300, PF3D7_1250600, PF3D7_1302600, PF3D7_1302800, PF3D7_1303100, PF3D7_1306400, PF3D7_1308300, PF3D7_1309100, PF3D7_1311500, PF3D7_1313000, PF3D7_1317800, PF3D7_1320600, PF3D7_1323100, PF3D7_1323400, PF3D7_1324700, PF3D7_1328100, PF3D7_1331800, PF3D7_1332000, PF3D7_1332300, PF3D7_1341200, PF3D7_1341300, PF3D7_1341600, PF3D7_1342000, PF3D7_1343600, PF3D7_1345500, PF3D7_1348100, PF3D7_1350400, PF3D7_1351400, PF3D7_1353800, PF3D7_1353900, PF3D7_1356300, PF3D7_1357000, PF3D7_1357800, PF3D7_1358800, PF3D7_1361100, PF3D7_1365900, PF3D7_1368100, PF3D7_1402300, PF3D7_1405200, PF3D7_1407500, PF3D7_1408600, PF3D7_1408700, PF3D7_1410600, PF3D7_1412600, PF3D7_1412900.1, PF3D7_1413800, PF3D7_1414000, PF3D7_1414300, PF3D7_1421200, PF3D7_1421600, PF3D7_1424100, PF3D7_1424400, PF3D7_1426000, PF3D7_1429500, PF3D7_1431700, PF3D7_1432600, PF3D7_1432700, PF3D7_1433500, PF3D7_1437800, PF3D7_1439100, PF3D7_1441200, PF3D7_1442500, PF3D7_1443000, PF3D7_1444500, PF3D7_1445900, PF3D7_1447000, PF3D7_1450100, PF3D7_1451100, PF3D7_1455900, PF3D7_1460300, PF3D7_1460700, PF3D7_1461300, PF3D7_1465900, PF3D7_1466300, PF3D7_1468700, PF3D7_1469100, PF3D7_1470900, PF3D7_1474800, PFDN1, PFDN2, PFDN4, PFDN5, PFDN6, PFSEC23, PGA3, PGA4, PGA5, PGANT1, PGANT10, PGANT11, PGANT12, PGANT13, PGANT2, PGANT3, PGANT35A, PGANT4, PGANT5, PGANT6, PGANT7, PGANT8, PGANT9, PGAP1, PGCL, PGM3, PGR, PH-D, PH-P, PHC1, PHC2, PHC2B, PHC3, PHD1, PHF8, PHM-2, PHO, PHO23, PHO81, PHYHDLA.3, PI31, PIAS1, PIAS1A, PIAS1B, PIAS2, PIAS3, PIAS4, PIAS4A, PIC, PIG-A, PIG-B, PIG-C, PIG-F, PIG-G, PIG-H, PIG-N, PIG-P, PIG-Q, PIG-V, PIG-WB, PIG-Z, PIGA, PIGB, PIGB-1, PIGC, PIGF, PIGG, PIGH, PIGK, PIGL, PIGL-1, PIGM, PIGN, PIGN-1, PIGO, PIGP, PIGQ, PIGS, PIGT, PIGU, PIGV, PIGV-1, PIGW, PIGW-1, PIGX, PIGY, PIGZ, PINLYP, PIP1, PIS-1, PISA, PKIB, PLA2G4B, PLA2G7, PLAA, PLAP, PLAUR, PLDG, PLET1, PLG, PLI1, PMI40, PML, PMM1, PMM2, PMMA, PMMB, PMT3, PNG-1, PNG1, PNG2, PNGL, PNPLA2, PNR, POC1B, POC1BL, POF11, POF7, POFUT2, POL30, POLB, POM121, POM121C, POMC, POMCA, POMGNT1, POMGNT2, POMK, POMT1, POMT2, PONT, PORIN, PORIN2, PPA1, PPA1A, PPA1B, PPA2, PPARA, PPARAB, PPARG, PPARGC1A, PPK15, PPN, PPP2R5B, PPP3R1, PPP6C, PPP6R1, PPP6R3, PQN-22, PRE1, PRE10, PRE2, PRE3, PRE4, PRE5, PRE6, PRE7, PRE8, PRE9, PREB, PRICKLE2, PRKCSH, PRKDC, PRKG1, PRKN, PRL, PRL-1, PRMT2, PRMT3, PRN, PRND, PRNP, PROC, PROCA, PRORP, PROS1, PROSALPHA1, PROSALPHA2, PROSALPHA3, PROSALPHA4, PROSALPHA4T1, PROSALPHA4T2, PROSALPHA5, PROSALPHA6, PROSALPHA7, PROSBETA1, PROSBETA2, PROSBETA2R1, PROSBETA2R2, PROSBETA3, PROSBETA4, PROSBETA5, PROSBETA5R1, PROSBETA5R2, PROSBETA6, PROSBETA7, PROZ, PROZA, PRPSAP1, PRSS21, PRSS23, PRSS27, PRSS41, PRSS55, PRT1, PRX-12, PRX-13, PRX-14, PRX-5, PRZ1, PSAP, PSC, PSC3, PSCA, PSENEN, PSM1, PSM3, PSMA1, PSMA2, PSMA3, PSMA4, PSMA5, PSMA6, PSMA6A, PSMA6B, PSMA7, PSMA8, PSMB1, PSMB10, PSMB11, PSMB11A, PSMB2, PSMB3, PSMB4, PSMB4-1; PSMB4-2, PSMB5, PSMB6, PSMB7, PSMB7.2, PSMB8, PSMB8A, PSMB9, PSMB9A, PSMC1, PSMC1A, PSMC2, PSMC3, PSMC4, PSMC5, PSMC6, PSMD-9, PSMD1, PSMD10, PSMD11, PSMD11B, PSMD12, PSMD13, PSMD14, PSMD2, PSMD3, PSMD4, PSMD4A, PSMD5, PSMD6, PSMD7, PSMD8, PSMD8-1; PSMD8-2, PSMD9, PSME1, PSME2, PSME3, PSME4, PSME4B, PSMF1, PSY1, PTB1, PTCD3, PTEN, PTENA, PTIP, PTP4A2, PTP4A2B, PTPRVP, PTRH2, PTS1, PUC1, PUF, PUM2, PUP1, PUP2, PUP3, PWN, PXR, PYCR3, PYP-1, Q06AB2, Q19831, Q21673, Q23123, Q7K6A8, Q86BM6, Q8IAV0, Q8IDV0, Q8IDV1, Q8IDV2, Q95XE4, Q9VA71, Q9VIT8, QARS1, QRI1, QSOX1, R05D3.12, R08D7.4, R4GFI5, RAB-1, RAB-10, RAB-11.1, RAB-11.2, RAB-14, RAB-18, RAB-19, RAB-21, RAB-3, RAB-30, RAB-33, RAB-35, RAB-37, RAB-39, RAB-5, RAB-6.1, RAB-6.2, RAB-7, RAB-8, RAB1, RAB10, RAB11, RAB11A, RAB11B, RAB11B.1, RAB11B.2, RAB11BB, RAB12, RAB13, RAB14, RAB14L, RAB15, RAB17, RAB18, RAB18B, RAB19, RAB1A, RAB1AB, RAB1B, RAB1C, RAB1D, RAB1E, RAB2, RAB20, RAB21, RAB22A, RAB23, RAB24, RAB25, RAB25A, RAB26, RAB27, RAB27A, RAB27B, RAB29, RAB2A, RAB2B, RAB3, RAB30, RAB31, RAB32, RAB32A, RAB32C, RAB32D, RAB33A, RAB33B, RAB33BA, RAB34, RAB34A, RAB35, RAB35B, RAB36, RAB37, RAB38, RAB38B, RAB39, RAB39A, RAB39B, RAB39BA, RAB3A, RAB3AB, RAB3B, RAB3C, RAB3D, RAB3DA, RAB4, RAB40, RAB40A, RAB40B, RAB40C, RAB41, RAB42, RAB43, RAB44, RAB4A, RAB4B, RAB5, RAB5A, RAB5AA, RAB5B, RAB5C, RAB6, RAB6A, RAB6B, RAB6BA, RAB7, RAB7A, RAB7B, RAB8, RAB8A, RAB8B, RAB9, RAB9A, RAB9B, RAB9D, RAB9DB, RAB9E, RAB9FA, RAB9FB, RABA, RABC, RABF1-1; RABF1-2, RABG1, RABG2, RABGGTA, RABGGTALPHA, RABGGTB, RABH, RABJ, RABK1, RABK2, RABK3, RABL, RABM, RABN1, RABN2, RABO, RABQ, RABR, RABS, RABT1, RABT2, RABU, RABV, RABW, RABX2, RABX4, RABX5, RABY, RABZ, RAD-23, RAD18, RAD21, RAD21B, RAD22, RAD23, RAD23A, RAD23AA, RAD23AB, RAD23B, RAD34, RAD4, RAD51, RAD52, RAE-1, RAE1, RAET1G, RAET1L, RAGA, RAN-2, RANBP2, RANGAP, RANGAP1, RANGAP1A, RARA, RARAA, RARS1, RARS2, RAS-2, RAS1, RAS2, RASB, RASC, RASD, RASG, RASGRP4, RASS, RBBD, RBBE, RBBP-5, RBBP5, RBBP7, RBX-1, RBX-2, RBX1, RCD5, RCE1, RCE1A, RCG_34378, RCG_52671, RCN1, REC11, RECK, REG, RELA, REN, REN1, RENBP, REP, REP-1, REPE, RER2, RET2, RET3, RFL-1, RFT1, RFWD2, RGD1564606, RGD1566373, RGS11, RGS6, RGS7, RGS7A, RGS9, RGS9B, RHO-1, RHO1, RHOA, RHOAB, RHOT1, RHOT1A, RHP42, RIB2, RING1, RIPK1, RIPK1L, RIPK2, RKM5, RLA-0, RLA-1, RMT3, RNA1, RNF103, RNF123, RNF128, RNF128A, RNF135, RNF139, RNF144A, RNF144AA, RNF146, RNF152, RNF168, RNF181, RNF185, RNF2, RNF20, RNF38, RNF40, RNF5, RNF7, ROC1A, ROC2, RORA, RORAA, RP11-385D13.1, RPA1, RPD3, RPG1, RPL-1, RPL-10, RPL-11.1, RPL-11.2, RPL-12, RPL-13, RPL-14, RPL-15, RPL-16, RPL-17, RPL-18, RPL-19, RPL-2, RPL-20, RPL-21, RPL-22, RPL-23, RPL-24.1, RPL-25.2, RPL-26, RPL-27, RPL-28, RPL-29, RPL-3, RPL-30, RPL-31, RPL-32, RPL-33, RPL-35, RPL-36, RPL-36.A, RPL-38, RPL-39, RPL-4, RPL-43, RPL-5, RPL-6, RPL-7, RPL-7A, RPL-9, RPL10, RPL1001, RPL1002, RPL101, RPL10A, RPL10AB, RPL10L, RPL11, RPL1101, RPL1102, RPL11A, RPL11B, RPL12, RPL1201, RPL1202, RPL12A, RPL12B, RPL13, RPL13A, RPL13B, RPL14, RPL14B, RPL15, RPL15-1; RPL15-2, RPL1502, RPL15A, RPL15B, RPL1601, RPL1602, RPL16A, RPL17, RPL17-C18ORF32, RPL1701, RPL17A, RPL18, RPL1802, RPL18A, RPL18B, RPL19, RPL1902, RPL19A, RPL19B, RPL1A, RPL1B, RPL2001, RPL2002, RPL20A, RPL20B, RPL21, RPL2101, RPL2102, RPL21A, RPL22, RPL22-LIKE, RPL22A, RPL22B, RPL22L1, RPL23, RPL2301, RPL2302, RPL23A, RPL23B, RPL24, RPL2401, RPL2402, RPL24A, RPL25, RPL2502, RPL26, RPL26A, RPL26B, RPL26L1, RPL27, RPL2701, RPL27A, RPL28, RPL2801, RPL2802, RPL29, RPL2A, RPL2B, RPL3, RPL30, RPL3002, RPL301, RPL302, RPL31, RPL31B, RPL32, RPL3202, RPL33A, RPL34, RPL35, RPL35A, RPL35B, RPL36, RPL3602, RPL36A, RPL36A-HNRNPH2, RPL36AL, RPL36B, RPL37, RPL3702, RPL3703, RPL37A, RPL37B, RPL38, RPL3801, RPL39, RPL39L, RPL3L, RPL4, RPL40, RPL401, RPL402, RPL41, RPL42, RPL42A, RPL42B, RPL44, RPL4A, RPL4B, RPL5, RPL502, RPL5B, RPL6, RPL6-PS1, RPL6B, RPL7, RPL701, RPL7A, RPL8, RPL801, RPL802, RPL803, RPL8A, RPL8B, RPL9, RPL9; RPL9P7; RPL9P8; RPL9P9, RPL901, RPL9B, RPLP0, RPLP1, RPLP2, RPN-1, RPN-10, RPN-11, RPN-12, RPN-13, RPN-2, RPN-3, RPN-5, RPN-6.1, RPN-7, RPN-8, RPN-9, RPN1, RPN10, RPN11, RPN12, RPN13, RPN1301, RPN2, RPN3, RPN5, RPN501, RPN502, RPN6, RPN7, RPN8, RPN9, RPP0, RPP101, RPP102, RPP1B, RPP203, RPP2A, RPS-0, RPS-1, RPS-10, RPS-11, RPS-12, RPS-13, RPS-15, RPS-16, RPS-17, RPS-18, RPS-19, RPS-2, RPS-20, RPS-21, RPS-22, RPS-23, RPS-24, RPS-25, RPS-26, RPS-27, RPS-28, RPS-29, RPS-3, RPS-4, RPS-5, RPS-6, RPS-7, RPS-8, RPS-9, RPS001, RPS0A, RPS0B, RPS10, RPS1002, RPS101, RPS102, RPS10B, RPS11, RPS1101, RPS1102, RPS11A, RPS11B, RPS12, RPS1201, RPS13, RPS14, RPS15, RPS1501, RPS1502, RPS15A, RPS15AA, RPS16, RPS1601, RPS1602, RPS16A, RPS16B, RPS17, RPS1701, RPS1702, RPS17A, RPS17B, RPS18, RPS1801, RPS1802, RPS18A, RPS18B, RPS19, RPS1901, RPS19A, RPS19B, RPS1B, RPS2, RPS20, RPS21, RPS21A, RPS21B, RPS2201, RPS2202, RPS22A, RPS23, RPS2302, RPS23A, RPS23B, RPS24, RPS2402, RPS24A, RPS24B, RPS25, RPS2501, RPS25A, RPS26, RPS2601, RPS2602, RPS26A, RPS26B, RPS26L, RPS27, RPS27.2, RPS27A, RPS27B, RPS27L, RPS28, RPS2801, RPS2802, RPS28B, RPS28P9, RPS29, RPS29A, RPS3, RPS3A, RPS4, RPS401, RPS402, RPS403, RPS4A, RPS4B, RPS4X, RPS4Y1, RPS4Y2, RPS5, RPS502, RPS5A, RPS6, RPS602, RPS6A, RPS6B, RPS7, RPS7A, RPS8, RPS802, RPS8A, RPS8B, RPS9, RPS901, RPS902, RPS9B, RPSA, RPT-1, RPT-2, RPT-3, RPT-4, RPT-5, RPT-6, RPT1, RPT2, RPT3, RPT4, RPT5, RPT6, RRAGA, RRG1, RRS1, RSG7, RSMB, RSMM, RSMN, RTF1, RTN4R, RTN4RL1, RTN4RL2, RUB1, RUVB-1, RUVBL1, RWDD3, RXRA, RXRAA, RXRB, RYH1, S1P, S2P, SAA1, SAE1, SAF-B, SAFB, SAFB2, SAMUEL, SAP-R, SAP155, SAP185, SAP190, SAP4, SAPA, SAPA1, SAR-1, SAR1, SAR1A, SAR1B, SARA, SARS1, SARS2, SATB1, SATB1B, SATB2, SBSP-1, SBSPON, SCC-1, SCC-3, SCCRO3, SCCRO4, SCE, SCE3, SCF, SCFD1, SCG2, SCG2B, SCG3, SCL1, SCM, SCMH1, SCNY, SCOSPONDIN, SCYL3, SDC, SDC2, SDIC1, SDIC2, SDIC3, SDIC4, SDICC, SDN-1, SDS3, SEC-10, SEC-12, SEC-15, SEC-16A.1, SEC-16A.2, SEC-22, SEC-23, SEC-24.1, SEC-24.2, SEC-3, SEC-31, SEC-5, SEC-6, SEC-8, SEC10, SEC11A, SEC11C, SEC12, SEC13, SEC15, SEC16, SEC16A, SEC16B, SEC17, SEC18, SEC21, SEC22, SEC22A, SEC22B, SEC22BB, SEC22C, SEC23, SEC231, SEC23A, SEC23IP, SEC24, SEC24A, SEC24AB, SEC24B, SEC24C, SEC24CD, SEC24D, SEC24L, SEC26, SEC27, SEC28, SEC3, SEC31, SEC31A, SEC4, SEC5, SEC53, SEC59, SEC6, SEC61A1, SEC61A2, SEC61B, SEC61G, SEC65, SEC8, SED5, SEDL-1, SEE1, SEH1, SEH1L, SEL-9, SEL1L, SELENOS, SELR, SEM1, SEMA5A, SEMA5B, SEMA5BA, SEMA5C, SEMG1, SENP1, SENP2, SENP3, SENP5, SENP8, SEP1, SERP1, SERPINA1, SERPINA10, SERPINA10A, SERPINA1C, SERPINA1L, SERPINA3K-PROV, SERPINB14, SERPINB1L1, SERPINB1L3, SERPINB1L4, SERPINC1, SERPIND1, SERPINH2, SEX-1, SF1, SFB2, SFB3, SFC2, SFMBT, SFT1, SFTA3, SFTPA, SFTPA1, SFTPA2, SFTPB, SFTPBA, SFTPC, SFTPD, SGKA, SHC1, SHISA5, SHPRH, SI:BUSM1-169I8.11, SI:CABZ01007802.1, SI:CABZ01007807.1, SI:CH1073-205C8.3, SI:CH1073-342H5.2, SI:CH1073-416D2.3, SI:CH211-113A14.11, SI:CH211-113A14.19, SI:CH211-113A14.22, SI:CH211-114C12.5, SI:CH211-114N24.6, SI:CH211-120G10.1, SI:CH211-12P12.2, SI:CH211-14A17.10, SI:CH211-155E24.3, SI:CH211-15E22.3, SI:CH211-175L6.2, SI:CH211-190P8.2, SI:CH211-191D2.2, SI:CH211-196F2.3, SI:CH211-226H7.3, SI:CH211-226H7.5, SI:CH211-226H7.6, SI:CH211-226H7.8, SI:CH211-253P14.2, SI:CH211-262E15.1, SI:CH211-269C21.2, SI:CH211-285C6.5, SI:CH73-100L22.3, SI:CH73-122K23.1, SI:CH73-142C19.1, SI:CH73-144L3.1, SI:CH73-299H12.6, SI:CH73-364H19.2, SI:CH73-373M9.1, SI:CH73-49O8.1, SI:CH73-54F23.4, SI:DKEY-108K21.11, SI:DKEY-119F1.1, SI:DKEY-125E8.3, SI:DKEY-147F3.8, SI:DKEY-156N14.5, SI:DKEY-17E16.10, SI:DKEY-182I3.10, SI:DKEY-183C6.9, SI:DKEY-18P12.4, SI:DKEY-192D15.1, SI:DKEY-202E17.1, SI:DKEY-20I20.10, SI:DKEY-211G8.9, SI:DKEY-218F9.10, SI:DKEY-219E21.2, SI:DKEY-219E21.4, SI:DKEY-21C19.3, SI:DKEY-239B22.2, SI:DKEY-23A13.22, SI:DKEY-261M9.19, SI:DKEY-261M9.6, SI:DKEY-37F18.2, SI:DKEY-37M8.11, SI:DKEY-42I9.6, SI:DKEY-82J4.2, SI:DKEYP-110A12.4, SI:RP71-1G18.13, SI:RP71-1K13.6, SI:RP71-45K5.4, SI:RP71-68N21.9, SIAH-1, SIAH1, SIAH2, SIAH2L, SIAL-T2, SIAT, SIAT4B, SIAT8C, SIBA, SIBB, SIBC, SIBD, SIDL, SIMA, SIN3A, SIN3AA, SINA, SIT4, SK1, SKI7, SKIV2L, SKP1, SKP2, SKPA, SKPT-1, SKR-1, SLC-17.1, SLC-30A5, SLC17A5, SLC2A15A, SLC30A5, SLC30A6, SLC30A7, SLC30A8, SLC34A1, SLC34A1A, SLC34A2, SLC34A2B, SLC35A1, SLC35C1, SLC52A1, SLCO2B1, SLCO5A1, SLH, SLURP1, SLY-1, SLY1, SMA-1, SMA-2, SMAD1, SMAD2, SMAD3, SMAD3A, SMAD4, SMAD4.1, SMAD4A, SMAD7, SMAD9, SMC-3, SMC-5, SMC-6, SMC1, SMC1A, SMC3, SMC5, SMC6, SMO-1, SMOX, SMP3, SMT3, SMURF, SMURF2, SNAP-1, SNCA, SNCAIP, SNPA, SNPC, SNX3, SOCS16D, SOCS2, SOCS3, SOCS36E, SOCS3B, SOCS44A, SOCS5, SOCS5B, SOCS6, SOCS6A, SOCS6B, SOF1, SONA, SORL1, SP-A, SP1, SP100, SP100.1, SP100.3, SP100.4, SP3, SP3A, SPAC13C5.01C, SPAC13C5.05C, SPAC13G6.05C, SPAC15A10.12C, SPAC15E1.10, SPAC17A5.08, SPAC17G8.12, SPAC17H9.07, SPAC1A6.02, SPAC1F5.02, SPAC20G8.02, SPAC22F8.04, SPAC25G10.08, SPAC31A2.04C, SPAC3A11.03, SPAC3A12.02, SPAC3A12.13C, SPAC3H8.09C, SPAC4D7.04C, SPAC4F10.18, SPAC56F8.03, SPAC6G10.04C, SPAC7D4.09C, SPAC890.06, SPACA4, SPARC, SPARCL1, SPBC1271.04C, SPBC12C2.11, SPBC15C4.03, SPBC16E9.09C, SPBC17D11.08, SPBC211.03C, SPBC4.03C, SPBC577.10, SPBC646.16, SPBPJ4664.04, SPC-1, SPCC1450.15, SPCC18.17C, SPCC285.14, SPCS1, SPCS2, SPCS3, SPH161, SPHK-1, SPHK1, SPIA1, SPN100A, SPN27A, SPN28B, SPN28DA, SPN28DB, SPN28DC, SPN28F, SPN31A, SPN38F, SPN42DA, SPN42DB, SPN42DD, SPN42DE, SPN43AA, SPN43AB, SPN43AD, SPN47C, SPN53F, SPN55B, SPN75F, SPN77BA, SPN77BB, SPN77BC, SPN85F, SPN88EA, SPN88EB, SPO11, SPO14, SPON-1, SPON1, SPON1A, SPON2, SPON2B, SPP-10, SPP-8, SPP1, SPP2, SPRN, SPS18, SPSB-1, SPSB-2, SPSB1, SPSB2, SPSB3, SPSB3A, SPSB4, SPSB4A, SPTA1, SPTAN1, SPTB, SPTBN1, SPTBN2, SPTBN4, SPTBN5, SPTF-2, SPTF-3, SRAS, SRD5A3, SRF-3, SRP, SRP-1, SRP-2, SRP-3, SRP-6, SRP-7, SRP-8, SRP14, SRP14-1; SRP14-2, SRP19, SRP21, SRP54, SRP54K, SRP68, SRP72, SRP9, SRPA-68, SRPA-72, SRPRA, SRPRB, SSA1, SSA2, SSA3, SSA4, SSO1, SSO2, SSPO, SSPOP, SSR1, SSR2, SSR3, SSR4, SST2, ST3GAL1, ST3GAL1L3, ST3GAL2, ST3GAL2.1, ST3GAL3, ST3GAL3A, ST3GAL3B, ST3GAL4, ST3GAL5, ST3GAL6, ST3GAL7, ST6GAL1, ST6GAL2, ST6GAL2B, ST6GALNAC1, ST6GALNAC1.1, ST6GALNAC2, ST6GALNAC3, ST6GALNAC4, ST6GALNAC5, ST6GALNAC5A, ST6GALNAC6, ST8SIA1, ST8SIA2, ST8SIA3, ST8SIA4, ST8SIA5, ST8SIA6, ST8SIA7.1, STA, STAB2, STAG1, STAG1A, STAG2, STAG2B, STAM, STAM2, STAMBP, STAMBPB, STAMBPL1, STARD8, STAT3, STC2, STC2A, STE14, STE4, STG, STK36, STL, STOPS, STPG4, STS, STT3A, STX17, STX1A, STX5, STX5A, SU(VAR)2-10, SU(Z)2, SUDS3, SUI2, SUI3, SULT1A3, SUM1, SUMF1, SUMF2, SUMO, SUMO1, SUMO2, SUMO3, SUP-17, SUP35, SUP45, SUPERDEATH, SURO-1, SUZ12, SUZ12B, SVBP, SW, SWAH-1, SWAN-2, SWD1, SWD3, SWIP-1, SXC, SYN5, SYN7A, SYN8, SYT4, SYVN1, SYX-5, SYX17, SYX5, T23G5.3, TAB1, TADA2B, TADA3, TAF10, TAF10B, TAF9, TAF9B, TAG-225, TAG-30, TAKL1, TANC2, TANGO1, TANGO14, TAPT1, TARS1, TARS2, TATDN2, TBA-2, TBA-5, TBA-6, TBA-9, TBB-1, TBB-2, TBB-4, TBB-6, TBB5, TBB6, TBC-20, TBC1D20, TBCA, TBCB, TBCC, TBCD, TBCE, TBL2, TBP1, TCEANC, TCEB1, TCEB2, TCF25, TCF7L2, TCP-1 DELTA, TCP1, TDG, TDG.2, TECTA, TECTB, TEF1; TEF2, TEF101, TEF102, TEF103, TEF3, TEF5, TEP-1, TEP1, TEP2, TEP3, TEP4, TER94, TERB1, TES, TES-1, TEX101, TF, TFA, TFAP2A, TFAP2B, TFAP2C, TFG, TFG-1, TFPT, TGAS006M08.1, TGFA, TGFB1, TGFBI, TGFBR1, TGFBR1B, TGFBR2, TGOLN1, TGOLN2, TGY, THBS1, THBS1B, THBS2, THBS2A, THD1, THOR, THP1, THRA, THRAB, THRB, THSD1, THSD4, THSD7A, THSD7AA, THSD7B, THSD7BA, THY1, TICAM2, TIF1, TIF1; TIF2, TIF11, TIF211, TIF212, TIF213, TIF221, TIF222, TIF223, TIF224, TIF225, TIF32, TIF34, TIF35, TIF451, TIF452, TIF4631, TIF4632, TIF471, TIF5, TIF51A, TIF51B, TIMP, TIMP-1, TIMP1, TIPARP, TKO, TLN1, TM6SF2, TMED-10, TMED-12, TMED-3, TMED-4, TMED10, TMED2, TMED3, TMED7, TMED9, TMEM115, TMEM129, TMEM132A, TMEM132E, TMEM159, TMEM170A, TMEM170B, TMEM19, TMEM208, TMEM26, TMEM97, TMPRSS4, TNC, TNFAIP3, TNIP1, TNIP2, TNIP3, TNKS, TNKS2, TNKSA, TOC-1, TOM20, TOM70, TOMM-20, TOMM-70, TOMM20, TOMM20B, TOMM70, TOMM70A, TOP-1, TOP-2, TOP1, TOP1.2, TOP1L, TOP2, TOP2A, TOP2B, TOP2MT, TOPORS, TOPORSA, TOPORSB, TP53, TP53BP1, TPGS1, TPGS2, TPP1, TPR, TPRB, TPST1, TPST2, TRAF-LIKE, TRAF2, TRAF2B, TRAF3, TRAF6, TRAM1, TRAPCC10-1; TRAPCC10-2, TRAPPC1, TRAPPC1-1; TRAPPC1-2, TRAPPC10, TRAPPC2, TRAPPC2L, TRAPPC3, TRAPPC4, TRAPPC5, TRAPPC6A, TRAPPC6B, TRAPPC6BL, TRAPPC9, TRBD, TRF-2, TRIM 35-44, TRIM107, TRIM108, TRIM109, TRIM13, TRIM25, TRIM27, TRIM27.1, TRIM27.2, TRIM28, TRIM29, TRIM33L, TRIM35-1, TRIM35-12, TRIM35-13, TRIM35-14, TRIM35-19, TRIM35-20, TRIM35-21, TRIM35-22, TRIM35-23, TRIM35-24, TRIM35-27, TRIM35-28, TRIM35-29, TRIM35-3, TRIM35-31, TRIM35-33, TRIM35-34, TRIM35-39, TRIM35-40, TRIM35-7, TRIM35-9, TRIM39.2, TRIM47, TRIM58, TRIM69, TRIP11, TRM112, TRMT112, TRP-4, TRPP-1, TRPP-10, TRPP-3, TRPP-4, TRPP-5, TRPP-6, TRPP-9, TRRAP, TRS120, TRS130, TRS20, TRS23, TRS31, TRS33, TRX-1, TRX-2, TSF1, TSF2, TSF3, TSFM, TSHB, TSHBA, TSNARE1, TSP, TSPAN14, TSPAN15, TSPAN33, TSPAN5, TSPYL2, TSTA3, TTC19, TTF1, TTF1.6, TTGN1, TTL, TTLL1, TTLL10, TTLL11, TTLL12, TTLL13, TTLL13P, TTLL2, TTLL3, TTLL4, TTLL5, TTLL6, TTLL7, TTLL8, TTLL9, TTR, TUB-2, TUBA, TUBA1A, TUBA1B, TUBA1C, TUBA2, TUBA3A; TUBA3B, TUBA3C, TUBA3D, TUBA3E, TUBA4A, TUBA4B, TUBA4L, TUBA8, TUBA8A, TUBA8L3, TUBA8L4, TUBA8L5, TUBAL3, TUBAL3.1, TUBAL3.2, TUBB1, TUBB2, TUBB2A, TUBB2B, TUBB3, TUBB4A, TUBB4B, TUBB4BL, TUBB6, TUBB8, TUBB8B, TUBGCP5, TUFM, TULP4, TULP4A, TULP4B, TUSC3, TWE, TXN, UAP1, UBA-1, UBA-2, UBA1, UBA2, UBA3, UBA5, UBA52, UBA6, UBB, UBC, UBC-1, UBC-14, UBC-16, UBC-18, UBC-20, UBC-25, UBC-3, UBC-7, UBC-8, UBC-9, UBC10, UBC12, UBC2, UBC4, UBC6, UBC7, UBC9, UBCB, UBCC, UBCE2H, UBCE2M, UBD, UBE2A, UBE2B, UBE2C, UBE2D1, UBE2D1A, UBE2D1B, UBE2D2, UBE2D3, UBE2E1, UBE2E2, UBE2E3, UBE2F, UBE2G1, UBE2G1A, UBE2G2, UBE2H, UBE2I, UBE2IA, UBE2J2, UBE2K, UBE2KA, UBE2L3, UBE2L3A, UBE2L3B, UBE2L6, UBE2M, UBE2N, UBE2NA, UBE2NB, UBE2Q2, UBE2Q2L, UBE2R2, UBE2S, UBE2T, UBE2V, UBE2V2, UBE2W, UBE2WB, UBE2Z, UBH-1, UBH-2, UBH-3, UBH-4, UBI-P5E, UBI-P63E, UBL1, UBP6, UBPA, UBQ-1, UBQB, UBQC, UBQD, UBQF, UBQG, UBQH, UBQI, UBQJ, UBQO, UBXD7, UBXN-1, UBXN1, UBXN2A, UBXN7, UCH, UCH-L1, UCH-L5, UCH1, UCH2, UCH54, UCHL1, UCHL3, UCHL5, UCN, UEV-2, UEV-3, UFD-1, UFD-3, UFD1, UFD1-LIKE, UFD1L, UFO1, UGALT, UGGT1, UGGT2, UGTP-1, UHRF2, UIMC1, ULA-1, ULA1, ULBP1, ULBP2, ULK3, ULP-1, ULP-3, ULP-5, ULP1, UMOD, UMOD.1, UMODL1, UNC-108, UNC-55, UNC-70, UPK2, US11, USO-1, USO1, USP-14, USP-3, USP-46, USP-48, USP-5, USP10, USP11, USP12, USP12-46, USP12A, USP13, USP14, USP15, USP16, USP17L1, USP17L10, USP17L11, USP17L12, USP17L13, USP17L15, USP17L17, USP17L18, USP17L19, USP17L2, USP17L20, USP17L21, USP17L22, USP17L24; USP17L25; USP17L26; USP17L27; USP17L28; USP17L29; USP17L30, USP17L3, USP17L4, USP17L5, USP17L8, USP17LA, USP17LB, USP17LC, USP17LD, USP17LE, USP18, USP19, USP2, USP20, USP20-33, USP21, USP22, USP24, USP25, USP26, USP28, USP2A, USP3, USP30, USP33, USP34, USP37, USP4, USP41, USP42, USP44, USP47, USP48, USP49, USP5, USP7, USP8, USP9, USP9X, USP9Y, V9GZ90, VAMP2, VARS1, VARS2, VASH1, VASH2, VBP1, VCAN, VCANA, VCP, VCPIP1, VCPKMT, VDAC-1, VDAC1, VDAC2, VDAC3, VDAC5P, VDR, VDRA, VEZF1, VGF, VHL, VHL-1, VIH, VIP, VLET, VNN1, VNN2, VNN3, VPS21, VSIG10, VWA1, W03F8.3, W03G11.3, WAC, WACA, WAP, WARS1, WARS2, WBSCR17, WCY, WDR-12, WDR-20, WDR-23, WDR-48, WDR-5.1, WDR12, WDR20, WDR20A, WDR21, WDR48, WDR5, WDR5B, WDR61, WDR68, WDS, WDSOF1, WDTC1, WFS1, WFS1B, WIMA, WIPI1, WIPI2, WNT2B, WOL, WRAP53, WRN, WSB1, WSB2, WU:FB30F12, XB22169562, XB5756113, XB5813854 [PROVISIONAL:ANKRD52], XB5820455 [PROVISIONAL:MR1], XB5884180, XB5894692 [PROVISIONAL:AZGP1], XB5909790, XB5946283 [PROVISIONAL:FCRL5], XB5960289 [PROVISIONAL:FCRL4], XB995277 [PROVISIONAL:ADAMTSL5], XBP1, XIT, XPC, XPC-1, XPNPEP1, XPNPEP2, XPO4, XPO6, XRCC4, XRN2, YAK1, YAKA, YARS1, YARS2, YF5, YIF1A, YJL132W, YKT-6, YKT6, YLR352W, YMR085W, YNG1, YNG2, YOD1, YOR022C, YPT1, YPT10, YPT11, YPT2, YPT3, YPT31, YPT32, YPT4, YPT5, YPT52, YPT53, YPT6, YPT7, YPT71, YRB1, YRB2, YTM1, YUH1, YY1, YY1A, YY2, ZBED1, ZBTB1, ZBTB16, ZBTB16B, ZBTB17, ZBTB5, ZC3H12B, ZDA, ZDHHC2, ZETACOP, ZFP131, ZG16, ZGC:100832, ZGC:112234;, ZGC:114081, ZGC:114174, ZGC:114188, ZGC:136767, ZGC:136929, ZGC:153151, ZGC:153722, ZGC:153932, ZGC:158376, ZGC:161973, ZGC:162608, ZGC:171480, ZGC:171927, ZGC:173570, ZGC:173585, ZGC:173729, ZGC:174574, ZGC:194252, ZGC:194990, ZGC:55461, ZGC:55733, ZGC:56493, ZGC:56525, ZGC:65894, ZGC:77287, ZGC:92594, ZGC:92907, ZK1098.4, ZK512.4, ZK632.4, ZLD, ZMP:0000000729, ZMP:0000001316, ZNF1007, ZNF1013, ZNF1132, ZNF131, ZNF350, ZNF615, ZNFL2A, ZNHIT3, ZNT33D, ZNT35C, ZRANB1, ZRANB1B | 0.377728112 | |||||
| Metabolism of RNA | 0.625141846 | 4 | 0.625141846 | |||
| Metabolism of serotonin | 1.89838407 | 2.303196057 | 6.099964197 | -0.438630137 | 0.096060398 | 0.009954374 |
| Metabolism of steroid hormones | 0.368745703 | 0.938894818 | 0.086553357 | |||
| Metabolism of steroids | 0.281890027 | 1.060914825 | 0.074765327 | |||
| Metabolism of vitamin K | 2.299964402 | 0 | 4.599928804 | 0.281496913 | ||
| Metabolism of vitamins and cofactors | 0.150275479 | 0.015906764 | 0.000597599 | |||
| Metabolism of water-soluble vitamins and cofactors | 0.126042868 | 0.005253626 | 0.000165546 | |||
| Metal ion SLC transporters | 2.237327828 | 1.568201724 | 6.042857379 | 0.419506007 | 0.129386561 | 0.013003241 |
| Metal sequestration by antimicrobial proteins | 1.747401969 | 0 | 3.494803938 | 0.184279363 | ||
| Metalloprotease DUBs | 2.521903634 | 1.968482949 | 7.012290217 | 0.439024807 | 0.633973156 | 0.073187174 |
| Methionine salvage pathway | 1.679808197 | 0 | 3.359616394 | -0.849273774 | 1.987688462 | 0.23775457 |
| Methylation | 2.177256417 | 2.318063335 | 6.672576168 | -0.292066978 | 0.396163751 | 0.036720795 |
| Methylation of MeSeH for excretion | 1.992650243 | 0 | 3.985300487 | 0.227428218 | ||
| MGMT-mediated DNA damage reversal | 1.959268645 | 0.301029996 | 4.219567285 | 0.248036608 | ||
| MHC class II antigen presentation | 2.659755722 | 3.243286146 | 8.56279759 | 0.340212145 | 1.55197251 | 0.191028929 |
| Microbial modulation of RIPK1-mediated regulated necrosis | 0.422697368 | 0.239760339 | 0.025336516 | |||
| MicroRNA (miRNA) biogenesis | 2.375055279 | 1.633468456 | 6.383579014 | -0.323739035 | 0.080254904 | 0.007457501 |
| Microtubule-dependent trafficking of connexons from Golgi to the plasma membrane | 2.808551722 | 3.021189299 | 8.638292744 | 0.687122022 | 4 | 0.669700991 |
| Mineralocorticoid biosynthesis | 2.62004867 | 2.167317335 | 7.407414675 | 0.567493251 | 0.962027766 | 0.13140978 |
| Minus-strand DNA synthesis | 3.297254698 | 2.133538908 | 8.728048305 | 0.97863599 | 1.277057136 | 0.261824933 |
| Miscellaneous substrates | 2.667336478 | 2.46686762 | 7.801540577 | -0.177454745 | 0.00526424 | 0.00048918 |
| Miscellaneous transport and binding events | 1.622556579 | 0 | 3.245113157 | -0.93260274 | 1.90863956 | 0.245361223 |
| Mismatch Repair | -0.350328947 | 0.137633007 | 0.012054207 | |||
| Mismatch repair (MMR) directed by MSH2:MSH3 (MutSbeta) | 2.475252395 | 1.414973348 | 6.365478138 | -0.350328947 | 0.137633007 | 0.013214553 |
| Mismatch repair (MMR) directed by MSH2:MSH6 (MutSalpha) | 2.41521053 | 1.146128036 | 5.976549096 | -0.350328947 | 0.137633007 | 0.012524168 |
| Mitochondrial ABC transporters | 2.066747458 | 0 | 4.133494915 | 0.240464844 | ||
| Mitochondrial biogenesis | 0.256816923 | 0.355126899 | 0.022800649 | |||
| Mitochondrial calcium ion transport | 1.238808437 | 0 | 2.477616873 | -0.748355263 | 2.049833445 | 0.196012005 |
| Mitochondrial Fatty Acid Beta-Oxidation | 2.961414908 | 0.698970004 | 6.62179982 | 0.214778379 | 0.042316665 | 0.003485835 |
| mitochondrial fatty acid beta-oxidation of saturated fatty acids | -0.343745082 | 0.280588835 | 0.024112758 | |||
| mitochondrial fatty acid beta-oxidation of unsaturated fatty acids | 1.455863467 | 0 | 2.911726935 | -0.533424658 | 0.269331554 | 0.02002639 |
| Mitochondrial protein import | 2.811318121 | 1.84509804 | 7.467734282 | 0.176080882 | 0.011155022 | 0.00098664 |
| Mitochondrial transcription initiation | 2.057120711 | 0 | 4.114241423 | 0.238771119 | ||
| Mitochondrial translation | 0.689022666 | 0.671116718 | 0.115603658 | |||
| Mitochondrial translation elongation | 2.162099208 | 0 | 4.324198416 | 0.689022666 | 0.671116718 | 0.075180309 |
| Mitochondrial translation initiation | 1.00903784 | 0 | 2.018075681 | 0.054371958 | ||
| Mitochondrial translation termination | 1.00903784 | 0 | 2.018075681 | 0.054371958 | ||
| Mitochondrial tRNA aminoacylation | 2.484711601 | 0.477121255 | 5.446544456 | -0.194242718 | 0.007920484 | 0.000512063 |
| Mitophagy | 0.619400886 | 2.079263576 | 0.321974425 | |||
| Mitotic Anaphase | 0.60286378 | 4 | 0.60286378 | |||
| Mitotic G1 phase and G1/S transition | 0.511791684 | 4 | 0.511791684 | |||
| Mitotic G2-G2/M phases | 0.548165833 | 4 | 0.548165833 | |||
| Mitotic Metaphase and Anaphase | 0.60286378 | 4 | 0.60286378 | |||
| Mitotic Metaphase/Anaphase Transition | 2.361136006 | 2.31386722 | 7.036139233 | 0.819775404 | 0.438907956 | 0.071692754 |
| Mitotic Prometaphase | 2.890084191 | 3.209515015 | 8.989683397 | 0.498925614 | 4 | 0.593730736 |
| Mitotic Prophase | 2.95493488 | 2.522444234 | 8.432313994 | 0.440572889 | 1.947317942 | 0.260843314 |
| Mitotic Spindle Checkpoint | 0.410464215 | 1.181117343 | 0.121201601 | |||
| Mitotic Telophase/Cytokinesis | 1.942096995 | 2.31386722 | 6.19806121 | 0.388074344 | 0.161740087 | 0.01594303 |
| Modulation by Mtb of host immune system | 3.017021114 | 1.770852012 | 7.80489424 | 0.674782684 | 0.956143145 | 0.148330548 |
| Molecules associated with elastic fibres | 2.108919679 | 2.103803721 | 6.321643078 | 0.314031417 | 0.294876401 | 0.026807338 |
| MPS IV - Morquio syndrome B | 2.075038795 | 0.77815125 | 4.92822884 | 0.310377383 | ||
| MPS VII - Sly syndrome | 2.364710299 | 1 | 5.729420599 | 0.619282388 | 0.144939815 | 0.017599857 |
| mRNA 3'-end processing | 3.596268175 | 0.903089987 | 8.095626337 | 0.507140322 | 0.153201392 | 0.021130855 |
| mRNA Capping | 2.737273932 | 2.225309282 | 7.699857146 | 0.468337064 | 0.447819898 | 0.057309237 |
| mRNA decay by 3' to 5' exoribonuclease | 3.056728102 | 0.477121255 | 6.590577459 | 0.672692413 | 0.219870184 | 0.030608473 |
| mRNA decay by 5' to 3' exoribonuclease | 2.321350466 | 0 | 4.642700933 | 0.631881676 | 0.161878237 | 0.017642769 |
| mRNA Editing: C to U Conversion | 2.162473389 | 0.301029996 | 4.625976774 | 0.283788346 | ||
| mRNA Splicing | 0.229879141 | 0.093328897 | 0.005363592 | |||
| mRNA Splicing - Major Pathway | 2.940221547 | 1.397940009 | 7.278383104 | 0.229879141 | 0.093328897 | 0.008654458 |
| mRNA Splicing - Minor Pathway | 3.445387795 | 0.903089987 | 7.793865577 | 0.354604714 | 0.193852495 | 0.022287175 |
| MTOR signalling | 3.108922657 | 3.323664536 | 9.54150985 | 0.584349495 | 4 | 0.664910939 |
| mTORC1-mediated signalling | 2.957237604 | 3.229425848 | 9.143901055 | 0.597294962 | 2.374012235 | 0.386294148 |
| Mucopolysaccharidoses | -0.381030702 | 0.198085553 | 0.018869169 | |||
| Muscarinic acetylcholine receptors | 1.968403789 | 3.422097163 | 7.358904742 | 0.707979161 | 1.86726631 | 0.286684669 |
| Muscle contraction | 0.341962174 | 2.925427664 | 0.250096401 | |||
| Mycothiol biosynthesis | 1.858951487 | 0 | 3.717902974 | 0.203905328 | ||
| Mycothiol metabolism | 1.939323637 | 0 | 3.878647274 | 0.218045962 | ||
| MyD88 cascade initiated on plasma membrane | 2.244949266 | 2.206825876 | 6.696724409 | 0.371919355 | 2.483573285 | 0.255768175 |
| MyD88 deficiency (TLR2/4) | 3.352237199 | 2.480006943 | 9.184481342 | 0.476223853 | 0.915098894 | 0.135532853 |
| MyD88 deficiency (TLR5) | 1.927119524 | 0 | 3.854239048 | -0.791232877 | 1.026896273 | 0.121931382 |
| MyD88 dependent cascade initiated on endosome | 2.273275685 | 2.227886705 | 6.774438075 | 0.374393251 | 2.817231584 | 0.293824514 |
| MyD88:MAL(TIRAP) cascade initiated on plasma membrane | 2.992259408 | 2.678518379 | 8.663037195 | 0.398779751 | 3.38467959 | 0.445767719 |
| Myoclonic epilepsy of Lafora | 3.014914482 | 1.041392685 | 7.07122165 | 0.416706525 | 0.123466194 | 0.01400259 |
| Myogenesis | 2.925801268 | 3.515741417 | 9.367343953 | 0.611698206 | 2.049366478 | 0.343064062 |
| N-Glycan antennae elongation | 2.223489263 | 0 | 4.446978526 | -0.622088243 | 0.720159303 | 0.075789156 |
| N-glycan antennae elongation in the medial/trans-Golgi | -0.216364635 | 0.117385599 | 0.006349523 | |||
| N-glycan trimming and elongation in the cis-Golgi | 1.875929262 | 0 | 3.751858524 | -0.881917808 | 1.423719191 | 0.183308065 |
| N-glycan trimming in the ER and Calnexin/Calreticulin cycle | 3.217704479 | 2.021189299 | 8.456598257 | 0.53953633 | 1.244807071 | 0.182530629 |
| Na+/Cl- dependent neurotransmitter transporters | 2.68293052 | 3.539452492 | 8.905313532 | 0.46053838 | 1.420662877 | 0.202510005 |
| NADE modulates death signalling | 2.214739237 | 1.672097858 | 6.101576331 | -0.373183438 | 0.301029996 | 0.028738177 |
| NADPH regeneration | 2.269525144 | 2.456366033 | 6.995416321 | 0.894275541 | 0.659202877 | 0.113469108 |
| NCAM signaling for neurite out-growth | 2.423839518 | 3.243534102 | 8.091213138 | 0.61289307 | 4 | 0.604363139 |
| NCAM1 interactions | 3.49089337 | 2.769377326 | 9.751164067 | 0.853869033 | 4 | 0.810486593 |
| Neddylation | 3.206122343 | 3.108565024 | 9.52080971 | 0.574837251 | 4 | 0.659086914 |
| Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters | 0.490835592 | 0.681657029 | 0.083645383 | |||
| Nef and signal transduction | 2.522276397 | 2.602059991 | 7.646612785 | 0.502127379 | 0.731874579 | 0.096249509 |
| Nef Mediated CD4 Down-regulation | 3.195113868 | 2.29666519 | 8.686892927 | 0.549616228 | 0.642195673 | 0.096884102 |
| Nef Mediated CD8 Down-regulation | 0.583401808 | 0.08932757 | 0.013028466 | |||
| Nef mediated downregulation of CD28 cell surface expression | 1.505109573 | 0 | 3.010219146 | 0.141650564 | ||
| Nef mediated downregulation of MHC class I complex cell surface expression | 0.553273076 | 0.052322698 | 0.007237185 | |||
| Negative epigenetic regulation of rRNA expression | 0.26875013 | 0.185946014 | 0.012493254 | |||
| Negative feedback regulation of MAPK pathway | 2.654484462 | 3.35755372 | 8.666522644 | 0.737794844 | 2.517900174 | 0.43842544 |
| Negative regulation of activity of TFAP2 (AP-2) family transcription factors | 2.24180643 | 1.230448921 | 5.714061781 | -0.531506849 | 0.263052488 | 0.029003825 |
| Negative regulation of FGFR1 signaling | 2.770773715 | 3.136086097 | 8.677633526 | 0.484950064 | 1.677047205 | 0.239249471 |
| Negative regulation of FGFR2 signaling | 2.774629669 | 3.108903128 | 8.658162465 | 0.463637585 | 1.504681736 | 0.210273192 |
| Negative regulation of FGFR3 signaling | 2.837249796 | 3.113609151 | 8.788108743 | 0.525194813 | 2.158619745 | 0.321885966 |
| Negative regulation of FGFR4 signaling | 2.823008322 | 3.125481266 | 8.771497909 | 0.525158433 | 2.158619745 | 0.321413698 |
| Negative regulation of MAPK pathway | 2.628374404 | 3.321184027 | 8.577932835 | 0.364023082 | 1.052592235 | 0.132899665 |
| Negative regulation of MET activity | 2.84260177 | 3.008600172 | 8.693803712 | 0.417526256 | 0.540551471 | 0.072672668 |
| Negative regulation of NMDA receptor-mediated neuronal transmission | 2.407093564 | 3.098297536 | 7.912484664 | 0.597503403 | 2.747130446 | 0.403448971 |
| Negative regulation of NOTCH4 signaling | 3.332383183 | 3.143951116 | 9.808717482 | 0.797454808 | 4 | 0.785248613 |
| Negative regulation of TCF-dependent signaling by DVL-interacting proteins | 3.857110097 | 0.698970004 | 8.413190198 | 0.437106056 | 0.16393583 | 0.021847735 |
| Negative regulation of TCF-dependent signaling by WNT ligand antagonists | 2.008172725 | 0.602059991 | 4.618405442 | -0.498629386 | 0.586265724 | 0.053947009 |
| Negative regulation of the PI3K/AKT network | 2.519905128 | 3.028571253 | 8.068381508 | 0.435819778 | 4 | 0.514648628 |
| Negative regulators of DDX58/IFIH1 signaling | 2.352158904 | 2.519827994 | 7.224145802 | 0.29296711 | 0.165940342 | 0.016580298 |
| NEP/NS2 Interacts with the Cellular Export Machinery | 2.384008289 | 1.799340549 | 6.567357128 | 0.542000762 | 0.447790653 | 0.054888223 |
| Nephrin family interactions | 2.63366634 | 3.076640444 | 8.343973123 | 0.535096489 | 1.227432975 | 0.177518882 |
| Nervous system development | 0.588268502 | 4 | 0.588268502 | |||
| Netrin-1 signaling | 2.215731795 | 2.602059991 | 7.033523582 | 0.32439633 | 0.547226564 | 0.055481859 |
| Netrin mediated repulsion signals | 2.547293084 | 2.550228353 | 7.644814521 | 0.620273973 | 0.428078669 | 0.062609102 |
| Neurexins and neuroligins | 2.171996903 | 3.068185862 | 7.412179667 | 0.466253468 | 0.966141733 | 0.119804612 |
| Neurodegenerative Diseases | 0.293306767 | 0.272305844 | 0.019967287 | |||
| Neuronal System | 0.312798316 | 4 | 0.312798316 | |||
| Neurophilin interactions with VEGF and VEGFR | 2.659433757 | 3.209783015 | 8.528650528 | 0.587076266 | 0.584103706 | 0.089663097 |
| Neurotoxicity of clostridium toxins | -0.721643836 | 0.7770188 | 0.140182707 | |||
| Neurotransmitter clearance | 2.574196699 | 3.123198075 | 8.271591474 | 0.492278095 | 1.020203386 | 0.141135247 |
| Neurotransmitter receptors and postsynaptic signal transmission | 2.384104297 | 2.582063363 | 7.350271957 | 0.409386836 | 4 | 0.46438549 |
| Neurotransmitter release cycle | 1.813230024 | 0.477121255 | 4.103581303 | 0.241815823 | 0.185281834 | 0.009871165 |
| Neurotransmitter uptake and metabolism In glial cells | -0.610852288 | 0.698970004 | 0.106741857 | |||
| Neutrophil degranulation | 2.548658388 | 3.963504499 | 9.060821275 | 0.24786566 | 1.031951129 | 0.121736876 |
| NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 | 2.48930435 | 2.230448921 | 7.209057622 | 0.465050416 | 0.583749784 | 0.07076976 |
| NF-kB is activated and signals survival | 2.373172729 | 2.63748973 | 7.383835188 | 0.363770426 | 0.396962968 | 0.043994293 |
| NFG and proNGF binds to p75NTR | 1.158211139 | 0 | 2.316422278 | -0.967945205 | 2.558083107 | 0.309510535 |
| NGF-independant TRKA activation | 2.743474733 | 2.382017043 | 7.868966509 | 0.721963257 | 1.993215788 | 0.322618278 |
| NGF-stimulated transcription | 2.322302954 | 2.494154594 | 7.138760502 | 0.288968111 | 0.252749974 | 0.024849394 |
| NGF processing | 2.851244769 | 2.382017043 | 8.084506581 | 0.731719671 | 1.753466155 | 0.290825712 |
| Nicotinamide salvaging | 2.357636129 | 2.409933123 | 7.12520538 | -0.252500601 | 0.278753601 | 0.026086555 |
| Nicotinate metabolism | 2.255776032 | 1.230448921 | 5.742000984 | 0.245944366 | 0.097983966 | 0.007341324 |
| NIK-->noncanonical NF-kB signaling | 3.256052039 | 2.974511693 | 9.486615771 | 0.756710511 | 4 | 0.748259508 |
| Nilotinib-resistant KIT mutants | 2.617771423 | 2.158362492 | 7.393905339 | 0.918104629 | 0.749379508 | 0.135074794 |
| Nitric oxide stimulates guanylate cyclase | 2.261105917 | 2.243038049 | 6.765249882 | 0.403137039 | 1.026959972 | 0.110675423 |
| NOD1/2 Signaling Pathway | 2.903233659 | 3.444513206 | 9.250980524 | 0.449323754 | 2.258660242 | 0.328866524 |
| Non-integrin membrane-ECM interactions | 2.856734572 | 2.127104798 | 7.840573943 | 0.402031137 | 1.205475037 | 0.146465792 |
| Noncanonical activation of NOTCH3 | 2.980054286 | 3.122215878 | 9.082324449 | 0.913580247 | 4 | 0.805837326 |
| Nonhomologous End-Joining (NHEJ) | 2.123142736 | 3.003891166 | 7.250176638 | 0.189602497 | 0.037282685 | 0.003255985 |
| Nonsense-Mediated Decay (NMD) | 0.868826591 | 4 | 0.868826591 | |||
| Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | 4.230487718 | 2.910624405 | 11.37159984 | 0.868826591 | 4 | 0.901562291 |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | 4.357323431 | 2.903089987 | 11.61773685 | 0.902943941 | 4 | 0.931318967 |
| NoRC negatively regulates rRNA expression | 2.456337085 | 2.617000341 | 7.529674512 | 0.287792553 | 0.228334419 | 0.023566599 |
| Norepinephrine Neurotransmitter Release Cycle | 2.458887905 | 2.624282096 | 7.542057906 | 0.465185705 | 0.438644321 | 0.055069492 |
| NOSIP mediated eNOS trafficking | 2.332021621 | 1.602059991 | 6.266103234 | 0.592714325 | 0.102039535 | 0.012757949 |
| NOSTRIN mediated eNOS trafficking | 2.814870926 | 1.653212514 | 7.282954365 | 0.396771129 | 0.175233817 | 0.019915533 |
| Notch-HLH transcription pathway | 2.68485169 | 3.479719235 | 8.849422615 | 0.633943834 | 4 | 0.65400391 |
| NOTCH1 Intracellular Domain Regulates Transcription | 2.655708685 | 3.252367514 | 8.563784884 | 0.557300006 | 4 | 0.600946186 |
| NOTCH2 Activation and Transmission of Signal to the Nucleus | 3.041855907 | 3.124178055 | 9.207889869 | 0.736456789 | 4 | 0.723753412 |
| NOTCH2 intracellular domain regulates transcription | 3.135732684 | 1.431363764 | 7.702829133 | 0.271096986 | 0.069455919 | 0.007178767 |
| NOTCH3 Activation and Transmission of Signal to the Nucleus | 3.12229916 | 3.400710637 | 9.645308956 | 0.803562908 | 4 | 0.779872554 |
| NOTCH3 Intracellular Domain Regulates Transcription | 2.905521862 | 2.507855872 | 8.318899595 | 0.369302581 | 0.418213268 | 0.051682173 |
| NOTCH4 Activation and Transmission of Signal to the Nucleus | 3.048063313 | 3.124178055 | 9.220304682 | 0.846777717 | 4 | 0.779554345 |
| NOTCH4 Intracellular Domain Regulates Transcription | 3.044466266 | 2.677606953 | 8.766539484 | 0.397896924 | 0.603032656 | 0.080158807 |
| NR1D1 (REV-ERBA) represses gene expression | 2.219976507 | 2.053078443 | 6.493031457 | 0.495024355 | 0.291649621 | 0.03375698 |
| NR1H2 & NR1H3 regulate gene expression linked to lipogenesis | 2.471884838 | 2.491361694 | 7.43513137 | 0.548359676 | 1.104218025 | 0.148586265 |
| NR1H2 & NR1H3 regulate gene expression linked to triglyceride lipolysis in adipose | 2.470204195 | 2.344392274 | 7.284800664 | 0.481098689 | 0.507360133 | 0.063022222 |
| NR1H2 & NR1H3 regulate gene expression to control bile acid homeostasis | 2.167975937 | 2.725911632 | 7.061863506 | 0.380934645 | 0.385999889 | 0.04200457 |
| NR1H2 and NR1H3-mediated signaling | 0.35755333 | 0.823609124 | 0.073621046 | |||
| NR1H3 & NR1H2 regulate gene expression linked to cholesterol transport and efflux | 2.42604244 | 3.088844563 | 7.940929442 | 0.409259789 | 0.980308 | 0.121262504 |
| NRAGE signals death through JNK | 2.65660582 | 2.667452953 | 7.980664593 | 0.306499097 | 0.221363233 | 0.024652285 |
| NrCAM interactions | 1.379396584 | 0.301029996 | 3.059823164 | 0.146014216 | ||
| NRIF signals cell death from the nucleus | 2.378497064 | 3.156851901 | 7.913846029 | 0.702206172 | 4 | 0.63986947 |
| NS1 Mediated Effects on Host Pathways | 2.281151328 | 1.681241237 | 6.243543894 | 0.579788889 | 0.786820091 | 0.096875521 |
| NTF3 activates NTRK2 (TRKB) signaling | 2.274010625 | 1.755874856 | 6.303896105 | 0.749931526 | 0.319254816 | 0.046345917 |
| NTF3 activates NTRK3 signaling | 2.129005556 | 1.944482672 | 6.202493784 | 0.606409203 | 0.125222126 | 0.015768088 |
| NTF4 activates NTRK2 (TRKB) signaling | 2.274010625 | 1.755874856 | 6.303896105 | 0.749931526 | 0.319254816 | 0.046345917 |
| NTRK2 activates RAC1 | 2.158067982 | 2.418301291 | 6.734437256 | 0.530282269 | 0.403987446 | 0.049797797 |
| NTRK3 as a dependence receptor | 2.444106414 | 1.491361694 | 6.379574522 | 0.475890411 | 0.11317227 | 0.01266284 |
| Nuclear Envelope (NE) Reassembly | 0.500188825 | 3.468041843 | 0.433668943 | |||
| Nuclear Envelope Breakdown | 3.445841946 | 0.301029996 | 7.192713888 | 0.601134499 | 2.821916494 | 0.389516917 |
| Nuclear Events (kinase and transcription factor activation) | 0.343087549 | 1.014387882 | 0.087005963 | |||
| Nuclear import of Rev protein | 2.525955299 | 2.167317335 | 7.219227932 | 0.47371286 | 0.384697438 | 0.047105052 |
| Nuclear Pore Complex (NPC) Disassembly | 2.777644825 | 2.64738297 | 8.20267262 | 0.597256516 | 1.476603929 | 0.222337777 |
| Nuclear Receptor transcription pathway | 2.54227957 | 3.780893109 | 8.865452249 | 0.329414496 | 1.250507131 | 0.157115653 |
| Nuclear signaling by ERBB4 | 2.790882594 | 3.590395947 | 9.172161135 | 0.695744646 | 4 | 0.701554125 |
| Nucleobase biosynthesis | 0.326569593 | 0.281091666 | 0.022948998 | |||
| Nucleobase catabolism | -0.552341598 | 4 | 0.552341598 | |||
| Nucleosome assembly | -0.280504524 | 0.079993771 | 0.005609654 | |||
| Nucleotide-binding domain, leucine rich repeat containing receptor (NLR) signaling pathways | 0.449344775 | 3.123515891 | 0.350883886 | |||
| Nucleotide-like (purinergic) receptors | 0.301263173 | 0.295906891 | 0.022286462 | |||
| Nucleotide Excision Repair | 0.242777686 | 0.203151066 | 0.012330136 | |||
| Nucleotide salvage | -0.233351679 | 0.585541409 | 0.034159268 | |||
| O-glycosylation of TSR domain-containing proteins | 1.916026484 | 0.477121255 | 4.309174222 | -0.519736842 | 0.661330404 | 0.05996164 |
| O-linked glycosylation | 3.004007236 | 0 | 6.008014472 | -0.419278784 | 2.698591122 | 0.269916814 |
| O-linked glycosylation of mucins | 3.842528513 | 0.477121255 | 8.16217828 | -0.423845708 | 1.720513223 | 0.220871096 |
| O2/CO2 exchange in erythrocytes | 0.508681617 | 0.511025918 | 0.064987373 | |||
| OAS antiviral response | 3.207860572 | 1.278753601 | 7.694474744 | 0.711585867 | 0.272258253 | 0.04310135 |
| Olfactory Signaling Pathway | 1.976339192 | 0.301029996 | 4.25370838 | -0.602465753 | 0.427862737 | 0.042912048 |
| Oncogene Induced Senescence | 2.527837928 | 3.433769834 | 8.48944569 | 0.540766825 | 1.988683401 | 0.29275632 |
| Oncogenic MAPK signaling | 0.460724614 | 3.940332641 | 0.453852059 | |||
| Opioid Signalling | 2.162765652 | 3.27600199 | 7.601533294 | 0.364090309 | 2.24324963 | 0.25500117 |
| Opsins | 1.534313284 | 0 | 3.068626567 | 0.146788649 | ||
| Orc1 removal from chromatin | 3.267445944 | 3.097951071 | 9.632842959 | 0.765019786 | 4 | 0.759957883 |
| Orexin and neuropeptides FF and QRFP bind to their respective receptors | 2.346495033 | 2.755874856 | 7.448864922 | 0.425708567 | 0.337817647 | 0.040338187 |
| Organelle biogenesis and maintenance | 0.3410528 | 2.672149085 | 0.227835982 | |||
| Organic anion transport | 2.566940017 | 1.707570176 | 6.841450209 | -0.462846175 | 0.61441043 | 0.071404549 |
| Organic anion transporters | 1.999912231 | 0 | 3.999824462 | -0.51000274 | 0.422763592 | 0.036130082 |
| Organic cation transport | 2.310076522 | 2.389166084 | 7.009319128 | 0.332462175 | 0.18103683 | 0.018480859 |
| Organic cation/anion/zwitterion transport | 0.356537984 | 0.459296733 | 0.040939183 | |||
| Other interleukin signaling | 2.514526184 | 2.621176282 | 7.650228651 | 0.317880267 | 0.206495346 | 0.022410254 |
| Other semaphorin interactions | 2.009201357 | 1.176091259 | 5.194493974 | -0.311592217 | 0.019744058 | 0.001501898 |
| Ovarian tumor domain proteases | 2.872868451 | 3.300378065 | 9.046114968 | 0.460158202 | 1.420662877 | 0.205022355 |
| Oxidative Stress Induced Senescence | 2.544811846 | 3.708250889 | 8.79787458 | 0.454664373 | 3.661009519 | 0.51410171 |
| Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | 3.215023448 | 3.093771781 | 9.523818678 | 0.703968955 | 4 | 0.723807996 |
| p130Cas linkage to MAPK signaling for integrins | 2.98524571 | 3.216693599 | 9.187185019 | 0.570471082 | 1.643537283 | 0.262839582 |
| P2Y receptors | 2.477481334 | 1.995635195 | 6.950597863 | -0.303063036 | 0.818244023 | 0.079902498 |
| p38MAPK events | 2.769634597 | 3.361160995 | 8.900430189 | 0.730576189 | 4 | 0.704951525 |
| p53-Dependent G1 DNA Damage Response | 2.260934682 | 2.752816431 | 7.274685796 | 0.758426989 | 4 | 0.635006139 |
| p53-Dependent G1/S DNA damage checkpoint | 0.758426989 | 4 | 0.758426989 | |||
| p53-Independent DNA Damage Response | 0.765606497 | 4 | 0.765606497 | |||
| p53-Independent G1/S DNA damage checkpoint | 0.765606497 | 4 | 0.765606497 | |||
| p75 NTR receptor-mediated signalling | 0.371051787 | 1.905542315 | 0.176763721 | |||
| p75NTR negatively regulates cell cycle via SC1 | 1.879326873 | 2.615950052 | 6.374603797 | 0.741826596 | 2.208499699 | 0.320382342 |
| p75NTR recruits signalling complexes | 2.400226429 | 2.683947131 | 7.484399988 | 0.502887163 | 1.267171728 | 0.164116228 |
| p75NTR regulates axonogenesis | -0.455842019 | 0.80513474 | 0.091753561 | |||
| p75NTR signals via NF-kB | 0.407315085 | 0.747330773 | 0.076099774 | |||
| Packaging of Eight RNA Segments | 2.943361751 | 1.51851394 | 7.405237442 | 0.528279191 | ||
| Packaging Of Telomere Ends | 4.182684107 | 1.397940009 | 9.763308222 | 0.716360647 | 1.137986733 | 0.211198647 |
| PAOs oxidise polyamines to amines | 1.64589607 | 0 | 3.29179214 | -0.902191781 | 1.56005569 | 0.195558597 |
| Paradoxical activation of RAF signaling by kinase inactive BRAF | 2.812505299 | 3.255031163 | 8.880041761 | 0.512626574 | 4 | 0.594924896 |
| Parasite infection | 0.471666562 | 2.487978692 | 0.293374089 | |||
| Passive transport by Aquaporins | 2.53085884 | 0 | 5.06171768 | -0.652054795 | 0.541806089 | 0.063344565 |
| Pausing and recovery of HIV elongation | 3.159791186 | 2.471291711 | 8.790874084 | 0.564968965 | 1.077108237 | 0.166008232 |
| Pausing and recovery of Tat-mediated HIV elongation | 3.055459709 | 2.472756449 | 8.583675867 | 0.709364615 | 1.918797679 | 0.325238437 |
| PCNA-Dependent Long Patch Base Excision Repair | 2.524804096 | 2.598790507 | 7.648398699 | 0.364437256 | 0.266706108 | 0.030490563 |
| PCP/CE pathway | 3.394516564 | 1.342422681 | 8.131455809 | 0.659158095 | 4 | 0.629571737 |
| PD-1 signaling | 2.68412335 | 2.718501689 | 8.086748389 | 0.439063202 | 0.80228493 | 0.103739026 |
| PDE3B signalling | 2.268962307 | 2.220108088 | 6.758032702 | 0.657260274 | 0.563803944 | 0.078618102 |
| PDGFR mutants bind TKIs | 2.318185123 | 2.017033339 | 6.653403585 | 0.882497946 | 0.618515789 | 0.102826733 |
| PECAM1 interactions | 2.659623236 | 3.295127085 | 8.614373556 | 0.496127802 | 1.288579687 | 0.184579347 |
| Pentose phosphate pathway | 2.24597678 | 0 | 4.49195356 | -0.642817982 | 1.39441043 | 0.151169018 |
| Peptide chain elongation | 4.408152663 | 2.894316063 | 11.71062139 | 0.907230923 | 4 | 0.938254293 |
| Peptide hormone biosynthesis | 3.771816824 | 0 | 7.543633649 | -0.216068001 | 0.010267487 | 0.000969511 |
| Peptide hormone metabolism | 0.274915837 | 0.511014772 | 0.035121513 | |||
| Peptide ligand-binding receptors | 2.872019159 | 4.15730563 | 9.901343947 | 0.420855168 | 4 | 0.601727314 |
| PERK regulates gene expression | 2.298428468 | 1.934498451 | 6.531355387 | 0.184906208 | 0.012817794 | 0.000993052 |
| Peroxisomal lipid metabolism | 1.786094194 | 0 | 3.572188389 | -0.157072296 | 0.035715552 | 0.001279689 |
| Peroxisomal protein import | 2.400686658 | 2.942008053 | 7.743381368 | 0.11595085 | 0.002216924 | 0.000187301 |
| Pervasive developmental disorders | 0.548637099 | 1.301534696 | 0.178517555 | |||
| Pexophagy | 3.08258445 | 1.681241237 | 7.846410137 | 0.546326754 | 0.637203727 | 0.088961733 |
| Phase 0 - rapid depolarisation | 3.232540162 | 3.534406899 | 9.999487224 | 0.587858233 | 4 | 0.690291976 |
| Phase 1 - inactivation of fast Na+ channels | 2.975678736 | 0.602059991 | 6.553417463 | -0.438596491 | 0.372769049 | 0.040807146 |
| Phase 2 - plateau phase | 3.675087837 | 3.162265614 | 10.51244129 | 0.812879458 | 4 | 0.829265457 |
| Phase 3 - rapid repolarisation | 2.823483983 | 2.731588765 | 8.378556732 | 0.478213209 | 0.253757963 | 0.035008894 |
| Phase 4 - resting membrane potential | 2.807693681 | 1.544068044 | 7.159455407 | 0.21845124 | 0.018023104 | 0.001617905 |
| Phase I - Functionalization of compounds | 2.145034828 | 1.919078092 | 6.209147748 | 0.158282692 | 0.025462083 | 0.001782115 |
| Phase II - Conjugation of compounds | -0.443178496 | 4 | 0.443178496 | |||
| Phenylalanine and tyrosine metabolism | -0.529181244 | 0.883255742 | 0.116850593 | |||
| Phenylalanine metabolism | 1.575395522 | 0 | 3.150791044 | -0.927103316 | 2.984897273 | 0.378034285 |
| Phenylketonuria | 1.924053407 | 0 | 3.848106814 | 0.215359326 | ||
| Phosphate bond hydrolysis by NTPDase proteins | 2.10934126 | 0.77815125 | 4.996833771 | -0.62709076 | 1.419060853 | 0.160291989 |
| Phosphate bond hydrolysis by NUDT proteins | -0.888980263 | 3.558319149 | 0.790818873 | |||
| phospho-PLA2 pathway | 2.617047551 | 2.992111488 | 8.226206589 | 0.818630137 | 1.158222193 | 0.206799377 |
| Phospholipase C-mediated cascade: FGFR1 | 2.660564583 | 2.367355921 | 7.688485086 | 0.423270489 | 0.392006579 | 0.047900798 |
| Phospholipase C-mediated cascade; FGFR2 | 2.669561809 | 2.176091259 | 7.515214878 | 0.382942497 | 0.326066775 | 0.037470989 |
| Phospholipase C-mediated cascade; FGFR3 | 2.811360567 | 2.214843848 | 7.837564983 | 0.515013993 | 0.673809746 | 0.091358175 |
| Phospholipase C-mediated cascade; FGFR4 | 2.782445333 | 2.301029996 | 7.865920661 | 0.514367072 | 0.663530107 | 0.090153419 |
| Phospholipid metabolism | 0.263823207 | 0.788246473 | 0.051989428 | |||
| Phosphorylation of CD3 and TCR zeta chains | 2.59246457 | 2.671172843 | 7.856101983 | 0.319837788 | 0.212088912 | 0.023632342 |
| Phosphorylation of Emi1 | 2.836175105 | 2.730782276 | 8.403132486 | 0.820224719 | 1.795715722 | 0.325078696 |
| Phosphorylation of proteins involved in G1/S transition by active Cyclin E:Cdk2 complexes | 2.189297789 | 2.635483747 | 7.014079325 | 0.646150738 | 1.116813782 | 0.157868202 |
| Phosphorylation of proteins involved in the G2/M transition by Cyclin A:Cdc2 complexes | 2.348324186 | 2.374748346 | 7.071396718 | 0.825431625 | 1.873126511 | 0.308139119 |
| Phosphorylation of the APC/C | 2.609220561 | 2.730782276 | 7.949223397 | 0.582224371 | 1.197930408 | 0.174210125 |
| Physiological factors | 2.496067425 | 2.167317335 | 7.159452186 | 0.344894244 | 0.294175149 | 0.031057171 |
| PI-3K cascade:FGFR1 | 3.098769943 | 3.363799945 | 9.561339831 | 0.46073972 | 0.807762105 | 0.121998141 |
| PI-3K cascade:FGFR2 | 3.103268556 | 3.347915187 | 9.554452299 | 0.428203976 | 0.730929022 | 0.107356273 |
| PI-3K cascade:FGFR3 | 3.207150423 | 3.350635608 | 9.764936455 | 0.525093495 | 1.179128296 | 0.190667796 |
| PI-3K cascade:FGFR4 | 3.159710318 | 3.35755372 | 9.676974355 | 0.525034507 | 1.179128296 | 0.189321412 |
| PI Metabolism | 0.330010009 | 0.92701043 | 0.07648068 | |||
| PI3K Cascade | 2.966248282 | 3.5402043 | 9.472700865 | 0.466886525 | 2.136326484 | 0.321853424 |
| PI3K events in ERBB2 signaling | 3.132339663 | 3.583085366 | 9.847764693 | 0.726887841 | 3.573564922 | 0.671811928 |
| PI3K events in ERBB4 signaling | 3.459005803 | 3.304490528 | 10.22250213 | 0.829437779 | 2.714429434 | 0.558213524 |
| PI3K/AKT activation | 2.745222732 | 3.43806745 | 8.928512915 | 0.511013037 | 1.083744803 | 0.161645608 |
| PI3K/AKT Signaling in Cancer | 0.451445873 | 4 | 0.451445873 | |||
| PI5P Regulates TP53 Acetylation | 2.521901932 | 2.818225894 | 7.862029758 | 0.517563117 | 1.128823177 | 0.153766821 |
| PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling | 2.784035236 | 4.128560807 | 9.69663128 | 0.45101432 | 4 | 0.606245936 |
| PINK1-PRKN Mediated Mitophagy | 3.224718271 | 1.113943352 | 7.563379894 | 0.47574678 | 0.24589213 | 0.031262696 |
| PIP3 activates AKT signaling | 2.822818376 | 4.142764775 | 9.788401528 | 0.522191567 | 4 | 0.646568909 |
| PIWI-interacting RNA (piRNA) biogenesis | 2.648882824 | 2.519827994 | 7.817593643 | 0.77127445 | 0.922458929 | 0.154382272 |
| PKA-mediated phosphorylation of CREB | 2.16770205 | 2.252853031 | 6.58825713 | 0.576838436 | 3.332166794 | 0.423851271 |
| PKA-mediated phosphorylation of key metabolic factors | 2.408393159 | 2.117271296 | 6.934057613 | 0.557017544 | 0.676121793 | 0.087342883 |
| PKA activation | 3.539833206 | 2.409933123 | 9.489599536 | 0.607067963 | 3.199679684 | 0.53881979 |
| PKA activation in glucagon signalling | 3.310656446 | 2.558708571 | 9.180021462 | 0.637639649 | 3.697636215 | 0.622041447 |
| PKB-mediated events | 0.657260274 | 0.563803944 | 0.092641484 | |||
| PKMTs methylate histone lysines | 2.239179921 | 3.179838928 | 7.658198771 | -0.159311562 | 0.15296746 | 0.013584793 |
| Plasma lipoprotein assembly | 0.552437903 | 1.426791101 | 0.197053371 | |||
| Plasma lipoprotein assembly, remodeling, and clearance | 0.42552909 | 2.259989038 | 0.24042277 | |||
| Plasma lipoprotein clearance | 0.404851332 | 0.900272932 | 0.091119174 | |||
| Plasma lipoprotein remodeling | 0.387183578 | 0.621061883 | 0.060116241 | |||
| Plasmalogen biosynthesis | 1.535436621 | 0 | 3.070873242 | 0.146986289 | ||
| Platelet activation, signaling and aggregation | 0.316531236 | 2.81853512 | 0.223038602 | |||
| Platelet Adhesion to exposed collagen | 3.167516553 | 2.587710965 | 8.922744072 | 0.295256497 | 0.170261715 | 0.020790779 |
| Platelet Aggregation (Plug Formation) | 2.916911627 | 2.818885415 | 8.652708668 | 0.538752773 | 4 | 0.596260073 |
| Platelet calcium homeostasis | 0.298103997 | 0.341968635 | 0.025485554 | |||
| Platelet homeostasis | 2.313902973 | 2.640481437 | 7.268287383 | 0.330930672 | 1.335501507 | 0.140537459 |
| Platelet sensitization by LDL | 3.005170391 | 3.124830149 | 9.135170932 | 0.54299244 | 1.482084674 | 0.230934628 |
| PLC-gamma1 signalling | 1.768711725 | 2.075546961 | 5.612970412 | 0.425469428 | 0.138856191 | 0.013288567 |
| PLC beta mediated events | 2.882141161 | 1.477121255 | 7.241403576 | 0.401091794 | 2.474303024 | 0.281217863 |
| PLCG1 events in ERBB2 signaling | 2.652542165 | 2.954724791 | 8.259809122 | 0.646215624 | 1.116813782 | 0.175820592 |
| Plus-strand DNA synthesis | 3.297254698 | 2.133538908 | 8.728048305 | 0.97863599 | 1.277057136 | 0.261824933 |
| POLB-Dependent Long Patch Base Excision Repair | 2.263031944 | 2.653212514 | 7.179276402 | 0.32718499 | 0.208275942 | 0.021580671 |
| Polo-like kinase mediated events | 2.303416676 | 2.5289167 | 7.135750053 | 0.366043279 | 0.368141322 | 0.039726747 |
| Polymerase switching | 2.373873483 | 0.84509804 | 5.592845007 | -0.420504386 | 0.313724266 | 0.029747342 |
| Polymerase switching on the C-strand of the telomere | 3.424730095 | 1.477121255 | 8.326581445 | 0.309611291 | 0.108625289 | 0.012624016 |
| Positive epigenetic regulation of rRNA expression | 0.300255296 | 0.299177991 | 0.022457444 | |||
| Post-chaperonin tubulin folding pathway | 2.81460137 | 2.920645001 | 8.549847741 | 0.680672557 | 4 | 0.661913456 |
| Post-translational modification: synthesis of GPI-anchored proteins | 1.937327506 | 1.255272505 | 5.129927517 | -0.625480505 | 3.44101844 | 0.393899077 |
| Post-translational protein modification | 0.237399677 | 1.276158208 | ||||
| , RABK1, RABK2, RABK3, RABL, RABM, RABN1, RABN2, RABO, RABQ, RABR, RABS, RABT1, RABT2, RABU, RABV, RABW, RABX2, RABX4, RABX5, RABY, RABZ, RAD-23, RAD18, RAD21, RAD21B, RAD22, RAD23, RAD23A, RAD23AA, RAD23AB, RAD23B, RAD34, RAD4, RAD51, RAD52, RAE-1, RAE1, RAET1G, RAET1L, RAGA, RAN-2, RANBP2, RANGAP, RANGAP1, RANGAP1A, RARA, RARAA, RAS-2, RAS1, RAS2, RASB, RASC, RASD, RASG, RASGRP4, RASS, RBBD, RBBE, RBBP-5, RBBP5, RBBP7, RBX-1, RBX-2, RBX1, RCD5, RCE1, RCE1A, RCG_34378, RCG_52671, RCN1, REC11, RECK, REG, RELA, RENBP, REP, REP-1, REPE, RER2, RET2, RET3, RFL-1, RFT1, RFWD2, RHO-1, RHO1, RHOA, RHOAB, RHOT1, RHOT1A, RHP42, RIB2, RING1, RIPK1, RIPK1L, RIPK2, RKM5, RMT3, RNA1, RNF103, RNF123, RNF128, RNF128A, RNF135, RNF139, RNF144A, RNF144AA, RNF146, RNF152, RNF168, RNF181, RNF185, RNF2, RNF20, RNF38, RNF40, RNF5, RNF7, ROC1A, ROC2, RORA, RORAA, RP11-385D13.1, RPA1, RPD3, RPL37, RPL40, RPN-1, RPN-10, RPN-11, RPN-12, RPN-13, RPN-2, RPN-3, RPN-5, RPN-6.1, RPN-7, RPN-8, RPN-9, RPN1, RPN10, RPN11, RPN12, RPN13, RPN1301, RPN2, RPN3, RPN5, RPN501, RPN502, RPN6, RPN7, RPN8, RPN9, RPS2, RPS27A, RPS3A, RPT-1, RPT-2, RPT-3, RPT-4, RPT-5, RPT-6, RPT1, RPT2, RPT3, RPT4, RPT5, RPT6, RRAGA, RRG1, RRS1, RSMB, RSMM, RSMN, RTF1, RTN4R, RTN4RL1, RTN4RL2, RUB1, RUVB-1, RUVBL1, RWDD3, RXRA, RXRAA, RXRB, RYH1, S1P, SAE1, SAF-B, SAFB, SAFB2, SAMUEL, SAP155, SAP185, SAP190, SAP4, SAR-1, SAR1, SAR1B, SARA, SATB1, SATB1B, SATB2, SBSP-1, SBSPON, SCC-1, SCC-3, SCCRO3, SCCRO4, SCE, SCF, SCFD1, SCG2, SCG2B, SCG3, SCL1, SCM, SCMH1, SCNY, SCOSPONDIN, SDC, SDC2, SDIC1, SDIC2, SDIC3, SDIC4, SDICC, SDN-1, SDS3, SEC-12, SEC-16A.1, SEC-16A.2, SEC-22, SEC-23, SEC-24.1, SEC-24.2, SEC-31, SEC12, SEC13, SEC16, SEC16A, SEC16B, SEC17, SEC18, SEC21, SEC22, SEC22A, SEC22B, SEC22BB, SEC22C, SEC23, SEC231, SEC23A, SEC23IP, SEC24, SEC24A, SEC24AB, SEC24B, SEC24C, SEC24CD, SEC24D, SEC24L, SEC26, SEC27, SEC28, SEC31, SEC31A, SEC4, SEC53, SEC59, SED5, SEDL-1, SEE1, SEH1, SEH1L, SEL-9, SEL1L, SELENOS, SEM1, SEMA5A, SEMA5B, SEMA5BA, SEMA5C, SENP1, SENP2, SENP5, SENP8, SEP1, SERPINA1, SERPINA10, SERPINA10A, SERPINA1C, SERPINA1L, SERPINA3K-PROV, SERPINB14, SERPINB1L1, SERPINB1L3, SERPINB1L4, SERPINC1, SERPIND1, SERPINH2, SEX-1, SF1, SFB2, SFB3, SFC2, SFMBT, SFT1, SHISA5, SHPRH, SI:BUSM1-169I8.11, SI:CABZ01007802.1, SI:CABZ01007807.1, SI:CH1073-205C8.3, SI:CH1073-342H5.2, SI:CH1073-416D2.3, SI:CH211-113A14.11, SI:CH211-113A14.19, SI:CH211-113A14.22, SI:CH211-114C12.5, SI:CH211-114N24.6, SI:CH211-120G10.1, SI:CH211-12P12.2, SI:CH211-14A17.10, SI:CH211-155E24.3, SI:CH211-15E22.3, SI:CH211-175L6.2, SI:CH211-190P8.2, SI:CH211-191D2.2, SI:CH211-196F2.3, SI:CH211-226H7.3, SI:CH211-226H7.5, SI:CH211-226H7.6, SI:CH211-226H7.8, SI:CH211-253P14.2, SI:CH211-262E15.1, SI:CH211-269C21.2, SI:CH211-285C6.5, SI:CH73-100L22.3, SI:CH73-122K23.1, SI:CH73-142C19.1, SI:CH73-144L3.1, SI:CH73-299H12.6, SI:CH73-364H19.2, SI:CH73-373M9.1, SI:CH73-49O8.1, SI:CH73-54F23.4, SI:DKEY-108K21.11, SI:DKEY-119F1.1, SI:DKEY-125E8.3, SI:DKEY-147F3.8, SI:DKEY-156N14.5, SI:DKEY-182I3.10, SI:DKEY-183C6.9, SI:DKEY-18P12.4, SI:DKEY-192D15.1, SI:DKEY-202E17.1, SI:DKEY-20I20.10, SI:DKEY-218F9.10, SI:DKEY-219E21.2, SI:DKEY-219E21.4, SI:DKEY-21C19.3, SI:DKEY-239B22.2, SI:DKEY-23A13.22, SI:DKEY-261M9.19, SI:DKEY-261M9.6, SI:DKEY-37F18.2, SI:DKEY-37M8.11, SI:DKEY-42I9.6, SI:DKEY-82J4.2, SI:DKEYP-110A12.4, SI:RP71-1G18.13, SI:RP71-1K13.6, SI:RP71-45K5.4, SI:RP71-68N21.9, SIAH-1, SIAH2, SIAH2L, SIAL-T2, SIAT, SIAT4B, SIAT8C, SIBA, SIBB, SIBC, SIBD, SIDL, SIMA, SIN3A, SIN3AA, SINA, SIT4, SKI7, SKP1, SKP2, SKPA, SKPT-1, SKR-1, SLC-17.1, SLC17A5, SLC2A15A, SLC35A1, SLC35C1, SLC52A1, SLCO2B1, SLCO5A1, SLH, SLURP1, SLY-1, SLY1, SMA-1, SMA-2, SMAD1, SMAD2, SMAD3, SMAD3A, SMAD4, SMAD4.1, SMAD4A, SMAD7, SMAD9, SMC-3, SMC-5, SMC-6, SMC1, SMC1A, SMC3, SMC5, SMC6, SMO-1, SMOX, SMP3, SMT3, SMURF, SMURF2, SNAP-1, SNPA, SNPC, SNX3, SOCS16D, SOCS2, SOCS3, SOCS36E, SOCS3B, SOCS44A, SOCS5, SOCS5B, SOCS6, SOCS6A, SOCS6B, SOF1, SONA, SP1, SP100, SP100.1, SP100.3, SP100.4, SP3, SP3A, SPAC13C5.01C, SPAC13C5.05C, SPAC13G6.05C, SPAC15A10.12C, SPAC15E1.10, SPAC17A5.08, SPAC1A6.02, SPAC1F5.02, SPAC20G8.02, SPAC22F8.04, SPAC31A2.04C, SPAC3A11.03, SPAC3H8.09C, SPAC4D7.04C, SPAC4F10.18, SPAC6G10.04C, SPAC7D4.09C, SPAC890.06, SPACA4, SPARC, SPARCL1, SPBC1271.04C, SPBC12C2.11, SPBC15C4.03, SPBC16E9.09C, SPBC17D11.08, SPBC211.03C, SPBC4.03C, SPBC577.10, SPBC646.16, SPBPJ4664.04, SPC-1, SPCC1450.15, SPCC18.17C, SPCC285.14, SPH161, SPIA1, SPN100A, SPN27A, SPN28B, SPN28DA, SPN28DB, SPN28DC, SPN28F, SPN31A, SPN38F, SPN42DA, SPN42DB, SPN42DD, SPN42DE, SPN43AA, SPN43AB, SPN43AD, SPN47C, SPN53F, SPN55B, SPN75F, SPN77BA, SPN77BB, SPN77BC, SPN85F, SPN88EA, SPN88EB, SPO11, SPO14, SPON-1, SPON1, SPON1A, SPON2, SPON2B, SPP1, SPP2, SPRN, SPS18, SPSB-1, SPSB-2, SPSB1, SPSB2, SPSB3, SPSB3A, SPSB4, SPSB4A, SPTA1, SPTAN1, SPTB, SPTBN1, SPTBN2, SPTBN4, SPTBN5, SPTF-2, SPTF-3, SRAS, SRD5A3, SRF-3, SRP, SRP-1, SRP-2, SRP-3, SRP-6, SRP-7, SRP-8, SSA1, SSA2, SSA3, SSA4, SSO1, SSO2, SSPO, SSPOP, SST2, ST3GAL1, ST3GAL1L3, ST3GAL2, ST3GAL2.1, ST3GAL3, ST3GAL3A, ST3GAL3B, ST3GAL4, ST3GAL5, ST3GAL6, ST3GAL7, ST6GAL1, ST6GAL2, ST6GAL2B, ST6GALNAC1, ST6GALNAC1.1, ST6GALNAC2, ST6GALNAC3, ST6GALNAC4, ST6GALNAC5, ST6GALNAC5A, ST6GALNAC6, ST8SIA1, ST8SIA2, ST8SIA3, ST8SIA4, ST8SIA5, ST8SIA6, ST8SIA7.1, STAG1, STAG1A, STAG2, STAG2B, STAM, STAM2, STAMBP, STAMBPB, STAMBPL1, STARD8, STC2, STC2A, STE14, STG, STK36, STL, STOPS, STPG4, STS, STT3A, STX17, STX5, STX5A, SU(VAR)2-10, SU(Z)2, SUDS3, SUMF1, SUMF2, SUMO, SUMO1, SUMO2, SUMO3, SUP-17, SUZ12, SUZ12B, SVBP, SW, SWAH-1, SWAN-2, SWD1, SWD3, SWIP-1, SXC, SYN5, SYN7A, SYN8, SYT4, SYVN1, SYX-5, SYX17, SYX5, T23G5.3, TAB1, TADA2B, TADA3, TAF10, TAF10B, TAF9, TAF9B, TAG-225, TAG-30, TAKL1, TANC2, TANGO1, TANGO14, TBA-2, TBA-5, TBA-6, TBA-9, TBB-1, TBB-2, TBB-4, TBB-6, TBB5, TBB6, TBC-20, TBC1D20, TBP1, TCEANC, TCEB1, TCEB2, TCF25, TDG, TDG.2, TECTA, TECTB, TEF1; TEF2, TEF101, TEF102, TEF103, TEP-1, TEP1, TEP2, TEP3, TEP4, TER94, TERB1, TEX101, TF, TFA, TFAP2A, TFAP2B, TFAP2C, TFG, TFG-1, TFPT, TGAS006M08.1, TGFA, TGFB1, TGFBR1, TGFBR1B, TGFBR2, TGOLN1, TGOLN2, TGY, THBS1, THBS1B, THBS2, THBS2A, THD1, THP1, THRA, THRAB, THRB, THSD1, THSD4, THSD7A, THSD7AA, THSD7B, THSD7BA, THY1, TICAM2, TIF51A, TIF51B, TIMP, TIMP-1, TIMP1, TM6SF2, TMED-10, TMED-12, TMED-3, TMED-4, TMED10, TMED2, TMED3, TMED7, TMED9, TMEM115, TMEM129, TMEM132A, TMEM132E, TMEM159, TMEM170B, TMEM19, TMEM208, TMEM26, TMEM97, TMPRSS4, TNC, TNFAIP3, TNIP1, TNIP2, TNIP3, TNKS, TNKS2, TNKSA, TOM20, TOM70, TOMM-20, TOMM-70, TOMM20, TOMM20B, TOMM70, TOMM70A, TOP-1, TOP-2, TOP1, TOP1.2, TOP1L, TOP2, TOP2A, TOP2B, TOP2MT, TOPORS, TOPORSA, TOPORSB, TP53, TP53BP1, TPGS1, TPGS2, TPR, TPRB, TPST1, TPST2, TRAF-LIKE, TRAF2, TRAF2B, TRAF3, TRAF6, TRAPCC10-1; TRAPCC10-2, TRAPPC1, TRAPPC1-1; TRAPPC1-2, TRAPPC10, TRAPPC2, TRAPPC2L, TRAPPC3, TRAPPC4, TRAPPC5, TRAPPC6A, TRAPPC6B, TRAPPC6BL, TRAPPC9, TRBD, TRF-2, TRIM 35-44, TRIM107, TRIM108, TRIM109, TRIM13, TRIM25, TRIM27, TRIM27.1, TRIM27.2, TRIM28, TRIM29, TRIM33L, TRIM35-1, TRIM35-12, TRIM35-13, TRIM35-14, TRIM35-19, TRIM35-20, TRIM35-21, TRIM35-22, TRIM35-23, TRIM35-24, TRIM35-27, TRIM35-28, TRIM35-29, TRIM35-3, TRIM35-31, TRIM35-33, TRIM35-34, TRIM35-39, TRIM35-40, TRIM35-7, TRIM35-9, TRIM39.2, TRIM47, TRIM58, TRIM69, TRIP11, TRP-4, TRPP-1, TRPP-10, TRPP-3, TRPP-4, TRPP-5, TRPP-6, TRPP-9, TRRAP, TRS120, TRS130, TRS20, TRS23, TRS31, TRS33, TSF1, TSF2, TSF3, TSP, TSTA3, TTGN1, TTL, TTLL1, TTLL10, TTLL11, TTLL12, TTLL13, TTLL13P, TTLL2, TTLL3, TTLL4, TTLL5, TTLL6, TTLL7, TTLL8, TTLL9, TUB-2, TUBA, TUBA1A, TUBA1B, TUBA1C, TUBA2, TUBA3A; TUBA3B, TUBA3C, TUBA3D, TUBA3E, TUBA4A, TUBA4B, TUBA4L, TUBA8, TUBA8A, TUBA8L3, TUBA8L4, TUBA8L5, TUBAL3, TUBAL3.1, TUBAL3.2, TUBB1, TUBB2, TUBB2A, TUBB2B, TUBB3, TUBB4A, TUBB4B, TUBB4BL, TUBB6, TUBB8, TUBB8B, TUBGCP5, TULP4, TULP4A, TULP4B, TUSC3, TWE, UAP1, UBA-1, UBA-2, UBA1, UBA2, UBA3, UBA5, UBA52, UBA6, UBB, UBC, UBC-1, UBC-14, UBC-16, UBC-18, UBC-20, UBC-25, UBC-3, UBC-7, UBC-8, UBC-9, UBC10, UBC12, UBC2, UBC4, UBC6, UBC7, UBC9, UBCB, UBCC, UBCE2H, UBCE2M, UBD, UBE2A, UBE2B, UBE2C, UBE2D1, UBE2D1A, UBE2D1B, UBE2D2, UBE2D3, UBE2E1, UBE2E2, UBE2E3, UBE2F, UBE2G1, UBE2G1A, UBE2G2, UBE2H, UBE2I, UBE2IA, UBE2J2, UBE2K, UBE2KA, UBE2L3, UBE2L3A, UBE2L3B, UBE2M, UBE2N, UBE2NA, UBE2NB, UBE2Q2, UBE2Q2L, UBE2R2, UBE2S, UBE2T, UBE2V, UBE2V2, UBE2W, UBE2WB, UBE2Z, UBH-1, UBH-2, UBH-3, UBH-4, UBI-P5E, UBI-P63E, UBL1, UBP6, UBPA, UBQ-1, UBQB, UBQC, UBQD, UBQF, UBQG, UBQH, UBQI, UBQJ, UBQO, UBXD7, UBXN-1, UBXN1, UBXN2A, UBXN7, UCH, UCH-L1, UCH-L5, UCH1, UCH2, UCH54, UCHL1, UCHL3, UCHL5, UEV-2, UEV-3, UFD-1, UFD-3, UFD1, UFD1-LIKE, UFD1L, UFO1, UGALT, UGGT1, UGGT2, UGTP-1, UHRF2, UIMC1, ULA-1, ULA1, ULBP1, ULBP2, ULK3, ULP-1, ULP-3, ULP-5, ULP1, UMOD, UMOD.1, UMODL1, UNC-108, UNC-55, UNC-70, UPK2, US11, USO-1, USO1, USP-14, USP-3, USP-46, USP-48, USP-5, USP10, USP11, USP12, USP12-46, USP12A, USP13, USP14, USP15, USP16, USP17L1, USP17L10, USP17L11, USP17L12, USP17L13, USP17L15, USP17L17, USP17L18, USP17L19, USP17L2, USP17L20, USP17L21, USP17L22, USP17L24; USP17L25; USP17L26; USP17L27; USP17L28; USP17L29; USP17L30, USP17L3, USP17L4, USP17L5, USP17L8, USP17LA, USP17LB, USP17LC, USP17LD, USP17LE, USP18, USP19, USP2, USP20, USP20-33, USP21, USP22, USP24, USP25, USP26, USP28, USP2A, USP3, USP30, USP33, USP34, USP37, USP4, USP41, USP42, USP44, USP47, USP48, USP49, USP5, USP7, USP8, USP9, USP9X, USP9Y, V9GZ90, VASH1, VASH2, VCAN, VCANA, VCP, VCPIP1, VCPKMT, VDAC-1, VDAC1, VDAC2, VDAC3, VDAC5P, VDR, VDRA, VGF, VHL, VHL-1, VIH, VIP, VLET, VNN1, VNN2, VNN3, VPS21, VSIG10, VWA1, W03G11.3, WAC, WACA, WAP, WBSCR17, WCY, WDR-12, WDR-20, WDR-23, WDR-48, WDR-5.1, WDR12, WDR20, WDR20A, WDR21, WDR48, WDR5, WDR5B, WDR61, WDR68, WDS, WDSOF1, WDTC1, WFS1, WFS1B, WIMA, WIPI2, WNT2B, WOL, WRN, WSB1, WSB2, WU:FB30F12, XB22169562, XB5756113, XB5813854 [PROVISIONAL:ANKRD52], XB5820455 [PROVISIONAL:MR1], XB5884180, XB5894692 [PROVISIONAL:AZGP1], XB5946283 [PROVISIONAL:FCRL5], XB5960289 [PROVISIONAL:FCRL4], XB995277 [PROVISIONAL:ADAMTSL5], XIT, XPC, XPC-1, XPNPEP1, XPNPEP2, XPO4, XPO6, XRCC4, YAK1, YAKA, YF5, YJL132W, YKT-6, YKT6, YLR352W, YMR085W, YNG1, YNG2, YOD1, YOR022C, YPT1, YPT10, YPT11, YPT2, YPT3, YPT31, YPT32, YPT4, YPT5, YPT52, YPT53, YPT6, YPT7, YPT71, YRB1, YRB2, YTM1, YUH1, YY1, YY1A, YY2, ZBED1, ZBTB1, ZBTB16, ZBTB16B, ZBTB5, ZDA, ZETACOP, ZFP131, ZG16, ZGC:100832, ZGC:112234;, ZGC:114081, ZGC:114174, ZGC:136767, ZGC:136929, ZGC:153151, ZGC:153722, ZGC:153932, ZGC:158376, ZGC:161973, ZGC:162608, ZGC:171927, ZGC:173570, ZGC:173585, ZGC:173729, ZGC:174574, ZGC:194252, ZGC:194990, ZGC:55461, ZGC:55733, ZGC:56525, ZGC:65894, ZGC:77287, ZGC:92594, ZGC:92907, ZK632.4, ZLD, ZMP:0000001316, ZNF1007, ZNF1013, ZNF1132, ZNF131, ZNF350, ZNF615, ZNFL2A, ZRANB1, ZRANB1B | 0.075739887 | |||||
| Post-translational protein phosphorylation | 2.505602132 | 2.998259338 | 8.009463603 | 0.250597042 | 0.253330929 | 0.026536271 |
| Post NMDA receptor activation events | 0.541701252 | 4 | 0.541701252 | |||
| Postmitotic nuclear pore complex (NPC) reformation | 2.24814329 | 2.146128036 | 6.642414615 | 0.454495614 | 0.321533205 | 0.036206416 |
| Postsynaptic nicotinic acetylcholine receptors | 0.305112699 | 0.272173256 | 0.020760879 | |||
| Potassium Channels | 0.206428369 | 0.137706549 | 0.007106635 | |||
| Potassium transport channels | 2.180029537 | 0.954242509 | 5.314301583 | 0.457514564 | 0.090122664 | 0.008638603 |
| Potential therapeutics for SARS | 2.638709486 | 3.354684554 | 8.632103526 | 0.442709687 | 3.890270849 | 0.532165251 |
| POU5F1 (OCT4), SOX2, NANOG activate genes related to proliferation | 2.26718338 | 1.832508913 | 6.366875673 | 0.365368798 | 0.216342565 | 0.021182311 |
| POU5F1 (OCT4), SOX2, NANOG repress genes related to differentiation | 1.535279459 | 0 | 3.070558917 | -0.826575342 | 1.15183501 | 0.130212917 |
| PP2A-mediated dephosphorylation of key metabolic factors | 3.138945485 | 0.698970004 | 6.976860975 | 0.441790812 | 0.171375746 | 0.019764916 |
| PPARA activates gene expression | 2.386556087 | 2.989004616 | 7.76211679 | 0.227196992 | 0.207270863 | 0.020444022 |
| PRC2 methylates histones and DNA | 2.232417274 | 1.707570176 | 6.172404725 | 0.360341337 | 0.386120101 | 0.036594297 |
| Pre-NOTCH Expression and Processing | 0.337579959 | 0.750512854 | 0.063339525 | |||
| Pre-NOTCH Processing in Golgi | 3.17269982 | 2.238046103 | 8.583445743 | 0.394922516 | 0.538479842 | 0.070106269 |
| Pre-NOTCH Processing in the Endoplasmic Reticulum | 3.401746715 | 1.414973348 | 8.218466778 | 0.646268112 | 1.116813782 | 0.175232429 |
| Pre-NOTCH Transcription and Translation | 2.765538631 | 2.907411361 | 8.438488623 | 0.428997679 | 1.400674296 | 0.185705283 |
| Prefoldin mediated transfer of substrate to CCT/TriC | 2.860316277 | 2.854306042 | 8.574938596 | 0.590576106 | 3.058085895 | 0.472596324 |
| Pregnenolone biosynthesis | 2.612218147 | 2.472756449 | 7.697192743 | 0.372987081 | 0.13050546 | 0.01514134 |
| Presynaptic depolarization and calcium channel opening | 3.915403194 | 2.677606953 | 10.50841334 | 0.960989011 | 4 | 0.903112435 |
| Presynaptic function of Kainate receptors | 2.164980579 | 0.698970004 | 5.028931163 | -0.449410474 | 0.598020014 | 0.054515697 |
| Presynaptic nicotinic acetylcholine receptors | 0.304945055 | 0.231695655 | 0.017663611 | |||
| Presynaptic phase of homologous DNA pairing and strand exchange | 2.730948796 | 2.462397998 | 7.92429559 | 0.29436143 | 0.234828079 | 0.025624803 |
| Prevention of phagosomal-lysosomal fusion | 2.732764413 | 1.278753601 | 6.744282427 | 0.706977102 | 0.716143688 | 0.104184249 |
| Processing and activation of SUMO | -0.508781559 | 1.828553321 | 0.232583552 | |||
| Processing of Capped Intron-Containing Pre-mRNA | 2.802802272 | 0.301029996 | 5.90663454 | 0.186429903 | 0.046958029 | 0.003268643 |
| Processing of DNA double-strand break ends | 2.486928036 | 2.909556029 | 7.883412102 | 0.32214379 | 0.662403088 | 0.074233563 |
| Processing of SMDT1 | 2.716300782 | 0 | 5.432601564 | -0.747945205 | 0.86719543 | 0.115929649 |
| Processive synthesis on the C-strand of the telomere | 3.071661216 | 2.152288344 | 8.295610776 | 0.389068675 | 0.485090184 | 0.060999562 |
| Processive synthesis on the lagging strand | 2.386066301 | 0.903089987 | 5.67522259 | -0.331961591 | 0.386112802 | 0.03274801 |
| Programmed Cell Death | 0.510967744 | 4 | 0.510967744 | |||
| Progressive trimming of alpha-1,2-linked mannose residues from Man9/8/7GlcNAc2 to produce Man5GlcNAc2 | 1.154476434 | 0 | 2.308952867 | 0.07996035 | ||
| Prolactin receptor signaling | 2.506873813 | 2.836324116 | 7.850071742 | 0.21131868 | 0.013137556 | 0.001284642 |
| Prolonged ERK activation events | 0.716216581 | 3.864118488 | 0.691886433 | |||
| Prostacyclin signalling through prostacyclin receptor | 2.265879315 | 2.416640507 | 6.948399137 | 0.371162281 | 0.126658717 | 0.013442952 |
| Prostanoid ligand receptors | 2.154433008 | 2.269512944 | 6.578378959 | 0.449712847 | 0.745825585 | 0.082922228 |
| Protein-protein interactions at synapses | 0.397508124 | 0.891532773 | 0.08859788 | |||
| Protein folding | 0.496216125 | 4 | 0.496216125 | |||
| Protein localization | 0.095467688 | 0.000436258 | 1.04E-05 | |||
| Protein methylation | 2.530067496 | 2.045322979 | 7.105457971 | 0.209935617 | 0.029334873 | 0.00258169 |
| Protein repair | 2.313962689 | 0.301029996 | 4.928955373 | -0.56058114 | 0.794541667 | 0.082447395 |
| Protein ubiquitination | -0.150769924 | 0.141329153 | 0.005327046 | |||
| Proton-coupled monocarboxylate transport | 2.223210437 | 1.113943352 | 5.560364226 | -0.425164474 | 0.328701422 | 0.03122125 |
| Proton/oligopeptide cotransporters | 2.088313336 | 1.322219295 | 5.498845966 | -0.61890411 | 0.459347133 | 0.054390216 |
| PTEN Loss of Function in Cancer | 2.694705717 | 0 | 5.389411434 | 0.350947497 | ||
| PTEN Regulation | 0.639682814 | 4 | 0.639682814 | |||
| PTK6 Activates STAT3 | 2.435468835 | 2.064457989 | 6.93539566 | 0.603561644 | 0.378390885 | 0.05108937 |
| PTK6 Down-Regulation | 2.304018257 | 2.718501689 | 7.326538202 | 0.538777747 | 0.437283016 | 0.057705675 |
| PTK6 Expression | 2.462416121 | 2.646403726 | 7.571235968 | 0.684484649 | 1.403120521 | 0.21514529 |
| PTK6 promotes HIF1A stabilization | 2.554265987 | 3.195346058 | 8.303878032 | 0.64707694 | 1.405164524 | 0.222165742 |
| PTK6 Regulates Cell Cycle | 2.011266502 | 2.808885867 | 6.831418872 | 0.660205896 | 1.732272575 | 0.243829415 |
| PTK6 Regulates Proteins Involved in RNA Processing | 2.101069672 | 1.698970004 | 5.901109349 | 0.717611613 | 0.27866862 | 0.037880596 |
| PTK6 Regulates RHO GTPases, RAS GTPase and MAP kinases | 2.581244759 | 1.875061263 | 7.037550781 | 0.421154467 | 0.556605251 | 0.06319366 |
| PTK6 Regulates RTKs and Their Effectors AKT1 and DOK1 | 2.864425559 | 2.92890769 | 8.657758809 | 0.718004933 | 1.1557155 | 0.19824759 |
| Purine catabolism | 1.830359013 | 1.568201724 | 5.228919751 | -0.550975543 | 3.617742402 | 0.385055377 |
| Purine ribonucleoside monophosphate biosynthesis | 2.669860796 | 1.278753601 | 6.618475193 | 0.267188003 | 0.077670655 | 0.006903626 |
| Purine salvage | 2.006161007 | 1.785329835 | 5.79765185 | -0.232903049 | 0.309147886 | 0.022880439 |
| Purinergic signaling in leishmaniasis infection | 2.432873731 | 2.716837723 | 7.582585185 | 0.36445318 | 0.62268746 | 0.070660005 |
| Pyrimidine biosynthesis | 1.933582304 | 2.328379603 | 6.195544212 | 0.571828236 | 0.53376068 | 0.06485656 |
| Pyrimidine catabolism | 1.79709239 | 0.698970004 | 4.293154784 | -0.63987932 | 1.773546287 | 0.18707261 |
| Pyrimidine salvage | 2.045894688 | 1.653212514 | 5.745001891 | -0.232711306 | 0.115795357 | 0.008488761 |
| Pyruvate metabolism | 2.283342864 | 1.633468456 | 6.200154183 | 0.278933036 | 0.21387982 | 0.018170426 |
| Pyruvate metabolism and Citric Acid (TCA) cycle | 0.184499279 | 0.053418075 | 0.002463899 | |||
| RA biosynthesis pathway | 2.047336466 | 2.63748973 | 6.732162662 | -0.170282889 | 0.024823584 | 0.001942109 |
| RAB GEFs exchange GTP for GDP on RABs | 2.75738593 | 3.070407322 | 8.585179181 | 0.531310401 | 1.663242118 | 0.244935312 |
| RAB geranylgeranylation | 2.661436944 | 1.924279286 | 7.247153175 | 0.305659135 | 0.178814117 | 0.018203359 |
| Rab regulation of trafficking | 0.473436236 | 1.34673452 | 0.15939823 | |||
| RAF-independent MAPK1/3 activation | 2.317923269 | 3.028571253 | 7.66441779 | 0.414409894 | 1.143841066 | 0.138148417 |
| RAF activation | 2.714736487 | 3.497206181 | 8.926679155 | 0.53059419 | 3.610624996 | 0.547293742 |
| RAF/MAP kinase cascade | 2.643739586 | 4.07685881 | 9.364337983 | 0.496956535 | 4 | 0.612074312 |
| Rap1 signalling | 2.986703417 | 2.888740961 | 8.862147796 | 0.603085206 | 2.499061316 | 0.399369415 |
| Ras activation upon Ca2+ influx through NMDA receptor | 2.377980909 | 2.92992956 | 7.685891377 | 0.536935885 | 1.930258469 | 0.263226615 |
| RAS GTPase cycle mutants | 2.969250815 | 1.342422681 | 7.280924311 | 0.619612313 | 0.743535052 | 0.105195458 |
| RAS processing | 2.929302242 | 3.754654069 | 9.613258553 | 0.37216263 | 0.681522425 | 0.095842322 |
| RAS signaling downstream of NF1 loss-of-function variants | 2.87555 | 1.146128036 | 6.897228036 | 0.631286299 | 1.035143736 | 0.142840278 |
| Reactions specific to the complex N-glycan synthesis pathway | 2.336066896 | 2 | 6.672133793 | 0.345176997 | 0.176351082 | 0.017515895 |
| Reactions specific to the hybrid N-glycan synthesis pathway | 1.774293098 | 0 | 3.548586196 | 0.189010574 | ||
| Receptor-type tyrosine-protein phosphatases | 2.493136221 | 1.819543936 | 6.805816379 | 0.483146067 | 0.258317527 | 0.030557485 |
| Receptor Mediated Mitophagy | 2.393741484 | 3.164650216 | 7.952133184 | 0.782605706 | 3.099591804 | 0.528514838 |
| Recognition and association of DNA glycosylase with site containing an affected purine | 3.937025917 | 1.716003344 | 9.590055178 | 0.484716486 | 0.521777871 | 0.080562353 |
| Recognition and association of DNA glycosylase with site containing an affected pyrimidine | 4.182684107 | 1.397940009 | 9.763308222 | 0.549246385 | 0.843447774 | 0.138916223 |
| Recognition of DNA damage by PCNA-containing replication complex | 3.227280542 | 2.139879086 | 8.59444017 | 0.341377347 | 0.278901681 | 0.034483846 |
| Recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double strand breaks | 2.413066509 | 3.179838928 | 8.005971946 | 0.245145201 | 0.166091712 | 0.017277345 |
| Recruitment of mitotic centrosome proteins and complexes | 2.522937784 | 3.312600439 | 8.358476007 | 0.433052919 | 1.568652939 | 0.20715275 |
| Recruitment of NuMA to mitotic centrosomes | 2.607092783 | 3.440121603 | 8.654307169 | 0.514564994 | 3.993284469 | 0.583267763 |
| Recycling of bile acids and salts | 2.746028891 | 3.081347308 | 8.57340509 | 0.439551613 | 1.030099451 | 0.139724824 |
| Recycling of eIF2:GDP | 1.76519832 | 0 | 3.530396641 | 0.187410444 | ||
| Recycling pathway of L1 | 2.611857612 | 3.571941635 | 8.795656859 | 0.499824058 | 3.89977452 | 0.569533105 |
| Reduction of cytosolic Ca++ levels | 3.389803307 | 1.672097858 | 8.451704472 | 0.417570473 | 0.223185923 | 0.029309851 |
| Reelin signalling pathway | 2.103010259 | 2.243038049 | 6.449058567 | 0.725006847 | 0.288465152 | 0.041517538 |
| Regorafenib-resistant KIT mutants | 2.617771423 | 2.158362492 | 7.393905339 | 0.918104629 | 0.749379508 | 0.135074794 |
| Regorafenib-resistant PDGFR mutants | 2.318185123 | 2.017033339 | 6.653403585 | 0.882497946 | 0.618515789 | 0.102826733 |
| Regulated Necrosis | 0.37053296 | 0.718422818 | 0.066549833 | |||
| Regulated proteolysis of p75NTR | 2.518909588 | 3.196176185 | 8.233995362 | 0.669199162 | 3.444041062 | 0.550945143 |
| Regulation by c-FLIP | 2.670496925 | 1.681241237 | 7.022235087 | -0.325192097 | 0.433215769 | 0.043902625 |
| Regulation of actin dynamics for phagocytic cup formation | 2.944104158 | 3.350441857 | 9.238650174 | 0.349095522 | 0.863677893 | 0.114795677 |
| Regulation of activated PAK-2p34 by proteasome mediated degradation | 3.347523725 | 2.820857989 | 9.515905439 | 0.798814684 | 4 | 0.770822624 |
| Regulation of APC/C activators between G1/S and early anaphase | 2.430042507 | 2.895422546 | 7.755507561 | 0.770158572 | 4 | 0.665677122 |
| Regulation of Apoptosis | 2.361545896 | 0 | 4.723091791 | 0.779411411 | 4 | 0.513863762 |
| Regulation of beta-cell development | 0.311368087 | 0.419356509 | 0.032643559 | |||
| Regulation of cholesterol biosynthesis by SREBP (SREBF) | 2.368164749 | 1.963787827 | 6.700117325 | 0.314682581 | 0.687643729 | 0.065926541 |
| Regulation of commissural axon pathfinding by SLIT and ROBO | 2.594661744 | 2.51851394 | 7.707837427 | 0.96850178 | 1.106360909 | 0.210869475 |
| Regulation of Complement cascade | 2.349950504 | 2.914871818 | 7.614772825 | 0.2621604 | 0.125734002 | 0.012712262 |
| Regulation of cytoskeletal remodeling and cell spreading by IPP complex components | 1.713832099 | 0.954242509 | 4.381906707 | 0.494340329 | 0.134634796 | 0.011905983 |
| Regulation of expression of SLITs and ROBOs | 4.071040669 | 3.158060794 | 11.30014213 | 0.825543194 | 4 | 0.876234149 |
| Regulation of FOXO transcriptional activity by acetylation | 2.394224756 | 2.086359831 | 6.874809342 | -0.292261251 | 0.279987393 | 0.026689377 |
| Regulation of FZD by ubiquitination | 2.373276439 | 1.113943352 | 5.860496229 | 0.366173878 | 0.119065963 | 0.010892236 |
| Regulation of gap junction activity | 2.851245081 | 2.994756945 | 8.697247107 | 0.96850178 | 1.106360909 | 0.224987418 |
| Regulation of gene expression by Hypoxia-inducible Factor | 2.62671036 | 2.550228353 | 7.803649072 | 0.453619741 | 0.699474705 | 0.089164014 |
| Regulation of gene expression in beta cells | 2.759251477 | 1.278753601 | 6.797256555 | 0.366075495 | 0.450278257 | 0.046626338 |
| Regulation of gene expression in early pancreatic precursor cells | -0.570684932 | 0.345152671 | 0.049243357 | |||
| Regulation of gene expression in late stage (branching morphogenesis) pancreatic bud precursor cells | 3.033402138 | 2.187520721 | 8.254324997 | 0.347190568 | 0.265686271 | 0.031877483 |
| Regulation of Glucokinase by Glucokinase Regulatory Protein | 2.586681436 | 1.447158031 | 6.620520903 | 0.572211565 | 0.535926741 | 0.068082875 |
| Regulation of glycolysis by fructose 2,6-bisphosphate metabolism | 2.651857573 | 2.276461804 | 7.580176951 | 0.435055147 | 0.691629641 | 0.0845656 |
| Regulation of HSF1-mediated heat shock response | 2.484676325 | 3.236033147 | 8.205385796 | 0.327534489 | 0.452603512 | 0.0529064 |
| Regulation of IFNA signaling | 2.501476197 | 3.025305865 | 8.02825826 | 0.372663293 | 0.359815307 | 0.043267832 |
| Regulation of IFNG signaling | 2.554381592 | 3.0923697 | 8.201132883 | 0.392948765 | 0.495776352 | 0.061979686 |
| Regulation of innate immune responses to cytosolic DNA | 2.710667428 | 2.303196057 | 7.724530913 | 0.71129249 | 1.112680867 | 0.176539621 |
| Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) | 2.515492473 | 3.2219356 | 8.252920546 | 0.271925584 | 0.481051548 | 0.053182864 |
| Regulation of insulin secretion | 3.031855745 | 3.209783015 | 9.273494504 | 0.372639909 | 2.237609442 | 0.305002647 |
| Regulation of KIT signaling | 2.799740037 | 3.368286885 | 8.967766958 | 0.536469615 | 1.829441612 | 0.279617383 |
| Regulation of lipid metabolism by PPARalpha | 2.362699912 | 2.856124444 | 7.581524269 | 0.220323068 | 0.181791319 | 0.017351241 |
| Regulation of localization of FOXO transcription factors | 3.010521341 | 3.035029282 | 9.056071964 | 0.526895015 | 1.404344921 | 0.214562968 |
| Regulation of MECP2 expression and activity | 2.407565869 | 3.176669933 | 7.99180167 | 0.561730641 | 3.082143581 | 0.442020513 |
| Regulation of mitotic cell cycle | 0.723065546 | 4 | 0.723065546 | |||
| Regulation of mRNA stability by proteins that bind AU-rich elements | 0.72615994 | 4 | 0.72615994 | |||
| Regulation of necroptotic cell death | 2.98873014 | 2.525044807 | 8.502505088 | 0.37053296 | 0.718422818 | 0.090593349 |
| Regulation of ornithine decarboxylase (ODC) | 3.173743476 | 2.914871818 | 9.262358769 | 0.782192363 | 4 | 0.749431204 |
| Regulation of PAK-2p34 activity by PS-GAP/RHG10 | 2.633619552 | 1.602059991 | 6.869299095 | 0.782799233 | 0.37598244 | 0.058867449 |
| Regulation of PLK1 Activity at G2/M Transition | 2.648390876 | 3.409256652 | 8.706038403 | 0.469039544 | 2.449133601 | 0.345422554 |
| Regulation of PTEN gene transcription | 2.799032888 | 3.594060901 | 9.192126678 | 0.458697674 | 2.459121392 | 0.359069006 |
| Regulation of PTEN localization | 3.032442635 | 2.158362492 | 8.223247761 | 0.560083657 | 0.914254984 | 0.133657219 |
| Regulation of PTEN mRNA translation | 2.694705717 | 0 | 5.389411434 | 0.350947497 | ||
| Regulation of PTEN stability and activity | 3.078056082 | 3.458788882 | 9.614901046 | 0.743819234 | 4 | 0.748431999 |
| Regulation of pyruvate dehydrogenase (PDH) complex | 2.151849036 | 2.591064607 | 6.89476268 | 0.358300405 | 0.334888263 | 0.034773368 |
| Regulation of RAS by GAPs | 3.457400834 | 2.909556029 | 9.824357697 | 0.731662049 | 4 | 0.753159099 |
| Regulation of RUNX1 Expression and Activity | 2.442392 | 2.868056362 | 7.752840363 | 0.62129528 | 1.978292507 | 0.292346071 |
| Regulation of RUNX2 expression and activity | 3.269781455 | 3.41564098 | 9.955203889 | 0.712092304 | 4 | 0.750124471 |
| Regulation of RUNX3 expression and activity | 3.256568676 | 3.433769834 | 9.946907186 | 0.767288991 | 4 | 0.777294795 |
| Regulation of signaling by CBL | 2.986038142 | 3.73183042 | 9.703906703 | 0.643671507 | 3.812911898 | 0.670071473 |
| Regulation of signaling by NODAL | 2.58094946 | 1.716003344 | 6.877902263 | 0.536547859 | 0.608400169 | 0.076597102 |
| Regulation of the apoptosome activity | 2.520787843 | 3.089905111 | 8.131480797 | 0.471749863 | 0.61901022 | 0.082927083 |
| Regulation of thyroid hormone activity | 1.801655604 | 0 | 3.603311208 | 0.19382472 | ||
| Regulation of TLR by endogenous ligand | 2.928992496 | 2.276461804 | 8.134446796 | 0.350723711 | 0.392628948 | 0.046674635 |
| Regulation of TNFR1 signaling | 2.693936504 | 2.892094603 | 8.27996761 | 0.366609514 | 0.676809623 | 0.083071452 |
| Regulation of TP53 Activity | 0.459221061 | 4 | 0.459221061 | |||
| Regulation of TP53 Activity through Acetylation | 2.54073861 | 3.375114685 | 8.456591905 | 0.424577357 | 1.439558243 | 0.190401323 |
| Regulation of TP53 Activity through Association with Co-factors | 2.412488388 | 3.191451014 | 8.016427791 | 0.787828947 | 2.233938698 | 0.384222357 |
| Regulation of TP53 Activity through Methylation | 2.666782449 | 3.279438788 | 8.613003686 | 0.465985525 | 1.235242507 | 0.172263246 |
| Regulation of TP53 Activity through Phosphorylation | 2.474195322 | 3.783117137 | 8.731507782 | 0.518590385 | 4 | 0.590244059 |
| Regulation of TP53 Degradation | 2.582675489 | 3.648750213 | 8.814101192 | 0.557317258 | 3.508277071 | 0.538405125 |
| Regulation of TP53 Expression | 1.881688969 | 2.687528961 | 6.450906899 | 0.870172555 | 0.574881992 | 0.09318562 |
| Regulation of TP53 Expression and Degradation | 0.557317258 | 3.508277071 | 0.488805839 | |||
| Relaxin receptors | 2.785432703 | 1.342422681 | 6.913288087 | -0.467653509 | 0.451107822 | 0.053115149 |
| Release | 2.943361751 | 1.51851394 | 7.405237442 | 0.528279191 | ||
| Release of apoptotic factors from the mitochondria | 3.400726665 | 1.146128036 | 7.947581366 | 0.476020828 | 0.247007136 | 0.032636878 |
| Release of Hh-Np from the secreting cell | 2.351318903 | 1.832508913 | 6.535146719 | 0.440404079 | 0.16857082 | 0.018451867 |
| Removal of aminoterminal propeptides from gamma-carboxylated proteins | 2.723909021 | 3.114944416 | 8.562762458 | 0.751370963 | 2.676758572 | 0.467046804 |
| Removal of the Flap Intermediate | 2.403763111 | 1.908485019 | 6.716011242 | -0.331870543 | 0.256320261 | 0.025177466 |
| Removal of the Flap Intermediate from the C-strand | 3.088190964 | 2.071882007 | 8.248263936 | 0.44610539 | 0.579369687 | 0.076631985 |
| Replication of the SARS-CoV-1 genome | 3.559632631 | 2.674861141 | 9.794126403 | 0.558275836 | 0.676121793 | 0.112389424 |
| Replication of the SARS-CoV-2 genome | 0.558275836 | 0.676121793 | 0.094365615 | |||
| Repression of WNT target genes | 2.636152416 | 2.771587481 | 8.043892312 | 0.610204142 | 1.243313704 | 0.186676538 |
| Reproduction | 0.309822712 | 0.424917717 | 0.03291229 | |||
| Resistance of ERBB2 KD mutants to AEE788 | 3.346562468 | 2.812244697 | 9.505369633 | 0.708169711 | 1.096594048 | 0.198745774 |
| Resistance of ERBB2 KD mutants to afatinib | 3.346562468 | 2.812244697 | 9.505369633 | 0.708169711 | 1.096594048 | 0.198745774 |
| Resistance of ERBB2 KD mutants to lapatinib | 3.346562468 | 2.812244697 | 9.505369633 | 0.708169711 | 1.096594048 | 0.198745774 |
| Resistance of ERBB2 KD mutants to neratinib | 3.346562468 | 2.812244697 | 9.505369633 | 0.708169711 | 1.096594048 | 0.198745774 |
| Resistance of ERBB2 KD mutants to osimertinib | 3.346562468 | 2.812244697 | 9.505369633 | 0.708169711 | 1.096594048 | 0.198745774 |
| Resistance of ERBB2 KD mutants to sapitinib | 3.346562468 | 2.812244697 | 9.505369633 | 0.708169711 | 1.096594048 | 0.198745774 |
| Resistance of ERBB2 KD mutants to tesevatinib | 3.346562468 | 2.812244697 | 9.505369633 | 0.708169711 | 1.096594048 | 0.198745774 |
| Resistance of ERBB2 KD mutants to trastuzumab | 3.346562468 | 2.812244697 | 9.505369633 | 0.708169711 | 1.096594048 | 0.198745774 |
| Resolution of Abasic Sites (AP sites) | 2.294566098 | 2.243038049 | 6.832170246 | 0.183888032 | 0.015917071 | 0.001292895 |
| Resolution of AP sites via the multiple-nucleotide patch replacement pathway | 2.720154873 | 2.507855872 | 7.948165619 | 0.276177902 | 0.139762513 | 0.014976457 |
| Resolution of AP sites via the single-nucleotide replacement pathway | 2.862017798 | 2.170261715 | 7.894297312 | 0.671506849 | 0.606999218 | 0.094617719 |
| Resolution of D-Loop Structures | 0.302414929 | 0.137043508 | 0.010361001 | |||
| Resolution of D-loop Structures through Holliday Junction Intermediates | 2.659464602 | 2.238046103 | 7.556975308 | 0.302414929 | 0.137043508 | 0.014443124 |
| Resolution of D-loop Structures through Synthesis-Dependent Strand Annealing (SDSA) | 2.481364181 | 2.238046103 | 7.200774464 | 0.302414929 | 0.137043508 | 0.013813543 |
| Resolution of Sister Chromatid Cohesion | 2.880735599 | 3.322839273 | 9.08431047 | 0.530280801 | 4 | 0.614290061 |
| Respiratory electron transport | 2.124252536 | 1.477121255 | 5.725626327 | 0.19057709 | 0.01325753 | 0.00089875 |
| Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. | -0.181522766 | 0.039193721 | 0.001778638 | |||
| Response of EIF2AK1 (HRI) to heme deficiency | 1.905793828 | 1.255272505 | 5.066860161 | -0.308472717 | 0.134514121 | 0.009958388 |
| Response of EIF2AK4 (GCN2) to amino acid deficiency | 4.314707627 | 2.902002891 | 11.53141814 | 0.884185795 | 4 | 0.917486785 |
| Response of Mtb to phagocytosis | 0.398619485 | 0.552213619 | 0.055030777 | |||
| Response to elevated platelet cytosolic Ca2+ | 2.441101507 | 2.926342447 | 7.80854546 | 0.294389302 | 0.770372634 | 0.082916876 |
| RET signaling | 2.778461438 | 3.685204134 | 9.242127011 | 0.523266427 | 2.848724854 | 0.440786399 |
| Retinoid cycle disease events | 1.837511746 | 1.113943352 | 4.788966844 | -0.284813596 | 0.037301411 | 0.002517503 |
| Retinoid metabolism and transport | 2.319962484 | 1.812913357 | 6.452838324 | 0.240719705 | 0.090829323 | 0.007578664 |
| Retinoid metabolism disease events | 2.536604392 | 0.903089987 | 5.976298771 | 0.715420433 | 0.27866862 | 0.038074505 |
| Retrograde neurotrophin signalling | 2.077283885 | 2.559906625 | 6.714474395 | 0.402555586 | 0.312611868 | 0.033462725 |
| Retrograde transport at the Trans-Golgi-Network | 2.195868649 | 1.919078092 | 6.310815389 | -0.312287238 | 0.694962655 | 0.06293078 |
| Reuptake of GABA | 2.590533606 | 2.264817823 | 7.445885036 | 0.558114035 | 0.676121793 | 0.091898759 |
| Rev-mediated nuclear export of HIV RNA | 2.078945785 | 1.643452676 | 5.801344246 | 0.542000762 | 0.447790653 | 0.05046428 |
| Reversal of alkylation damage by DNA dioxygenases | 1.901906303 | 0 | 3.803812605 | -0.632776103 | 0.78230721 | 0.076885401 |
| Reverse Transcription of HIV RNA | 0.97863599 | 1.277057136 | 0.312443519 | |||
| Reversible hydration of carbon dioxide | 2.079183379 | 2.738780558 | 6.897147316 | 0.505342521 | 1.526243523 | 0.186578315 |
| Rho GTPase cycle | 3.356599082 | 2.053078443 | 8.766276608 | 0.28497694 | 0.239687084 | 0.028476685 |
| RHO GTPase Effectors | 0.382631275 | 4 | 0.382631275 | |||
| RHO GTPases activate CIT | 2.476483395 | 1.740362689 | 6.69332948 | -0.310982487 | 0.677353978 | 0.064567448 |
| RHO GTPases Activate Formins | 2.931997227 | 3.298853076 | 9.16284753 | 0.422328608 | 2.822962805 | 0.398295792 |
| RHO GTPases activate IQGAPs | 2.808457799 | 3.269512944 | 8.886428542 | 0.534317647 | 3.266166498 | 0.494905814 |
| RHO GTPases activate KTN1 | 3.03056692 | 1.414973348 | 7.476107188 | -0.251782776 | 0.04692182 | 0.00459922 |
| RHO GTPases Activate NADPH Oxidases | 2.532583277 | 3.568553712 | 8.633720266 | 0.423203679 | 1.132426758 | 0.152171524 |
| RHO GTPases activate PAKs | 2.548851173 | 2.677606953 | 7.775309299 | 0.395688532 | 0.587725539 | 0.070448259 |
| RHO GTPases activate PKNs | 2.732473013 | 2.252853031 | 7.717799056 | 0.325175845 | 0.684172969 | 0.07547119 |
| RHO GTPases Activate Rhotekin and Rhophilins | 3.005022469 | 0.903089987 | 6.913134925 | 0.457111537 | 0.206528862 | 0.024044932 |
| RHO GTPases Activate ROCKs | 2.431837516 | 2.770115295 | 7.633790326 | 0.393043035 | 0.621147553 | 0.073115288 |
| RHO GTPases Activate WASPs and WAVEs | 2.82276906 | 2.948901761 | 8.594439881 | 0.381169117 | 0.66790311 | 0.085902727 |
| RHO GTPases regulate CFTR trafficking | 2.920024602 | 1.041392685 | 6.88144189 | 0.47890411 | 0.114083696 | 0.013546229 |
| Ribosomal scanning and start codon recognition | 3.930750816 | 2.593286067 | 10.4547877 | 0.79966241 | 4 | 0.819682633 |
| RIP-mediated NFkB activation via ZBP1 | 2.416575001 | 2.722633923 | 7.555783924 | 0.310264961 | 0.23283583 | 0.024763647 |
| RIPK1-mediated regulated necrosis | 2.740241635 | 2.531478917 | 8.011962188 | 0.37053296 | 0.718422818 | 0.086048122 |
| RMTs methylate histone arginines | 2.332970072 | 2.772321707 | 7.438261851 | 0.31976509 | 0.414057115 | 0.043901849 |
| RNA Pol II CTD phosphorylation and interaction with CE | 2.737273932 | 2.225309282 | 7.699857146 | 0.468337064 | 0.447819898 | 0.057309237 |
| RNA Pol II CTD phosphorylation and interaction with CE during HIV infection | 2.605185205 | 2.025305865 | 7.235676276 | 0.468337064 | 0.447819898 | 0.054628285 |
| RNA Polymerase I Promoter Clearance | 0.429426502 | 1.286153899 | 0.138077143 | |||
| RNA Polymerase I Promoter Escape | 2.676797086 | 2.227886705 | 7.581480876 | 0.381549343 | 0.389696035 | 0.045048337 |
| RNA Polymerase I Promoter Opening | 2.378621285 | 2.281033367 | 7.038275938 | 0.572583952 | 0.535926741 | 0.070995346 |
| RNA Polymerase I Transcription | 0.429426502 | 1.286153899 | 0.138077143 | |||
| RNA Polymerase I Transcription Initiation | 2.668842081 | 3.22685757 | 8.564541733 | 0.42627967 | 1.053884317 | 0.141082193 |
| RNA Polymerase I Transcription Termination | 2.698592885 | 2.225309282 | 7.622495051 | 0.405047761 | 0.317980224 | 0.037860293 |
| RNA Polymerase II HIV Promoter Escape | 2.549132026 | 2.029383778 | 7.127647831 | 0.242677495 | 0.042908549 | 0.003964169 |
| RNA Polymerase II Pre-transcription Events | 2.783629119 | 2.762678564 | 8.329936802 | 0.380301652 | 0.592162456 | 0.074077004 |
| RNA Polymerase II Promoter Escape | 2.643419309 | 2.227886705 | 7.514725322 | 0.242677495 | 0.042908549 | 0.004178379 |
| RNA polymerase II transcribes snRNA genes | 2.498852894 | 2.922206277 | 7.919912066 | 0.273524639 | 0.114628591 | 0.012203407 |
| RNA Polymerase II Transcription | 0.345233877 | 4 | ||||
| ZNF564, ZNF565, ZNF566, ZNF567, ZNF568, ZNF569, ZNF570, ZNF571, ZNF572, ZNF573, ZNF574, ZNF575, ZNF577, ZNF582, ZNF583, ZNF584, ZNF585A, ZNF585B, ZNF586, ZNF587, ZNF589, ZNF595, ZNF596, ZNF597, ZNF599, ZNF600, ZNF605, ZNF606, ZNF607, ZNF610, ZNF611, ZNF613, ZNF614, ZNF615, ZNF616, ZNF619, ZNF620, ZNF621, ZNF624, ZNF625, ZNF626, ZNF627, ZNF641, ZNF648, ZNF649, ZNF655, ZNF658, ZNF658B, ZNF660, ZNF662, ZNF664, ZNF665, ZNF667, ZNF668, ZNF669, ZNF670, ZNF671, ZNF675, ZNF676, ZNF677, ZNF678, ZNF679, ZNF680, ZNF681, ZNF682, ZNF684, ZNF688, ZNF689, ZNF691, ZNF692, ZNF696, ZNF697, ZNF699, ZNF70, ZNF700, ZNF701, ZNF702P, ZNF703, ZNF704, ZNF705A, ZNF705D, ZNF705E, ZNF705F, ZNF705G, ZNF706, ZNF706L, ZNF707, ZNF708, ZNF709, ZNF71, ZNF710, ZNF710A, ZNF711, ZNF713, ZNF714, ZNF716, ZNF717, ZNF718, ZNF720, ZNF721, ZNF724, ZNF726, ZNF726P1, ZNF727, ZNF728, ZNF729, ZNF730, ZNF732, ZNF735, ZNF736, ZNF737, ZNF738, ZNF74, ZNF740, ZNF746, ZNF747, ZNF749, ZNF750, ZNF75A, ZNF75CP, ZNF75D, ZNF761, ZNF764, ZNF767P, ZNF77, ZNF770, ZNF771, ZNF772, ZNF773, ZNF774, ZNF775, ZNF776, ZNF777, ZNF778, ZNF782, ZNF784, ZNF785, ZNF786, ZNF79, ZNF790, ZNF791, ZNF792, ZNF793, ZNF799, ZNF804B, ZNF805, ZNF835, ZNF839, ZNF840P, ZNF852, ZNF860, ZNF875, ZNF879, ZNF92, ZNF985, ZNF988, ZNF99, ZNF996, ZSCAN25, ZSCAN32, ZW, ZWF1 | 0.345233877 | |||||
| RNA Polymerase II Transcription Elongation | 2.865608884 | 2.761927838 | 8.493145606 | 0.516537675 | 1.325163675 | 0.191128622 |
| RNA Polymerase II Transcription Initiation | 2.643419309 | 2.227886705 | 7.514725322 | 0.242677495 | 0.042908549 | 0.004178379 |
| RNA Polymerase II Transcription Initiation And Promoter Clearance | 2.643419309 | 2.227886705 | 7.514725322 | 0.242677495 | 0.042908549 | 0.004178379 |
| RNA Polymerase II Transcription Pre-Initiation And Promoter Opening | 2.643419309 | 2.227886705 | 7.514725322 | 0.242677495 | 0.042908549 | 0.004178379 |
| RNA Polymerase II Transcription Termination | 3.596268175 | 0.903089987 | 8.095626337 | 0.507140322 | 0.153201392 | 0.021130855 |
| RNA Polymerase III Abortive And Retractive Initiation | 1.666770972 | 0 | 3.333541944 | -0.894246575 | 1.507878456 | 0.188332374 |
| RNA Polymerase III Transcription | -0.894246575 | 1.507878456 | 0.337103786 | |||
| RNA Polymerase III Transcription Termination | 1.666770972 | 0 | 3.333541944 | 0.170093187 | ||
| ROBO receptors bind AKAP5 | 2.961948255 | 2.598790507 | 8.522687017 | 0.608414289 | 1.378138233 | 0.215121563 |
| Role of ABL in ROBO-SLIT signaling | 2.80329006 | 2.942999593 | 8.549579714 | 0.877808219 | 1.627733804 | 0.309459648 |
| Role of LAT2/NTAL/LAB on calcium mobilization | 2.988956137 | 3.525951341 | 9.503863616 | 0.625154234 | 2.856470509 | 0.488007399 |
| Role of phospholipids in phagocytosis | 2.541423358 | 3.535547279 | 8.618393996 | 0.335809728 | 0.505058268 | 0.06225074 |
| Role of second messengers in netrin-1 signaling | 2.286555002 | 0.77815125 | 5.351261254 | -0.381513988 | 0.420767539 | 0.036535433 |
| RORA activates gene expression | 2.288762554 | 2.238046103 | 6.815571211 | 0.346710694 | 0.344186913 | 0.034888735 |
| ROS and RNS production in phagocytes | 2.258498168 | 2.220108088 | 6.737104425 | -0.368406593 | 1.402644131 | 0.144564419 |
| rRNA modification in the nucleus and cytosol | 4.446701262 | 1.707570176 | 10.6009727 | 0.867526056 | 4 | 0.861156017 |
| rRNA processing | 0.84164511 | 4 | 0.84164511 | |||
| rRNA processing in the mitochondrion | 2.162841658 | 1.977723605 | 6.303406921 | 0.742810189 | 0.310480891 | 0.044793876 |
| rRNA processing in the nucleus and cytosol | 0.850635713 | 4 | 0.850635713 | |||
| RSK activation | 2.423328961 | 3.032618761 | 7.879276683 | 0.675171468 | 2.242416896 | 0.350135859 |
| RUNX1 and FOXP3 control the development of regulatory T lymphocytes (Tregs) | 2.279648349 | 2.133538908 | 6.692835606 | 0.405485018 | 0.271213767 | 0.029054995 |
| RUNX1 interacts with co-factors whose precise effect on RUNX1 targets is not known | 2.48454285 | 2.994317153 | 7.963402852 | 0.387302826 | 0.716856668 | 0.086914282 |
| RUNX1 regulates estrogen receptor mediated transcription | 2.882642462 | 3.072984745 | 8.838269669 | 0.441951716 | 0.378012278 | 0.05267908 |
| RUNX1 regulates expression of components of tight junctions | 2.368182458 | 2.123851641 | 6.860216557 | 0.708219178 | 0.716143688 | 0.105366241 |
| RUNX1 regulates genes involved in megakaryocyte differentiation and platelet function | 2.280707988 | 3.1775365 | 7.738952475 | 0.212495465 | 0.087745712 | 0.008467275 |
| RUNX1 regulates transcription of genes involved in BCR signaling | 2.270180905 | 2.278753601 | 6.81911541 | 0.708413264 | 1.096594048 | 0.160787221 |
| RUNX1 regulates transcription of genes involved in differentiation of HSCs | 3.209536235 | 3.405858399 | 9.82493087 | 0.698560421 | 4 | 0.736637854 |
| RUNX1 regulates transcription of genes involved in differentiation of keratinocytes | 2.373304722 | 2.747411808 | 7.494021252 | 0.555626352 | 0.887926046 | 0.120962421 |
| RUNX1 regulates transcription of genes involved in differentiation of myeloid cells | 2.558696442 | 2.413299764 | 7.530692649 | 0.589786024 | 1.232010197 | 0.173680431 |
| RUNX1 regulates transcription of genes involved in interleukin signaling | 2.368182458 | 2.123851641 | 6.860216557 | 0.545720965 | 0.456745495 | 0.057923437 |
| RUNX1 regulates transcription of genes involved in WNT signaling | 2.551575802 | 2.683947131 | 7.787098735 | 0.585160123 | 0.819543936 | 0.117770033 |
| RUNX2 regulates bone development | 2.66411994 | 1.826074803 | 7.154314683 | 0.498507903 | 2.106056752 | 0.262644628 |
| RUNX2 regulates chondrocyte maturation | 2.446110622 | 2.227886705 | 7.120107949 | 0.364105989 | 0.114573221 | 0.012312929 |
| RUNX2 regulates genes involved in cell migration | 2.393186465 | 2.771587481 | 7.55796041 | 0.531975068 | 0.98926351 | 0.132658772 |
| RUNX2 regulates genes involved in differentiation of myeloid cells | 2.504604957 | 2.1430148 | 7.152224713 | 0.543545765 | 0.634729108 | 0.082712798 |
| RUNX2 regulates osteoblast differentiation | 2.688724913 | 3.289365952 | 8.666815777 | 0.546558881 | 2.418323826 | 0.363287195 |
| RUNX3 regulates BCL2L11 (BIM) transcription | 2.721504808 | 2.385606274 | 7.828615889 | 0.515376716 | 0.348406021 | 0.047214049 |
| RUNX3 regulates CDKN1A transcription | 2.096123063 | 2.859738566 | 7.051984692 | 0.563852163 | 0.709980061 | 0.09340316 |
| RUNX3 Regulates Immune Response and Cell Migration | 2.465167905 | 2.365487985 | 7.295823795 | 0.698190789 | 1.482301767 | 0.224560762 |
| RUNX3 regulates NOTCH signaling | 3.033402138 | 2.487138375 | 8.553942652 | 0.351857506 | 0.28275681 | 0.035183231 |
| RUNX3 regulates p14-ARF | 2.463181186 | 2.723455672 | 7.649818045 | 0.524698383 | 1.179128296 | 0.158443771 |
| RUNX3 regulates RUNX1-mediated transcription | 2.366160662 | 1.826074803 | 6.558396128 | 0.708025199 | 0.265941131 | 0.038086258 |
| RUNX3 regulates WNT signaling | 2.703989477 | 2.593286067 | 8.001265022 | 0.584700201 | 1.197930408 | 0.175384905 |
| RUNX3 regulates YAP1-mediated transcription | 4.590895528 | 1.301029996 | 10.48282105 | 0.986849315 | 3.460322441 | 0.791308528 |
| S Phase | 0.526300795 | 4 | 0.526300795 | |||
| S33 mutants of beta-catenin aren't phosphorylated | 3.063502619 | 2.416640507 | 8.543645745 | 0.46828187 | 0.791988351 | 0.109967218 |
| S37 mutants of beta-catenin aren't phosphorylated | 3.063502619 | 2.416640507 | 8.543645745 | 0.46828187 | 0.791988351 | 0.109967218 |
| S45 mutants of beta-catenin aren't phosphorylated | 3.063502619 | 2.416640507 | 8.543645745 | 0.46828187 | 0.791988351 | 0.109967218 |
| SARS-CoV-1 Genome Replication and Transcription | 0.558275836 | 0.676121793 | 0.094365615 | |||
| SARS-CoV-1 Infection | 0.181847741 | 0.048616972 | 0.002210222 | |||
| SARS-CoV-2 Genome Replication and Transcription | 0.558275836 | 0.676121793 | 0.094365615 | |||
| SARS-CoV-2 Infection | 0.160889987 | 0.027152246 | 0.001092131 | |||
| SARS-CoV Infections | 0.337902374 | 2.510133331 | 0.212045003 | |||
| Scavenging by Class A Receptors | 2.317288888 | 1.579783597 | 6.214361372 | 0.362699896 | 0.261004129 | 0.024954693 |
| Scavenging by Class B Receptors | 1.931393123 | 1.255272505 | 5.118058752 | 0.359664492 | 0.109389764 | 0.008870589 |
| Scavenging by Class F Receptors | 2.717185718 | 2.825426118 | 8.259797555 | 0.692189977 | 1.417843778 | 0.23135975 |
| Scavenging by Class H Receptors | 3.295959823 | 0.698970004 | 7.29088965 | 0.914817858 | 0.730697295 | 0.130436321 |
| Scavenging of heme from plasma | 2.811786003 | 2.238046103 | 7.861618108 | 0.423321724 | 0.241752519 | 0.030082043 |
| SCF-beta-TrCP mediated degradation of Emi1 | 3.366310677 | 2.794488047 | 9.527109401 | 0.806611817 | 4 | 0.775299193 |
| SCF(Skp2)-mediated degradation of p27/p21 | 3.094581722 | 3.200029267 | 9.389192711 | 0.731181914 | 4 | 0.730469236 |
| Sealing of the nuclear envelope (NE) by ESCRT-III | 2.808551722 | 3.021189299 | 8.638292744 | 0.687122022 | 4 | 0.669700991 |
| Selective autophagy | 0.555635613 | 4 | 0.555635613 | |||
| Selenoamino acid metabolism | 2.816132866 | 1.176091259 | 6.808356991 | 0.847060847 | 4 | 0.655265558 |
| Selenocysteine synthesis | 4.454260569 | 2.893206753 | 11.80172789 | 0.914844895 | 4 | 0.946761387 |
| SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion | 2.41358085 | 2.527629901 | 7.354791602 | 0.530573074 | 0.75980565 | 0.099764718 |
| Sema3A PAK dependent Axon repulsion | 2.6826805 | 3.120244796 | 8.485605795 | 0.465807211 | 1.032259426 | 0.142236728 |
| Sema4D in semaphorin signaling | 0.426501323 | 0.895060388 | 0.09543611 | |||
| Sema4D induced cell migration and growth-cone collapse | 2.452761943 | 3.080987047 | 7.986510932 | 0.419458973 | 0.723450814 | 0.090837315 |
| Sema4D mediated inhibition of cell attachment and migration | 2.689331049 | 2.704150517 | 8.082812614 | 0.517676076 | 0.355514109 | 0.049445015 |
| Semaphorin interactions | 0.392352502 | 1.516314976 | 0.148732494 | |||
| Senescence-Associated Secretory Phenotype (SASP) | 2.417021868 | 3.489114369 | 8.323158106 | 0.423988319 | 1.697303838 | 0.221445793 |
| Sensing of DNA Double Strand Breaks | 2.496447391 | 1.113943352 | 6.106838134 | -0.449561404 | 0.398834539 | 0.041910039 |
| Separation of Sister Chromatids | 3.122033039 | 3.407050815 | 9.651116892 | 0.666166517 | 4 | 0.711473985 |
| Serine biosynthesis | 1.698227812 | 0.301029996 | 3.697485619 | -0.534794521 | 0.271444913 | 0.022980876 |
| Serotonin and melatonin biosynthesis | 2.522009099 | 1.041392685 | 6.085410883 | -0.476556073 | 0.686961097 | 0.074314934 |
| Serotonin clearance from the synaptic cleft | 3.059454983 | 3.549861188 | 9.668771155 | 0.498200033 | 0.301662617 | 0.047391311 |
| Serotonin Neurotransmitter Release Cycle | 3.050292241 | 1.875061263 | 7.975645745 | 0.561142599 | 0.494534056 | 0.070783386 |
| Serotonin receptors | 2.554274847 | 3.902383684 | 9.010933379 | 0.578713464 | 2.158967043 | 0.342585392 |
| SHC-mediated cascade:FGFR1 | 2.997348762 | 2.436162647 | 8.430860171 | 0.531901757 | 1.736815273 | 0.252441579 |
| SHC-mediated cascade:FGFR2 | 3.004096682 | 2.278753601 | 8.286946965 | 0.506663503 | 1.542633939 | 0.216487886 |
| SHC-mediated cascade:FGFR3 | 3.153172205 | 2.309630167 | 8.615974578 | 0.58241025 | 1.963898576 | 0.302535497 |
| SHC-mediated cascade:FGFR4 | 3.088759324 | 2.380211242 | 8.55772989 | 0.58218967 | 1.963898576 | 0.30100607 |
| SHC-related events triggered by IGF1R | 2.875493794 | 2.908485019 | 8.659472606 | 0.535524295 | 1.227432975 | 0.182579053 |
| SHC1 events in EGFR signaling | 2.987962395 | 3.086359831 | 9.062284621 | 0.459491337 | 0.91498921 | 0.132160821 |
| SHC1 events in ERBB2 signaling | 2.742410546 | 3.580924976 | 9.065746068 | 0.579298627 | 3.396815702 | 0.541657295 |
| SHC1 events in ERBB4 signaling | 3.287765008 | 1.748188027 | 8.323718043 | 0.539597215 | 1.244807071 | 0.180406759 |
| Sialic acid metabolism | 2.152068706 | 1.079181246 | 5.383318658 | -0.586263736 | 3.476073767 | 0.392231885 |
| Signal amplification | 0.368053093 | 0.502762427 | 0.046260817 | |||
| Signal attenuation | 2.94659208 | 3.21005085 | 9.103235009 | 0.459149878 | 0.798354636 | 0.115701725 |
| Signal regulatory protein family interactions | 3.321705151 | 2.488550717 | 9.131961018 | 0.509224647 | 1.071680291 | 0.16241857 |
| Signal transduction by L1 | 2.385733403 | 3.587149498 | 8.358616304 | 0.481818841 | 1.978573916 | 0.273350438 |
| Signaling by activated point mutants of FGFR1 | 3.04367831 | 2.36361198 | 8.450968599 | 0.659497479 | 1.217483944 | 0.196692087 |
| Signaling by activated point mutants of FGFR3 | 3.033432602 | 2.209515015 | 8.276380219 | 0.659637114 | 1.217483944 | 0.193971909 |
| Signaling by Activin | 2.503739523 | 2.484299839 | 7.491778885 | 0.38530729 | 0.334930235 | 0.038487331 |
| Signaling by BMP | 2.589095507 | 1.86923172 | 7.047422733 | 0.273528165 | 0.149958662 | 0.01427728 |
| Signaling by BRAF and RAF fusions | 2.761578054 | 3.315760491 | 8.838916599 | 0.483021826 | 3.920849007 | 0.566563573 |
| Signaling by cytosolic FGFR1 fusion mutants | 2.701347235 | 2.923761961 | 8.326456432 | 0.434801329 | 0.691629641 | 0.091200579 |
| Signaling by cytosolic PDGFRA and PDGFRB fusion proteins | 3.503516717 | 1.146128036 | 8.15316147 | 0.855382087 | 0.541103565 | 0.098589544 |
| Signaling by EGFR | 2.41424346 | 3.267171728 | 8.095658648 | 0.345497123 | 0.78990187 | 0.092990113 |
| Signaling by EGFR in Cancer | 0.495073206 | 2.115293378 | 0.261806269 | |||
| Signaling by EGFRvIII in Cancer | 0.508948849 | 1.652090981 | 0.210207451 | |||
| Signaling by ERBB2 | 2.732594543 | 3.60368555 | 9.068874636 | 0.486529271 | 3.738891926 | 0.552998917 |
| Signaling by ERBB2 ECD mutants | 3.109150693 | 3.302979937 | 9.521281322 | 0.531485356 | 2.218428802 | 0.353526205 |
| Signaling by ERBB2 in Cancer | 0.51501562 | 2.409913395 | 0.31028576 | |||
| Signaling by ERBB2 KD Mutants | 3.109150693 | 3.302979937 | 9.521281322 | 0.548591812 | 2.889587981 | 0.466660169 |
| Signaling by ERBB2 TMD/JMD mutants | 3.158565299 | 3.104487111 | 9.42161771 | 0.505970757 | 1.894908002 | 0.293491137 |
| Signaling by ERBB4 | 2.226042312 | 3.272537777 | 7.724622401 | 0.604724299 | 4 | 0.581366645 |
| Signaling by Erythropoietin | 2.652078505 | 2.472756449 | 7.77691346 | 0.506811048 | 2.633254294 | 0.352268644 |
| Signaling by extracellular domain mutants of KIT | 2.617771423 | 2.158362492 | 7.393905339 | 0.918104629 | 0.749379508 | 0.135074794 |
| Signaling by FGFR | 0.475118914 | 3.141417359 | 0.373136701 | |||
| Signaling by FGFR in disease | 0.416260665 | 1.574921216 | 0.163894438 | |||
| Signaling by FGFR1 | 0.52040744 | 3.019372017 | 0.392825915 | |||
| Signaling by FGFR1 amplification mutants | 0.915365653 | 0.735293047 | 0.1682655 | |||
| Signaling by FGFR1 in disease | 2.940476422 | 3.030599722 | 8.911552566 | 0.393733669 | 0.95516068 | 0.12825517 |
| Signaling by FGFR2 | 0.436609685 | 2.072862304 | 0.226257939 | |||
| Signaling by FGFR2 amplification mutants | 2.695850494 | 2.117271296 | 7.508972283 | 0.9326212 | 0.833359637 | 0.152961055 |
| Signaling by FGFR2 fusions | 2.695850494 | 2.117271296 | 7.508972283 | 0.9326212 | 0.833359637 | 0.152961055 |
| Signaling by FGFR2 IIIa TM | 2.893723407 | 2.170261715 | 7.95770853 | 0.654386055 | 1.190045696 | 0.183928179 |
| Signaling by FGFR2 in disease | 2.832466885 | 2.960470778 | 8.625404548 | 0.476981802 | 1.566790912 | 0.220904197 |
| Signaling by FGFR3 | 0.541863632 | 3.465531978 | 0.469461436 | |||
| Signaling by FGFR3 fusions in cancer | 2.894070113 | 2.958563883 | 8.746704109 | 0.616479413 | 1.930745081 | 0.308905958 |
| Signaling by FGFR3 in disease | 0.557566008 | 1.904706835 | 0.265499947 | |||
| Signaling by FGFR3 point mutants in cancer | 2.913281692 | 2.967079734 | 8.793643117 | 0.557566008 | 1.904706835 | 0.291866504 |
| Signaling by FGFR4 | 0.541755817 | 3.465531978 | 0.469368027 | |||
| Signaling by FGFR4 in disease | 2.822443796 | 2.932473765 | 8.577361358 | 0.550929459 | 1.426791101 | 0.213469818 |
| Signaling by GPCR | 0.322687392 | 4 | ||||
| S6KA1, RPS6KA2, RPS6KA3, RPS6KA3A, RRH, RSG7, RSKN-1, RTGEF, RTP1, RTP2, RTP3, RTP4, RTP5, RUT, RXFP1, RXFP2, RXFP2A, RXFP3, RXFP3.2B, RXFP4, RYA-R, S1PR1, S1PR2, S1PR3, S1PR3A, S1PR4, S1PR5, S1PR5A, S6KII, SAA1, SAG, SAGA, SAX-3, SCH9, SCK1, SCK2, SCT, SCTR, SCYC1, SDC1, SDC2, SDC3, SDC4, SDR9C7, SEM-5, SER-2, SER-4, SER-5, SER-7, SFTPBA, SHH, SI:CH1073-140O9.2, SI:CH211-103A14.5, SI:CH211-117L17.5, SI:CH211-132P1.2, SI:CH211-137J23.6, SI:CH211-137J23.7, SI:CH211-137J23.8, SI:CH211-196H16.12, SI:CH211-214P16.3, SI:CH211-231M23.4, SI:CH211-272N13.7, SI:CH73-15B2.5, SI:CH73-167I17.6, SI:CH73-334D15.2, SI:CH73-62L21.1, SI:CH73-90P23.1, SI:DKEY-117A8.1, SI:DKEY-117A8.2, SI:DKEY-117A8.3, SI:DKEY-117A8.4, SI:DKEY-172J4.3, SI:DKEY-211G8.6, SI:DKEY-211G8.7, SI:DKEY-211G8.9, SI:DKEY-33C9.6, SI:DKEY-38P12.3, SI:DKEY-42L23.2, SI:DKEY-42L23.3, SI:DKEY-42L23.4, SI:DKEY-42L23.5, SI:DKEY-42L23.6, SI:DKEY-42L23.7, SI:DKEY-42L23.9, SI:DKEY-63B1.1, SI:DKEY-73N8.3, SI:DKEY-94E7.2, SI:DKEYP-118H3.5, SIF, SIFAR, SLC24A1, SMA-5, SMO, SOS, SOS-1, SOS1, SOS2, SPAC1786.02, SPAC1A6.03C, SPAC22H12.03, SPAC977.09C, SPBC1348.10C, SPBC800.10C, SPCC1450.09C, SPE-41, SPK1, SPN100A, SPN27A, SPN28B, SPN28DA, SPN28DB, SPN28DC, SPN28F, SPN31A, SPN38F, SPN42DA, SPN42DB, SPN42DD, SPN42DE, SPN43AA, SPN43AB, SPN43AD, SPN47C, SPN53F, SPN55B, SPN75F, SPN77BA, SPN77BB, SPN77BC, SPN85F, SPN88EA, SPN88EB, SPO1, SRC, SRO-1, SRP-1, SRP-2, SRP-3, SRP-6, SRP-7, SRP-8, SS1R, SS2R, SS3R, SST, SST.1, SST1.1, SST2, SSTR1, SSTR1A, SSTR1B, SSTR2, SSTR2B, SSTR3, SSTR4, SSTR5, STE6, STI, STPG4, STRA6, SUCNR1, T2R10, T2R10C, T2R14, T2R15, T2R17, T2R19, T2R20, T2R22, T2R24, T2R32, T2R34, T2R37, T2R38, T2R43, T2R45, T2R46, T2R48, T2R49, T2R52, T2R53, T2R65A, T2R9, TAAR1, TAAR13A, TAAR13B, TAAR13C, TAAR13D, TAAR13E, TAAR19T, TAAR1B, TAAR2, TAAR3, TAAR3P, TAAR5, TAAR6, TAAR8, TAAR8B, TAAR9, TAC1, TAC3, TAC3A, TACR1, TACR1A, TACR2, TACR3, TACR3L, TAG-89, TAPBP, TAS1R1, TAS1R2, TAS1R2.1, TAS1R3, TAS2R1, TAS2R10, TAS2R106, TAS2R119, TAS2R120, TAS2R13, TAS2R131, TAS2R135, TAS2R136, TAS2R14, TAS2R140, TAS2R145, TAS2R16, TAS2R19, TAS2R20, TAS2R200.1, TAS2R200.2, TAS2R203, TAS2R3, TAS2R30, TAS2R31, TAS2R38, TAS2R39, TAS2R4, TAS2R40, TAS2R41, TAS2R42, TAS2R43, TAS2R45, TAS2R46, TAS2R5, TAS2R50, TAS2R60, TAS2R7, TAS2R8, TAS2R9, TAX-2, TAX-4, TBXA2R, TCF3, TDA5, THOP1, TIAM-1, TIAM1, TIAM1A, TIAM2, TIAM2A, TKR-1, TMEM26, TMIGD3, TOS3, TPA-1, TPK1, TPK2, TPK3, TRAP1, TRE1, TRH, TRHR, TRHR-1, TRHRA, TRIO, TRIOB, TRISSINR, TRP-1, TRPC3, TRPC6, TRPC6B, TRPC7, TRPC7A, TSHB, TSHBA, TSHR, TSPAN17, TTC19, TTR, TYRA-2, TYRA-3, TYRR, TYRRII, UBE2N, UCN, UCN2, UCN3, UCN3L, UIG-1, UNC-73, UTS1, UTS2, UTS2B, UTS2D, UTS2R, VAV, VAV-1, VAV1, VAV2, VAV3, VAV3B, VIP, VIPR1, VIPR1B, VIPR2, VPAC2, WNT1, WNT10A, WNT10B, WNT11, WNT16, WNT2, WNT2B, WNT3, WNT3A, WNT4, WNT5A, WNT6, WNT7A, WNT7B, WNT8A, WNT8B, WNT9A, WNT9B, WUN, WUN-LIKE, WUN2, XB5807236, XCL1, XCL2, XCR1, XCR1A.1, XENTR_V90028168MG, XENTR_V90028851MG, XK, YDL114W, YPK1, ZC3H12B, ZC4H2, ZGC:101744, ZGC:162608, ZK697.8, ZNF750 | 0.322687392 | |||||
| Signaling by Hedgehog | 0.586950853 | 4 | 0.586950853 | |||
| Signaling by high-kinase activity BRAF mutants | 2.88749796 | 3.131297797 | 8.906293716 | 0.495804176 | 2.83945208 | 0.417305763 |
| Signaling by Hippo | 3.26939044 | 2.155336037 | 8.694116917 | 0.427337408 | 0.717964044 | 0.097407727 |
| Signaling by Insulin receptor | 3.001919535 | 2.713490543 | 8.717329613 | 0.414634241 | 2.348453867 | 0.315593771 |
| Signaling by Interleukins | 0.360898812 | 4 | 0.360898812 | |||
| Signaling by juxtamembrane domain KIT mutants | 2.617771423 | 2.158362492 | 7.393905339 | 0.918104629 | 0.749379508 | 0.135074794 |
| Signaling by kinase domain mutants of KIT | 2.617771423 | 2.158362492 | 7.393905339 | 0.918104629 | 0.749379508 | 0.135074794 |
| Signaling by KIT in disease | 0.521679193 | 2.837065416 | 0.370009499 | |||
| Signaling by Leptin | 2.459449892 | 2.428134794 | 7.347034579 | 0.327919116 | 0.164850068 | 0.017452867 |
| Signaling by Ligand-Responsive EGFR Variants in Cancer | 0.531143323 | 2.212638209 | 0.293807003 | |||
| Signaling by MAP2K mutants | 2.470889264 | 2.730782276 | 7.672560804 | 0.737390351 | 1.794131117 | 0.289309858 |
| Signaling by MAPK mutants | 1.817129537 | 2.46834733 | 6.102606404 | 0.419288781 | 0.129386561 | 0.013099433 |
| Signaling by membrane-tethered fusions of PDGFRA or PDGFRB | 3.18480326 | 2.658964843 | 9.028571363 | 0.910684932 | 1.904926272 | 0.381755127 |
| Signaling by MET | 0.434568243 | 2.392853994 | 0.259964589 | |||
| Signaling by moderate kinase activity BRAF mutants | 2.812505299 | 3.255031163 | 8.880041761 | 0.512626574 | 4 | 0.594924896 |
| Signaling by MRAS-complex mutants | 3.164353873 | 2.53529412 | 8.864001867 | 0.652316973 | 1.449368186 | 0.240574324 |
| Signaling by MST1 | 2.55331666 | 2.146128036 | 7.252761356 | 0.530508852 | 0.403987446 | 0.052509885 |
| Signaling by NODAL | 2.894038582 | 2.850646235 | 8.6387234 | 0.456098003 | 0.777165337 | 0.107678433 |
| Signaling by Non-Receptor Tyrosine Kinases | 0.492010176 | 3.063694616 | 0.376842232 | |||
| Signaling by NOTCH | 0.564774877 | 4 | 0.564774877 | |||
| Signaling by NOTCH1 | 0.529502447 | 4 | 0.529502447 | |||
| Signaling by NOTCH1 HD Domain Mutants in Cancer | 0.504950302 | 0.617772617 | 0.077986117 | |||
| Signaling by NOTCH1 HD+PEST Domain Mutants in Cancer | 0.554377404 | 4 | 0.554377404 | |||
| Signaling by NOTCH1 in Cancer | 0.554377404 | 4 | 0.554377404 | |||
| Signaling by NOTCH1 PEST Domain Mutants in Cancer | 0.554377404 | 4 | 0.554377404 | |||
| Signaling by NOTCH1 t(7;9)(NOTCH1:M1580_K2555) Translocation Mutant | 0.40154604 | 0.183885387 | 0.018459612 | |||
| Signaling by NOTCH2 | 0.598198446 | 3.731676948 | 0.558070838 | |||
| Signaling by NOTCH3 | 0.5813053 | 4 | 0.5813053 | |||
| Signaling by NOTCH4 | 0.695218835 | 4 | 0.695218835 | |||
| Signaling by NTRK1 (TRKA) | 0.410249819 | 3.854751215 | 0.395352747 | |||
| Signaling by NTRK2 (TRKB) | 0.493269841 | 2.081233785 | 0.256652465 | |||
| Signaling by NTRK3 (TRKC) | 2.740747257 | 1.505149978 | 6.986644493 | 0.405704211 | 0.911652478 | 0.10114441 |
| Signaling by NTRKs | 0.411176827 | 4 | 0.411176827 | |||
| Signaling by Nuclear Receptors | 0.301680038 | 2.239348362 | 0.168891675 | |||
| Signaling by Overexpressed Wild-Type EGFR in Cancer | 0.510128905 | 0.514753791 | 0.065647697 | |||
| Signaling by PDGF | 2.767428096 | 3.027349608 | 8.562205799 | 0.36470741 | 1.067832202 | 0.134698603 |
| Signaling by PDGFR in disease | 0.55101575 | 2.381135869 | 0.328010842 | |||
| Signaling by PDGFRA extracellular domain mutants | 2.730689018 | 3.033825694 | 8.49520373 | 0.497493661 | 1.42612837 | 0.202333734 |
| Signaling by PDGFRA transmembrane, juxtamembrane and kinase domain mutants | 2.730689018 | 3.033825694 | 8.49520373 | 0.497493661 | 1.42612837 | 0.202333734 |
| Signaling by phosphorylated juxtamembrane, extracellular and kinase domain KIT mutants | 2.601966591 | 3.197556213 | 8.401489394 | 0.521679193 | 2.837065416 | 0.407660123 |
| Signaling by plasma membrane FGFR1 fusions | 2.548008907 | 2.330413773 | 7.426431587 | 0.915365653 | 0.735293047 | 0.132592437 |
| Signaling by PTK6 | 0.492010176 | 3.063694616 | 0.376842232 | |||
| Signaling by RAF1 mutants | 2.839605303 | 3.247482261 | 8.926692867 | 0.548719301 | 4 | 0.615377951 |
| Signaling by RAS mutants | 0.512626574 | 4 | 0.512626574 | |||
| Signaling by Receptor Tyrosine Kinases | 0.371673319 | 4 | 0.371673319 | |||
| Signaling by Retinoic Acid | 2.207133934 | 2.806179974 | 7.220447842 | 0.271151192 | 0.347473189 | 0.033754427 |
| Signaling by Rho GTPases | 0.373267041 | 4 | 0.373267041 | |||
| Signaling by ROBO receptors | 2.244791026 | 1.924279286 | 6.413861338 | 0.737935166 | 4 | 0.580351018 |
| Signaling by SCF-KIT | 2.748806444 | 3.783903579 | 9.281516467 | 0.436419391 | 2.659862366 | 0.384039604 |
| Signaling by TGF-beta Receptor Complex | 0.341959393 | 1.076118199 | 0.091997181 | |||
| Signaling by TGF-beta Receptor Complex in Cancer | 0.468478216 | 0.599986983 | 0.070270208 | |||
| Signaling by TGFB family members | 0.331081084 | 1.227843147 | 0.10162891 | |||
| Signaling by the B Cell Receptor (BCR) | 0.544822576 | 4 | 0.544822576 | |||
| Signaling by Type 1 Insulin-like Growth Factor 1 Receptor (IGF1R) | 2.482621244 | 2.877371346 | 7.842613835 | 0.47445583 | 2.898686136 | 0.378510066 |
| Signaling by VEGF | 2.826864726 | 0.698970004 | 6.352699455 | 0.447974043 | 4 | 0.432215167 |
| Signaling by WNT | 0.411404091 | 4 | 0.411404091 | |||
| Signaling by WNT in cancer | 0.384865108 | 0.788102787 | 0.075828316 | |||
| Signaling downstream of RAS mutants | 2.812505299 | 3.255031163 | 8.880041761 | 0.512626574 | 4 | 0.594924896 |
| Signalling to ERK5 | 2.769867521 | 1.62324929 | 7.162984331 | 0.700821918 | 0.691320104 | 0.103774237 |
| Signalling to ERKs | 0.666810646 | 4 | 0.666810646 | |||
| Signalling to p38 via RIT and RIN | 1.892403622 | 2.50242712 | 6.287234365 | 0.787842426 | 1.542591568 | 0.230915186 |
| Signalling to RAS | 2.844581582 | 2.301029996 | 7.99019316 | 0.647441292 | 4 | 0.616425714 |
| Signalling to STAT3 | 1.818874589 | 2.146128036 | 5.783877214 | 0.502514722 | 0.314512504 | 0.033821137 |
| SIRT1 negatively regulates rRNA expression | 2.508906605 | 1.84509804 | 6.86291125 | 0.337843727 | 0.075421097 | 0.007607564 |
| SLC-mediated transmembrane transport | 0.218370108 | 0.25302227 | 0.013813125 | |||
| SLC transporter disorders | 0.387398721 | 1.515873844 | 0.146811897 | |||
| SLIT2:ROBO1 increases RHOA activity | 2.900434195 | 0.602059991 | 6.402928381 | 0.517392495 | 0.014693951 | 0.00172476 |
| SMAC, XIAP-regulated apoptotic response | 2.36428013 | 2.28780173 | 7.01636199 | 0.471749863 | 0.61901022 | 0.074024472 |
| SMAD2/3 MH2 Domain Mutants in Cancer | 2.58456228 | 2.380211242 | 7.549335803 | 0.502465753 | 0.145286923 | 0.018930686 |
| SMAD2/3 Phosphorylation Motif Mutants in Cancer | 2.431345472 | 2.506505032 | 7.369195976 | 0.421723816 | 0.320184867 | 0.037744213 |
| SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription | 2.670892598 | 3.158060794 | 8.49984599 | 0.43869448 | 1.129860423 | 0.15206364 |
| SMAD4 MH2 Domain Mutants in Cancer | 2.58456228 | 2.380211242 | 7.549335803 | 0.502465753 | 0.145286923 | 0.018930686 |
| Small interfering RNA (siRNA) biogenesis | 2.576339247 | 0 | 5.152678493 | 0.330122161 | ||
| Smooth Muscle Contraction | 2.710285928 | 2.255272505 | 7.675844361 | 0.275626762 | 0.167881289 | 0.017388366 |
| snRNP Assembly | 2.489734847 | 2.496929648 | 7.476399343 | 0.624391248 | 1.507232929 | 0.21794381 |
| Sodium-coupled sulphate, di- and tri-carboxylate transporters | 1.830942376 | 0 | 3.661884752 | -0.932310222 | 3.064684778 | 0.41033554 |
| Sodium/Calcium exchangers | 2.579123801 | 1.62324929 | 6.781496893 | 0.513574758 | 0.345595043 | 0.042088018 |
| Sodium/Proton exchangers | 2.715332671 | 2.542825427 | 7.973490768 | 0.467774201 | 0.364367932 | 0.047889834 |
| Sorafenib-resistant KIT mutants | 2.617771423 | 2.158362492 | 7.393905339 | 0.918104629 | 0.749379508 | 0.135074794 |
| Sorafenib-resistant PDGFR mutants | 2.318185123 | 2.017033339 | 6.653403585 | 0.882497946 | 0.618515789 | 0.102826733 |
| SOS-mediated signalling | 3.287020586 | 1.763427994 | 8.337469165 | 0.499744116 | 0.715380401 | 0.100241392 |
| Sperm Motility And Taxes | 2.505187611 | 0 | 5.010375222 | 0.317603779 | ||
| Sphingolipid de novo biosynthesis | 2.729625263 | 2.596597096 | 8.055847621 | 0.420437279 | 0.989241482 | 0.125215903 |
| Sphingolipid metabolism | 0.185330818 | 0.057429996 | 0.002660887 | |||
| Spry regulation of FGF signaling | 2.631719312 | 3.34869419 | 8.612132815 | 0.543690646 | 1.604002626 | 0.239251321 |
| SRP-dependent cotranslational protein targeting to membrane | 4.405137004 | 3.198106999 | 12.00838101 | 0.904450262 | 4 | 0.952225131 |
| Stabilization of p53 | 2.804816869 | 3.063708559 | 8.673342298 | 0.77785279 | 4 | 0.716874553 |
| STAT5 Activation | 2.46454014 | 2.240549248 | 7.169629529 | 0.329037565 | 0.1006462 | 0.010439317 |
| STAT6-mediated induction of chemokines | 2.588852659 | 2.307496038 | 7.485201356 | 0.556438356 | 0.250111405 | 0.034069684 |
| Sterols are 12-hydroxylated by CYP8B1 | 2.579706132 | 0 | 5.159412263 | 0.330714529 | ||
| Stimulation of the cell death response by PAK-2p34 | 2.414446085 | 1.785329835 | 6.614222006 | 0.510684932 | 0.158222193 | 0.018870462 |
| Stimuli-sensing channels | 2.806089791 | 2.745855195 | 8.358034776 | 0.265191273 | 0.446026942 | 0.049539916 |
| STING mediated induction of host immune responses | 2.141802094 | 0 | 4.283604189 | 0.213009813 | 0.015402128 | 0.000800873 |
| Striated Muscle Contraction | 2.883643295 | 0.301029996 | 6.068316586 | -0.352344393 | 0.265955784 | 0.024582909 |
| Sulfur amino acid metabolism | 2.213928317 | 0.84509804 | 5.272954674 | -0.437791815 | 1.895821873 | 0.17603698 |
| SUMO E3 ligases SUMOylate target proteins | 2.24180643 | 0.954242509 | 5.437855369 | 0.346713547 | 2.332768427 | 0.195012956 |
| SUMO is conjugated to E1 (UBA2:SAE1) | 2.481352684 | 1.414973348 | 6.377678715 | -0.508363038 | 0.798934856 | 0.092616188 |
| SUMO is proteolytically processed | 2.590071549 | 0.903089987 | 6.083233086 | -0.508363038 | 0.798934856 | 0.089582193 |
| SUMO is transferred from E1 to E2 (UBE2I, UBC9) | 2.407832708 | 1.505149978 | 6.320815395 | -0.508502468 | 0.997689579 | 0.114942446 |
| SUMOylation | 0.339458921 | 2.105317883 | 0.178667234 | |||
| SUMOylation of chromatin organization proteins | 2.645711196 | 3.051152522 | 8.342574914 | 0.380086013 | 0.650615475 | 0.081477739 |
| SUMOylation of DNA damage response and repair proteins | 2.598709067 | 2.667452953 | 7.864871087 | 0.426363639 | 1.167739185 | 0.145798551 |
| SUMOylation of DNA methylation proteins | 2.503909706 | 1.51851394 | 6.526333352 | 0.404500549 | 0.472605154 | 0.049556909 |
| SUMOylation of DNA replication proteins | 2.439411246 | 3.026533265 | 7.905355757 | 0.385682502 | 0.611213514 | 0.073524345 |
| SUMOylation of immune response proteins | 2.358210221 | 2.08278537 | 6.799205813 | -0.35428101 | 0.605151068 | 0.061786459 |
| SUMOylation of intracellular receptors | 2.56989331 | 3.634376494 | 8.774163113 | 0.421549876 | 1.818007457 | 0.247214634 |
| SUMOylation of RNA binding proteins | 2.745241535 | 1.380211242 | 6.870694312 | 0.409150605 | 0.490958356 | 0.053947286 |
| SUMOylation of SUMOylation proteins | 2.673442671 | 1.176091259 | 6.5229766 | 0.454495614 | 0.321533205 | 0.035711118 |
| SUMOylation of transcription cofactors | 2.871447912 | 2.123851641 | 7.866747464 | 0.28754845 | 0.27134202 | 0.029176786 |
| SUMOylation of transcription factors | 2.490717963 | 2.745855195 | 7.727291121 | 0.33510309 | 0.226582251 | 0.025303217 |
| SUMOylation of ubiquitinylation proteins | 3.191859837 | 2.784617293 | 9.168336967 | 0.500592764 | 0.597673141 | 0.090215907 |
| Sunitinib-resistant KIT mutants | 2.617771423 | 2.158362492 | 7.393905339 | 0.918104629 | 0.749379508 | 0.135074794 |
| Sunitinib-resistant PDGFR mutants | 2.318185123 | 2.017033339 | 6.653403585 | 0.882497946 | 0.618515789 | 0.102826733 |
| Suppression of apoptosis | 2.208366671 | 2.644438589 | 7.061171931 | 0.557565789 | 0.676121793 | 0.088497673 |
| Suppression of autophagy | 1.960492661 | 0 | 3.920985322 | 0.22177043 | ||
| Suppression of phagosomal maturation | 0.44839299 | 0.301602566 | 0.033809119 | |||
| Surfactant metabolism | 2.50753341 | 3.082066934 | 8.097133755 | 0.387127659 | 0.482084674 | 0.059270608 |
| Switching of origins to a post-replicative state | 4.228483415 | 0.301029996 | 8.757996825 | 0.744393687 | 4 | 0.704512257 |
| Synaptic adhesion-like molecules | 2.169131469 | 3.398634325 | 7.736897262 | 0.428213361 | 0.766646088 | 0.094631808 |
| Syndecan interactions | 2.444720062 | 2.760422483 | 7.649862607 | 0.436973793 | 1.224368232 | 0.15109763 |
| Synthesis and processing of ENV and VPU | 3.305464684 | 2.161368002 | 8.772297369 | 0.997261024 | 2.000945472 | 0.416038423 |
| Synthesis And Processing Of GAG, GAGPOL Polyproteins | 0.486160592 | 0.267582827 | 0.032522056 | |||
| Synthesis of (16-20)-hydroxyeicosatetraenoic acids (HETE) | 2.355537539 | 2.81756537 | 7.528640448 | 0.418769022 | 0.544068044 | 0.065054007 |
| Synthesis of 12-eicosatetraenoic acid derivatives | 2.058284878 | 2.651278014 | 6.76784777 | 0.544767547 | 0.453141371 | 0.056872535 |
| Synthesis of 15-eicosatetraenoic acid derivatives | 2.10387602 | 2.903089987 | 7.110842027 | 0.59193322 | 0.831023753 | 0.112875192 |
| Synthesis of 5-eicosatetraenoic acids | 2.524555505 | 3.089905111 | 8.139016121 | 0.454724375 | 0.407975501 | 0.053826748 |
| Synthesis of active ubiquitin: roles of E1 and E2 enzymes | 2.472635992 | 1.380211242 | 6.325483226 | -0.403907208 | 2.218132656 | 0.226680784 |
| Synthesis of bile acids and bile salts | 2.883547595 | 2.549003262 | 8.316098453 | -0.185694635 | 0.196294645 | 0.019745551 |
| Synthesis of bile acids and bile salts via 24-hydroxycholesterol | 2.268790442 | 0.698970004 | 5.236550889 | -0.455942904 | 1.440056753 | 0.136308007 |
| Synthesis of bile acids and bile salts via 27-hydroxycholesterol | 2.567361738 | 2.553883027 | 7.688606502 | 0.244258813 | 0.083705689 | 0.008355416 |
| Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol | 2.616302143 | 2.723455672 | 7.956059958 | -0.185694635 | 0.196294645 | 0.018834053 |
| Synthesis of diphthamide-EEF2 | 3.540119672 | 0.477121255 | 7.557360599 | 0.541661426 | ||
| Synthesis of DNA | 0.664276622 | 4 | 0.664276622 | |||
| Synthesis of Dolichyl-phosphate | 1.703789624 | 0 | 3.407579248 | -0.903561644 | 1.578940704 | 0.200554164 |
| Synthesis of epoxy (EET) and dihydroxyeicosatrienoic acids (DHET) | 2.243495577 | 2.875061263 | 7.362052418 | 0.501918787 | 1.010723865 | 0.129185471 |
| Synthesis of GDP-mannose | 1.901637744 | 0.477121255 | 4.280396743 | -0.613424658 | 0.449659866 | 0.045868917 |
| Synthesis of Hepoxilins (HX) and Trioxilins (TrX) | 2.300526334 | 2.06069784 | 6.661750509 | 0.714324843 | 0.277057136 | 0.040265701 |
| Synthesis of IP2, IP, and Ins in the cytosol | 2.032574316 | 2.227886705 | 6.293035337 | 0.511708661 | 0.164324792 | 0.018938615 |
| Synthesis of IP3 and IP4 in the cytosol | 2.737240662 | 1.716003344 | 7.190484667 | -0.315471283 | 0.981312853 | 0.100384483 |
| Synthesis of Ketone Bodies | 3.013741675 | 0 | 6.027483351 | 0.407078519 | ||
| Synthesis of Leukotrienes (LT) and Eoxins (EX) | 2.444932508 | 3.340642378 | 8.230507393 | 0.3803715 | 0.592162456 | 0.073322803 |
| Synthesis of Lipoxins (LX) | 2.52069981 | 3.162564407 | 8.203964027 | 0.522658595 | 0.821835732 | 0.116097164 |
| Synthesis of PA | 2.749605197 | 2.50242712 | 8.001637515 | 0.250983722 | 0.183573416 | 0.019219555 |
| Synthesis of PC | 2.199852241 | 3.294906911 | 7.694611392 | 0.450408613 | 0.99381946 | 0.124888462 |
| Synthesis of PE | 2.349656149 | 0.698970004 | 5.398282301 | -0.481085526 | 0.51054501 | 0.050994937 |
| Synthesis of PG | 2.19929505 | 1.079181246 | 5.477771347 | -0.414086051 | 0.16993095 | 0.015724321 |
| Synthesis of PI | 2.289764563 | 0.301029996 | 4.880559122 | -0.457808219 | 0.10898251 | 0.009840733 |
| Synthesis of PIPs at the early endosome membrane | 2.21507991 | 1.556302501 | 5.986462321 | -0.333333333 | 0.386112802 | 0.034364131 |
| Synthesis of PIPs at the ER membrane | 1.872813254 | 0.698970004 | 4.444596512 | -0.607671233 | 0.440183974 | 0.045517921 |
| Synthesis of PIPs at the Golgi membrane | 2.42084935 | 1.838849091 | 6.680547791 | -0.333424883 | 0.856305576 | 0.083886693 |
| Synthesis of PIPs at the late endosome membrane | 2.314828675 | 1.491361694 | 6.121019044 | -0.333150535 | 0.216470421 | 0.019636639 |
| Synthesis of PIPs at the plasma membrane | 2.790511442 | 3.53058386 | 9.111606743 | 0.396736818 | 1.386498966 | 0.190271423 |
| Synthesis of PIPs in the nucleus | 2.527219256 | 1.491361694 | 6.545800206 | 0.516853933 | 0.354083153 | 0.042190514 |
| Synthesis of Prostaglandins (PG) and Thromboxanes (TX) | 2.381218855 | 2.977723605 | 7.740161316 | 0.299725275 | 0.207098892 | 0.022245982 |
| Synthesis of pyrophosphates in the cytosol | 2.049018232 | 0 | 4.098036464 | -0.874794521 | 1.368676528 | 0.181113282 |
| Synthesis of substrates in N-glycan biosythesis | -0.587555066 | 4 | 0.587555066 | |||
| Synthesis of UDP-N-acetyl-glucosamine | 1.9113644 | 0 | 3.822728799 | -0.787671233 | 1.01499705 | 0.119654121 |
| Synthesis of very long-chain fatty acyl-CoAs | 2.653644726 | 1.612783857 | 6.920073309 | 0.317305139 | 0.177607123 | 0.017589812 |
| Synthesis, secretion, and deacylation of Ghrelin | 2.636702805 | 3.048441804 | 8.321847413 | 0.415079057 | 0.640916928 | 0.082895292 |
| Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) | 2.395677315 | 2.703291378 | 7.494646009 | 0.46385563 | 0.750311721 | 0.093614147 |
| Synthesis, secretion, and inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) | 2.166681689 | 2.413299764 | 6.746663143 | 0.342559902 | 0.171697883 | 0.017162592 |
| t(4;14) translocations of FGFR3 | 2.517271785 | 2.161368002 | 7.195911571 | 0.916735141 | 0.744633073 | 0.132190301 |
| T41 mutants of beta-catenin aren't phosphorylated | 3.063502619 | 2.416640507 | 8.543645745 | 0.46828187 | 0.791988351 | 0.109967218 |
| Tachykinin receptors bind tachykinins | 3.211847186 | 3.467015818 | 9.890710191 | 0.806464393 | 2.416757507 | 0.479716299 |
| TAK1 activates NFkB by phosphorylation and activation of IKKs complex | 2.763561539 | 3.002597981 | 8.529721058 | 0.384326673 | 1.016494178 | 0.130289699 |
| Tandem pore domain potassium channels | 0.337616587 | 0.160278439 | 0.013528165 | |||
| Tat-mediated elongation of the HIV-1 transcript | 2.906266717 | 2.604226053 | 8.416759486 | 0.559254361 | 1.514537726 | 0.225037 |
| Tat-mediated HIV elongation arrest and recovery | 3.055459709 | 2.472756449 | 8.583675867 | 0.709364615 | 1.918797679 | 0.325238437 |
| TBC/RABGAPs | 2.767683396 | 2.322219295 | 7.857586086 | 0.376405231 | 0.134386581 | 0.015927047 |
| TCF dependent signaling in response to WNT | 2.720052108 | 2.469822016 | 7.909926232 | 0.539610795 | 4 | 0.558369562 |
| TCR signaling | 0.561940451 | 4 | 0.561940451 | |||
| Telomere C-strand (Lagging Strand) Synthesis | 3.631201884 | 1.380211242 | 8.64261501 | 0.368398064 | 0.455793181 | 0.058177669 |
| Telomere C-strand synthesis initiation | 4.200290989 | 1.477121255 | 9.877703232 | 0.48723579 | 0.42845534 | 0.067877825 |
| Telomere Extension By Telomerase | 2.654421385 | 2.742725131 | 8.051567901 | 0.481019134 | 1.125262716 | 0.150892359 |
| Telomere Maintenance | 0.328928851 | 0.566675835 | 0.046599008 | |||
| Termination of O-glycan biosynthesis | 2.650991311 | 0.301029996 | 5.603012618 | -0.58513847 | 1.218444921 | 0.140767449 |
| Termination of translesion DNA synthesis | 2.735859186 | 2.62838893 | 8.100107302 | 0.450924157 | 0.858507854 | 0.112429657 |
| Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation | 2.548528336 | 3.196452542 | 8.293509214 | 0.488792127 | 0.759409105 | 0.104940647 |
| TFAP2 (AP-2) family regulates transcription of cell cycle factors | 2.14088056 | 0.698970004 | 4.980731125 | -0.437106056 | 0.210508824 | 0.018735416 |
| TFAP2 (AP-2) family regulates transcription of growth factors and their receptors | 2.822329814 | 3.339053736 | 8.983713363 | 0.587408897 | 1.405248021 | 0.224019155 |
| TFAP2A acts as a transcriptional repressor during retinoic acid induced cell differentiation | 2.987333392 | 0.84509804 | 6.819764825 | -0.535890411 | 0.275702736 | 0.034481373 |
| TGF-beta receptor signaling activates SMADs | 2.515552394 | 2.756636108 | 7.787740897 | 0.451569953 | 1.133443574 | 0.143960284 |
| TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) | 2.65478967 | 2.742725131 | 8.052304471 | 0.460970071 | 0.733606027 | 0.096541521 |
| TGFBR1 KD Mutants in Cancer | 2.431345472 | 2.506505032 | 7.369195976 | 0.421723816 | 0.320184867 | 0.037744213 |
| TGFBR1 LBD Mutants in Cancer | 2.454165024 | 2.064457989 | 6.972788038 | 0.583431462 | 0.812055914 | 0.107989982 |
| TGFBR2 Kinase Domain Mutants in Cancer | 2.329200932 | 1.913813852 | 6.572215717 | 0.536659304 | 0.423656973 | 0.051673701 |
| TGFBR2 MSI Frameshift Mutants in Cancer | 2.341358402 | 1.255272505 | 5.93798931 | 0.418594251 | 0.047397666 | 0.004693918 |
| The activation of arylsulfatases | 2.183075319 | 1.995635195 | 6.361785832 | 0.410073766 | 0.204463434 | 0.021148358 |
| The AIM2 inflammasome | 2.037758172 | 1.361727836 | 5.437244181 | 0.544234456 | 0.044719975 | 0.004842254 |
| The canonical retinoid cycle in rods (twilight vision) | 2.133700726 | 1.505149978 | 5.77255143 | -0.285048011 | 0.212187604 | 0.017018655 |
| The citric acid (TCA) cycle and respiratory electron transport | -0.264925325 | 1.590179454 | 0.105319702 | |||
| The IPAF inflammasome | 2.037758172 | 1.361727836 | 5.437244181 | 0.544234456 | 0.044719975 | 0.004842254 |
| The NLRP1 inflammasome | 3.949225406 | 3.725503269 | 11.62395408 | 0.913099293 | 2.859700336 | 0.669682755 |
| The NLRP3 inflammasome | 2.433806137 | 2.893206753 | 7.760819027 | 0.466298169 | 0.966141733 | 0.124154269 |
| The phototransduction cascade | 0.337516117 | 0.513951981 | 0.043366769 | |||
| The retinoid cycle in cones (daylight vision) | 1.246960789 | 0 | 2.493921578 | 0.096232 | ||
| The role of GTSE1 in G2/M progression after G2 checkpoint | 3.104674845 | 3.566908655 | 9.776258345 | 0.740758243 | 4 | 0.755225791 |
| The role of Nef in HIV-1 replication and disease pathogenesis | 0.456097086 | 0.98285457 | 0.112069276 | |||
| Threonine catabolism | 0.7 | 0 | 1.4 | 0 | ||
| Thrombin signalling through proteinase activated receptors (PARs) | 2.512437507 | 3.486855355 | 8.51173037 | 0.508006652 | 1.559667278 | 0.22366177 |
| Thromboxane signalling through TP receptor | 2.105011375 | 1.079181246 | 5.289203996 | -0.453399123 | 0.398834539 | 0.03789555 |
| Thyroxine biosynthesis | 1.991863383 | 1.939519253 | 5.923246018 | -0.326072671 | 0.184285738 | 0.016083968 |
| TICAM1-dependent activation of IRF3/IRF7 | 2.613755312 | 2.161368002 | 7.388878627 | 0.416981646 | 0.30324015 | 0.035643953 |
| TICAM1 deficiency - HSE | 2.168541728 | 0 | 4.337083456 | 0.258374475 | ||
| TICAM1, RIP1-mediated IKK complex recruitment | 2.876255204 | 2.580924976 | 8.333435384 | 0.443631011 | 1.265760917 | 0.168418398 |
| TICAM1,TRAF6-dependent induction of TAK1 complex | 2.692104492 | 1.86923172 | 7.253440704 | 0.47651661 | 0.489986037 | 0.060385259 |
| Tie2 Signaling | 3.170885844 | 3.481155871 | 9.822927559 | 0.513743142 | 2.043120152 | 0.329006634 |
| Tight junction interactions | 2.964120481 | 2.075546961 | 8.003787923 | 0.892906053 | 0.659202877 | 0.121929367 |
| TLR3-mediated TICAM1-dependent programmed cell death | 2.411309681 | 1.602059991 | 6.424679354 | 0.405727137 | 0.194500037 | 0.020169902 |
| TLR3 deficiency - HSE | 2.168541728 | 0 | 4.337083456 | 0.258374475 | ||
| TNF receptor superfamily (TNFSF) members mediating non-canonical NF-kB pathway | 2.729356545 | 2.243038049 | 7.701751139 | 0.507446065 | 1.065771432 | 0.141627067 |
| TNF signaling | 2.483406768 | 2.23299611 | 7.199809646 | 0.374861458 | 0.950202532 | 0.104370383 |
| TNFR1-induced NFkappaB signaling pathway | 2.694860623 | 2.924279286 | 8.314000531 | 0.470306552 | 1.549757573 | 0.210985272 |
| TNFR1-induced proapoptotic signaling | 2.453739197 | 2.322219295 | 7.229697688 | 0.374759945 | 0.303625906 | 0.033463501 |
| TNFR1-mediated ceramide production | 2.655587733 | 1.707570176 | 7.018745641 | 0.415182242 | 0.119286234 | 0.01342507 |
| TNFR2 non-canonical NF-kB pathway | 3.307063475 | 3.04453976 | 9.65866671 | 0.670184584 | 4 | 0.713872507 |
| TNFs bind their physiological receptors | 2.776727022 | 0.84509804 | 6.398552084 | -0.392396708 | 0.655885104 | 0.066702203 |
| Toll-like Receptor Cascades | 0.331022236 | 2.650062084 | 0.219307369 | |||
| Toll Like Receptor 10 (TLR10) Cascade | 0.371919355 | 2.483573285 | 0.230922243 | |||
| Toll Like Receptor 2 (TLR2) Cascade | 0.3934254 | 3.531766314 | 0.347371643 | |||
| Toll Like Receptor 3 (TLR3) Cascade | 2.168541728 | 0 | 4.337083456 | 0.356189267 | 2.292958649 | 0.161847824 |
| Toll Like Receptor 4 (TLR4) Cascade | 3.119912434 | 2.217483944 | 8.457308812 | 0.362311576 | 3.177819604 | 0.395605936 |
| Toll Like Receptor 5 (TLR5) Cascade | 1.536729139 | 0 | 3.073458279 | 0.371919355 | 2.483573285 | 0.139710018 |
| Toll Like Receptor 7/8 (TLR7/8) Cascade | 2.237238985 | 1.230448921 | 5.704926891 | 0.362458954 | 2.415535489 | 0.215006561 |
| Toll Like Receptor 9 (TLR9) Cascade | 2.666010887 | 1.568201724 | 6.900223498 | 0.369867272 | 2.702593709 | 0.284723711 |
| Toll Like Receptor TLR1:TLR2 Cascade | 4.199096823 | 1.763427994 | 10.16162164 | 0.3934254 | 3.531766314 | 0.531036327 |
| Toll Like Receptor TLR6:TLR2 Cascade | 3.00690718 | 1.69019608 | 7.704010441 | 0.398779751 | 3.38467959 | 0.403903138 |
| TP53 Regulates Metabolic Genes | 2.740471819 | 3.582404298 | 9.063347936 | 0.479922269 | 3.561570516 | 0.523577004 |
| TP53 regulates transcription of additional cell cycle genes whose exact role in the p53 pathway remain uncertain | 2.355148202 | 2.522444234 | 7.232740638 | -0.182146729 | 0.151267675 | 0.013035571 |
| TP53 Regulates Transcription of Caspase Activators and Caspases | 2.401844494 | 2.478566496 | 7.282255484 | 0.474348422 | 0.706278017 | 0.087111866 |
| TP53 Regulates Transcription of Cell Cycle Genes | 0.308655095 | 0.643806625 | 0.049678549 | |||
| TP53 Regulates Transcription of Cell Death Genes | 0.377178259 | 0.803163711 | 0.075733972 | |||
| TP53 Regulates Transcription of Death Receptors and Ligands | 2.619509035 | 2.387389826 | 7.626407896 | 0.267311459 | 0.026122312 | 0.002661821 |
| TP53 Regulates Transcription of DNA Repair Genes | 2.60641193 | 3.006893708 | 8.219717568 | 0.458359863 | 1.847252706 | 0.246481686 |
| TP53 Regulates Transcription of Genes Involved in Cytochrome C Release | 3.01782988 | 2.45331834 | 8.4889781 | 0.476412507 | 0.627740051 | 0.08735681 |
| TP53 Regulates Transcription of Genes Involved in G1 Cell Cycle Arrest | 2.279118826 | 2.907948522 | 7.466186173 | 0.675664755 | 2.806880789 | 0.423491203 |
| TP53 Regulates Transcription of Genes Involved in G2 Cell Cycle Arrest | 2.74043836 | 3.003029471 | 8.48390619 | 0.292532637 | 0.18528558 | 0.021513584 |
| TP53 regulates transcription of several additional cell death genes whose specific roles in p53-dependent apoptosis remain uncertain | 2.265119754 | 2.392696953 | 6.922936462 | 0.39279653 | 0.169278657 | 0.018368616 |
| TRAF3-dependent IRF activation pathway | 2.2353545 | 2.23299611 | 6.703705111 | 0.494572811 | 0.580555872 | 0.068741101 |
| TRAF3 deficiency - HSE | 2.168541728 | 0 | 4.337083456 | 0.258374475 | ||
| TRAF6-mediated induction of TAK1 complex within TLR4 complex | 3.216216745 | 2.158362492 | 8.590795983 | 0.565483973 | 1.457881897 | 0.221026431 |
| TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation | 2.352080729 | 2.274157849 | 6.978319307 | 0.374393251 | 2.817231584 | 0.301232468 |
| TRAF6 mediated IRF7 activation | 2.204252968 | 2.23299611 | 6.641502046 | 0.425278534 | 0.402378284 | 0.043835744 |
| TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling | 2.949499163 | 2.350248018 | 8.249246345 | 0.417583478 | 0.616103069 | 0.079301883 |
| TRAF6 mediated NF-kB activation | 2.593094101 | 2.882524538 | 8.068712739 | 0.355156035 | 0.450315855 | 0.053400013 |
| Trafficking and processing of endosomal TLR | 2.329513132 | 2.757396029 | 7.416422292 | 0.386981598 | 0.544797338 | 0.062188005 |
| Trafficking of AMPA receptors | 2.503551448 | 3.033423755 | 8.040526652 | 0.466836346 | 2.038346413 | 0.26942858 |
| Trafficking of GluR2-containing AMPA receptors | 2.216811931 | 2.984527313 | 7.418151175 | 0.43752317 | 0.78406011 | 0.094470518 |
| TRAIL signaling | 2.906953739 | 0.77815125 | 6.592058729 | 0.288149586 | 0.076488259 | 0.006972885 |
| trans-Golgi Network Vesicle Budding | 3.642168736 | 0 | 7.284337472 | 0.250321738 | 0.109889342 | 0.010479362 |
| Transcription-Coupled Nucleotide Excision Repair (TC-NER) | 3.110284468 | 2.1430148 | 8.363583735 | 0.247711369 | 0.159500453 | 0.017378518 |
| Transcription of E2F targets under negative control by DREAM complex | 2.483720293 | 2.403120521 | 7.370561108 | 0.169297253 | 0.003437057 | 0.000296779 |
| Transcription of E2F targets under negative control by p107 (RBL1) and p130 (RBL2) in complex with HDAC1 | 2.497140672 | 2.57054294 | 7.564824284 | 0.530796947 | 0.85032427 | 0.113977278 |
| Transcription of SARS-CoV-1 sgRNAs | 5.18567279 | 2.396199347 | 12.76754493 | 1 | ||
| Transcription of the HIV genome | 2.549132026 | 2.029383778 | 7.127647831 | 0.380301652 | 0.592162456 | 0.064894774 |
| Transcriptional activation of mitochondrial biogenesis | 2.553131622 | 2.661812686 | 7.768075929 | 0.226233493 | 0.135263536 | 0.013335733 |
| Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer | 0.333521568 | 0.604765885 | 0.050425617 | |||
| Transcriptional Regulation by E2F6 | 2.436650486 | 2.926856709 | 7.800157681 | 0.201446264 | 0.05310596 | 0.005093186 |
| Transcriptional Regulation by MECP2 | 2.488726466 | 1.982271233 | 6.959724164 | 0.459793619 | 3.202736384 | 0.375873822 |
| Transcriptional regulation by RUNX1 | 0.498028575 | 4 | 0.498028575 | |||
| Transcriptional regulation by RUNX2 | 2.519688784 | 2.918030337 | 7.957407906 | 0.611292219 | 4 | 0.596659814 |
| Transcriptional regulation by RUNX3 | 0.63371731 | 4 | 0.63371731 | |||
| Transcriptional regulation by small RNAs | 2.643347324 | 1.681241237 | 6.967935885 | 0.569630588 | 0.743521209 | 0.097546817 |
| Transcriptional regulation by the AP-2 (TFAP2) family of transcription factors | 0.342459262 | 0.478529018 | 0.040969174 | |||
| Transcriptional Regulation by TP53 | 0.344157239 | 4 | 0.344157239 | |||
| Transcriptional Regulation by VENTX | 2.494226139 | 3.056904851 | 8.045357129 | 0.445716094 | 1.169977646 | 0.151631729 |
| Transcriptional regulation of granulopoiesis | 2.42931504 | 3.118925753 | 7.977555834 | 0.31433696 | 0.481186443 | 0.054039836 |
| Transcriptional regulation of pluripotent stem cells | 2.229702176 | 1.698970004 | 6.158374357 | 0.314920171 | 0.201757541 | 0.017939431 |
| Transcriptional regulation of white adipocyte differentiation | 2.503425482 | 3.101747074 | 8.108598038 | 0.316145967 | 0.79423807 | 0.090719155 |
| Transfer of LPS from LBP carrier to CD14 | 4.44949331 | 0.84509804 | 9.74408466 | 0.74938373 | 0.315723947 | 0.059820115 |
| Transferrin endocytosis and recycling | 2.243478108 | 0.477121255 | 4.964077471 | -0.459829997 | 0.606137904 | 0.055538189 |
| Translation | 0.774026186 | 4 | 0.774026186 | |||
| Translation initiation complex formation | 3.893008999 | 2.593286067 | 10.37930407 | 0.787218394 | 4 | 0.809566487 |
| Translation of Replicase and Assembly of the Replication Transcription Complex | 4.110874628 | 2.419955748 | 10.641705 | -0.35260274 | 0.002273798 | 0.000344364 |
| Translation of structural proteins | -0.20149392 | 0.562439536 | 0.028332037 | |||
| Translesion Synthesis by POLH | 3.038556833 | 2.161368002 | 8.238481669 | 0.569941517 | 0.972246931 | 0.143524264 |
| Translesion synthesis by POLI | 2.793528815 | 2.460897843 | 8.047955472 | 0.546303412 | 0.637203727 | 0.090616214 |
| Translesion synthesis by POLK | 2.950974368 | 2.071882007 | 7.973830744 | 0.545309179 | 0.637203727 | 0.089927851 |
| Translesion synthesis by REV1 | 3.20432309 | 1.361727836 | 7.770374015 | 0.50420412 | 0.321082893 | 0.042821769 |
| Translesion synthesis by Y family DNA polymerases bypasses lesions on DNA template | 0.476383534 | 1.118828606 | 0.133247881 | |||
| Translocation of SLC2A4 (GLUT4) to the plasma membrane | 2.87454609 | 3.200029267 | 8.949121447 | 0.549709419 | 4 | 0.617030082 |
| Translocation of ZAP-70 to Immunological synapse | 2.580893454 | 2.694605199 | 7.856392106 | 0.42544189 | 0.492398197 | 0.061367997 |
| Transmission across Chemical Synapses | 0.389709501 | 4 | 0.389709501 | |||
| Transport and synthesis of PAPS | 3.942700933 | 0 | 7.885401865 | -0.683835616 | 0.638716102 | 0.100472722 |
| Transport of bile salts and organic acids, metal ions and amine compounds | 2.826930608 | 1.77815125 | 7.432012466 | 0.276347082 | 0.480627183 | 0.048312949 |
| Transport of connexons to the plasma membrane | 0.687122022 | 4 | 0.687122022 | |||
| Transport of fatty acids | 2.048381941 | 0 | 4.096763883 | -0.804383562 | 1.064684834 | 0.13149872 |
| Transport of gamma-carboxylated protein precursors from the endoplasmic reticulum to the Golgi apparatus | 2.54957959 | 3.197831693 | 8.296990873 | 0.722352642 | 1.993215788 | 0.333718551 |
| Transport of HA trimer, NA tetramer and M2 tetramer from the endoplasmic reticulum to the Golgi Apparatus | 3.395472905 | 1.113943352 | 7.904889162 | 0.572233425 | ||
| Transport of inorganic cations/anions and amino acids/oligopeptides | 2.682926964 | 0.698970004 | 6.064823931 | -0.262557407 | 1.901513223 | 0.154334226 |
| Transport of Mature mRNA derived from an Intron-Containing Transcript | 3.537404292 | 1.079181246 | 8.153989831 | 0.614980955 | 0.726367027 | 0.110525038 |
| Transport of Mature mRNA Derived from an Intronless Transcript | 3.165092201 | 1.707570176 | 8.037754577 | 0.724109589 | 0.750297964 | 0.123276491 |
| Transport of Mature mRNAs Derived from Intronless Transcripts | 0.724109589 | 0.750297964 | 0.135824488 | |||
| Transport of Mature Transcript to Cytoplasm | 0.625531001 | 1.005180513 | 0.157192893 | |||
| Transport of nucleosides and free purine and pyrimidine bases across the plasma membrane | 3.13970818 | 1.380211242 | 7.659627602 | 0.218905027 | 0.018549203 | 0.001785843 |
| Transport of organic anions | 3.241958262 | 2.8162413 | 9.300157824 | 0.3874394 | 0.546503802 | 0.075691425 |
| Transport of Ribonucleoproteins into the Host Nucleus | 2.544336894 | 1.832508913 | 6.921182702 | 0.688219178 | 0.667839008 | 0.097114695 |
| Transport of small molecules | 0.297472555 | 4 | 0.297472555 | |||
| Transport of the SLBP Dependant Mature mRNA | 3.165092201 | 1.707570176 | 8.037754577 | 0.724109589 | 0.750297964 | 0.123276491 |
| Transport of the SLBP independent Mature mRNA | 3.165092201 | 1.707570176 | 8.037754577 | 0.724109589 | 0.750297964 | 0.123276491 |
| Transport of vitamins, nucleosides, and related molecules | 3.622125698 | 0.77815125 | 8.022402647 | 0.256544011 | 0.209238368 | 0.02210806 |
| Transport to the Golgi and subsequent modification | 0.292646352 | 0.80792764 | 0.059109269 | |||
| TRIF-mediated programmed cell death | 3.244309406 | 2.120573931 | 8.609192743 | 0.590784421 | 1.232010197 | 0.190971112 |
| Triglyceride biosynthesis | 2.41601167 | 1.716003344 | 6.548026684 | 0.406598193 | 0.275349936 | 0.029022159 |
| Triglyceride catabolism | 2.396890716 | 2.198657087 | 6.992438519 | 0.356275453 | 0.600207986 | 0.062927227 |
| Triglyceride metabolism | 0.337265094 | 0.636672263 | 0.053681833 | |||
| Tristetraprolin (TTP, ZFP36) binds and destabilizes mRNA | 3.447738891 | 2.193124598 | 9.088602381 | 0.786849315 | 1.020838637 | 0.189568646 |
| TRKA activation by NGF | 1.689724437 | 2.546542663 | 5.925991537 | 0.706537894 | 0.712509403 | 0.096096783 |
| tRNA Aminoacylation | -0.175553595 | 0.009340026 | 0.000409919 | |||
| tRNA modification in the mitochondrion | 2.162841658 | 1.977723605 | 6.303406921 | 0.742810189 | 0.310480891 | 0.044793876 |
| tRNA modification in the nucleus and cytosol | 2.963966285 | 0.77815125 | 6.70608382 | 0.390946808 | 0.083373678 | 0.008794513 |
| tRNA processing | 0.495005511 | 0.952731918 | 0.117901888 | |||
| tRNA processing in the mitochondrion | 2.162841658 | 1.977723605 | 6.303406921 | 0.742810189 | 0.310480891 | 0.044793876 |
| tRNA processing in the nucleus | 2.905859205 | 1.643452676 | 7.455171087 | 0.576279378 | 0.783331763 | 0.108343286 |
| TRP channels | 2.605678012 | 3.080265627 | 8.291621652 | 0.265664817 | 0.210571594 | 0.023220147 |
| Truncations of AMER1 destabilize the destruction complex | 2.914767738 | 2.164352856 | 7.993888332 | 0.444696194 | 0.577408986 | 0.0743766 |
| Tryptophan catabolism | 1.807507925 | 1.968482949 | 5.583498798 | -0.390552046 | 1.417343897 | 0.128915135 |
| TWIK-related spinal cord K+ channel (TRESK) | 2.590849114 | 0 | 5.181698228 | 0.332675019 | ||
| TWIK-releated acid-sensitive K+ channel (TASK) | 2.815531504 | 1.301029996 | 6.932093004 | 0.60630137 | 0.386856932 | 0.052348441 |
| TWIK related potassium channel (TREK) | 3.354037509 | 1.204119983 | 7.912195001 | 0.36681256 | 0.054484113 | 0.006430317 |
| Type I hemidesmosome assembly | 3.558515051 | 0.301029996 | 7.418060097 | -0.401479857 | 0.150005691 | 0.017398003 |
| Type II Na+/Pi cotransporters | 2.194461218 | 0 | 4.388922436 | 0.262934737 | ||
| Tyrosine catabolism | 2.487924246 | 0.477121255 | 5.452969747 | -0.499726027 | 0.201181731 | 0.020705365 |
| TYSND1 cleaves peroxisomal proteins | 3.021683198 | 0.84509804 | 6.888464436 | -0.331049603 | 0.035912556 | 0.003603759 |
| Ub-specific processing proteases | 2.916633651 | 3.62930764 | 9.462574942 | 0.50614871 | 4 | 0.621738361 |
| Ubiquitin-dependent degradation of Cyclin D | 3.220044332 | 3.089905111 | 9.529993775 | 0.793072971 | 4 | 0.768678572 |
| Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | 3.298840352 | 2.969415912 | 9.567096616 | 0.765606497 | 4 | 0.75685944 |
| UCH proteinases | 3.152959543 | 2.956648579 | 9.262567665 | 0.689501226 | 4 | 0.703096412 |
| Unblocking of NMDA receptors, glutamate binding and activation | 2.219759166 | 3.616055195 | 8.055573526 | 0.502803352 | 1.878620262 | 0.257126597 |
| UNC93B1 deficiency - HSE | 2.168541728 | 0 | 4.337083456 | 0.258374475 | ||
| Uncoating of the HIV Virion | 3.297254698 | 2.133538908 | 8.728048305 | 0.97863599 | 1.277057136 | 0.261824933 |
| Uncoating of the Influenza Virion | 2.943361751 | 1.51851394 | 7.405237442 | 0.528279191 | ||
| Unfolded Protein Response (UPR) | 0.269483557 | 0.317727009 | 0.021405551 | |||
| Unwinding of DNA | 3.504540571 | 0.301029996 | 7.310111137 | 0.420810859 | 0.129386561 | 0.015139057 |
| Uptake and actions of bacterial toxins | 0.518018682 | 2.418462703 | 0.313202216 | |||
| Uptake and function of anthrax toxins | 2.676809855 | 2.678518379 | 8.032138088 | 0.649012075 | 2.288700074 | 0.354390907 |
| Uptake and function of diphtheria toxin | 3.232335057 | 2.80140371 | 9.266073823 | 0.58782022 | 1.064017305 | 0.173551008 |
| Urea cycle | 1.962906643 | 0.602059991 | 4.527873277 | -0.585913949 | 0.611819829 | 0.062259378 |
| Utilization of Ketone Bodies | 3.013741675 | 0 | 6.027483351 | 0.407078519 | ||
| Vasopressin-like receptors | 2.781286305 | 2.942504106 | 8.505076717 | 0.842653509 | 2.855126109 | 0.528622482 |
| Vasopressin regulates renal water homeostasis via Aquaporins | 3.009872089 | 2.937517892 | 8.95726207 | 0.545228585 | 3.056761612 | 0.470137297 |
| VEGF binds to VEGFR leading to receptor dimerization | 2.541540752 | 3.228400359 | 8.311481864 | 0.614477653 | 1.263057762 | 0.194674748 |
| VEGF ligand-receptor interactions | 2.501160204 | 0.84509804 | 5.847418449 | 0.614477653 | 1.263057762 | 0.154535101 |
| VEGFA-VEGFR2 Pathway | 2.676214165 | 4.028408565 | 9.380836895 | 0.450953783 | 4 | 0.5899241 |
| VEGFR2 mediated cell proliferation | 2.488043125 | 3.56937391 | 8.54546016 | 0.463170495 | 1.723708251 | 0.238275118 |
| VEGFR2 mediated vascular permeability | 2.8289612 | 3.543571424 | 9.201493824 | 0.496548626 | 2.119864899 | 0.319818381 |
| Vesicle-mediated transport | 0.282734134 | 2.043723433 | 0.144457594 | |||
| Vif-mediated degradation of APOBEC3G | 3.411498386 | 2.813580989 | 9.63657776 | 0.763053134 | 4 | 0.759167232 |
| Viral Messenger RNA Synthesis | 2.555470015 | 1.322219295 | 6.433159324 | 0.533930185 | 0.217659381 | 0.026083435 |
| Viral mRNA Translation | 4.363242568 | 2.910624405 | 11.63710954 | 0.918316087 | 4 | 0.940004461 |
| Viral RNP Complexes in the Host Cell Nucleus | 2.574906547 | 1.361727836 | 6.51154093 | -0.628219178 | 0.475988804 | 0.063131851 |
| Virus Assembly and Release | 0.881128458 | 0.618515789 | 0.136247966 | |||
| Visual phototransduction | 0.238489603 | 0.220239979 | 0.013131236 | |||
| Vitamin B1 (thiamin) metabolism | 4.941542721 | 0.301029996 | 10.18411544 | -0.41369863 | 0.06337078 | 0.009707405 |
| Vitamin B2 (riboflavin) metabolism | 2.922648827 | 0.602059991 | 6.447357646 | 0.378390069 | 0.066841442 | 0.006722687 |
| Vitamin B5 (pantothenate) metabolism | 2.563265336 | 2.489958479 | 7.616489151 | 0.155595117 | 0.001960701 | 0.000172161 |
| Vitamin C (ascorbate) metabolism | 2.891468988 | 2.029383778 | 7.812321753 | 0.325498525 | 0.131217802 | 0.014639908 |
| Vitamin D (calciferol) metabolism | 3.023739354 | 2.713490543 | 8.760969251 | 0.367425101 | 0.334030256 | 0.043105044 |
| Vitamin E | 2.064615572 | 0 | 4.129231143 | 0.240089761 | ||
| Vitamins | 3.235010104 | 1.602059991 | 8.0720802 | 0.646107456 | 1.116813782 | 0.173101468 |
| Vitamins B6 activation to pyridoxal phosphate | 2.605200446 | 1.301029996 | 6.511430888 | 0.401813066 | 0.102957451 | 0.010741641 |
| VLDL assembly | 2.899409638 | 1.414973348 | 7.213792624 | 0.611788214 | 0.719685441 | 0.100494235 |
| VLDL clearance | 3.295959823 | 0.954242509 | 7.546162155 | 0.914817858 | 0.730697295 | 0.132842012 |
| VLDLR internalisation and degradation | 2.303965187 | 2.475671188 | 7.083601563 | 0.502175943 | 0.606491606 | 0.07536003 |
| Voltage gated Potassium channels | 2.797557615 | 3.181843588 | 8.776958818 | 0.194749129 | 0.077178079 | 0.008309536 |
| Vpr-mediated induction of apoptosis by mitochondrial outer membrane permeabilization | 3.635750365 | 0.301029996 | 7.572530726 | 0.660821918 | 0.568974178 | 0.085569331 |
| Vpr-mediated nuclear import of PICs | 2.884935624 | 1.77815125 | 7.548022499 | 0.522202382 | 0.192900142 | 0.025607255 |
| Vpu mediated degradation of CD4 | 3.384230579 | 2.797267541 | 9.565728699 | 0.793024981 | 4 | 0.770498112 |
| vRNA Synthesis | 2.49350187 | 1.62324929 | 6.61025303 | 0.741166804 | 0.308747179 | 0.045702187 |
| vRNP Assembly | 2.73190287 | 2.545307116 | 8.009112857 | 0.985209532 | 1.407475511 | 0.276669532 |
| VxPx cargo-targeting to cilium | 2.002193473 | 0 | 4.004386946 | -0.620339912 | 0.938448242 | 0.09319979 |
| Wax and plasmalogen biosynthesis | -0.852836394 | 2.040394163 | 0.4350306 | |||
| Wax biosynthesis | -0.85260274 | 1.282253385 | 0.273313187 | |||
| WNT ligand biogenesis and trafficking | 2.486149059 | 1.322219295 | 6.294517412 | 0.357828352 | 0.153016453 | 0.014694964 |
| WNT mediated activation of DVL | 2.534986933 | 2.913813852 | 7.983787719 | 0.577628803 | 1.30051267 | 0.188960921 |
| WNT5:FZD7-mediated leishmania damping | 3.219575594 | 2.120573931 | 8.55972512 | 0.300481666 | 0.130863719 | 0.015452636 |
| WNT5A-dependent internalization of FZD2, FZD5 and ROR2 | 2.698340489 | 0.477121255 | 5.873802233 | -0.402686404 | 0.278396304 | 0.026786279 |
| WNT5A-dependent internalization of FZD4 | 2.616458383 | 2.92890769 | 8.161824456 | 0.592081005 | 1.232010197 | 0.184062288 |
| XBP1(S) activates chaperone genes | 2.760781474 | 2.571708832 | 8.093271779 | 0.268779163 | 0.141292483 | 0.015274155 |
| Xenobiotics | 2.410385416 | 3.101403351 | 7.922174183 | 0.281757682 | 0.250483607 | 0.026931676 |
| YAP1- and WWTR1 (TAZ)-stimulated gene expression | 2.680473394 | 2.045322979 | 7.406269767 | 0.468773146 | 0.979930017 | 0.121748349 |
| ZBP1(DAI) mediated induction of type I IFNs | 0.312525238 | 0.29113246 | 0.02274656 | |||
| RNF mutants show enhanced WNT signaling and proliferation | 1.906585335 | 0.301029996 | 4.114200667 | -0.552054795 | 0.295397044 | 0.027233634 |
| Defective ABCA1 causes Tangier disease | 0.7 | 0 | 1.4 | 0 | ||
| Cyclin E associated events during G1/S transition | 2.466988248 | 3.25358029 | 8.187556786 | 0.621555327 | 4 | 0.613664555 |
| Toxicity of botulinum toxin type E (BoNT/E) | 2.387863218 | 0 | 4.775726437 | -0.721643836 | 0.7770188 | 0.094737114 |
| Defective ABCB11 causes progressive familial intrahepatic cholestasis 2 and benign recurrent intrahepatic cholestasis 2 | 3.296148644 | 1.763427994 | 8.355725282 | 0.611893362 | ||
| SMAC (DIABLO) binds to IAPs | 2.455607959 | 2.507855872 | 7.41907179 | 0.471749863 | 0.61901022 | 0.077239527 |
| Biosynthesis of electrophilic ω-3 PUFA oxo-derivatives | 2.231477307 | 2.688419822 | 7.151374435 | 0.505946928 | ||
| Toxicity of tetanus toxin (TeNT) | 2.389557456 | 0 | 4.779114912 | 0.297259869 | ||
| AMER1 mutants destabilize the destruction complex | 0.444696194 | 0.577408986 | 0.064192895 | |||
| Defective ABCC6 causes pseudoxanthoma elasticum (PXE) | 3.435951327 | 0.301029996 | 7.17293265 | 0.507843399 | ||
| phosphorylation site mutants of CTNNB1 are not targeted to the proteasome by the destruction complex | 0.46828187 | 0.791988351 | 0.092718447 | |||
| NR1H2 & NR1H3 regulate gene expression linked to gluconeogenesis | 2.58088664 | 2.1430148 | 7.304788081 | 0.463815789 | 0.351325076 | 0.042971755 |
| Defective ABCB6 causes isolated colobomatous microphthalmia 7 (MCOPCB7) | 2.066747458 | 0 | 4.133494915 | 0.240464844 | ||
| Formation of RNA Pol II elongation complex | 2.865608884 | 2.761927838 | 8.493145606 | 0.516537675 | 1.325163675 | 0.191128622 |
| TCF7L2 mutants don't bind CTBP | 2.914382067 | 2.049218023 | 7.877982157 | 0.820009188 | 1.168441359 | 0.203577769 |
| Defective CSF2RB causes pulmonary surfactant metabolism dysfunction 5 (SMDP5) | 3.108908384 | 0 | 6.217816768 | 0.4238221 | ||
| GRB2:SOS provides linkage to MAPK signaling for Integrins | 2.875396769 | 3.223495941 | 8.974289479 | 0.512705988 | 1.304960289 | 0.195687527 |
| AXIN mutants destabilize the destruction complex, activating WNT signaling | 0.444696194 | 0.577408986 | 0.064192895 | |||
| NR1H2 & NR1H3 regulate gene expression to limit cholesterol uptake | 2.524072232 | 2.328379603 | 7.376524068 | 0.619778418 | 0.977151789 | 0.139472687 |
| Transcriptional activation of p53 responsive genes | 0.612655269 | 0.411479407 | 0.063023757 | |||
| Toxicity of botulinum toxin type F (BoNT/F) | 1.939217739 | 0 | 3.878435478 | 0.218027331 | ||
| Toxicity of botulinum toxin type A (BoNT/A) | 2.635186719 | 1.230448921 | 6.500822359 | -0.721643836 | 0.7770188 | 0.112025054 |
| DEx/H-box helicases activate type I IFN and inflammatory cytokines production | 2.264238834 | 2.315970345 | 6.844448014 | 0.354848369 | 0.143709786 | 0.014766943 |
| Cytosolic sensors of pathogen-associated DNA | 2.203712586 | 1.556302501 | 5.963727673 | 0.325140145 | 0.624406579 | 0.054749763 |
| truncated APC mutants destabilize the destruction complex | 0.444696194 | 0.577408986 | 0.064192895 | |||
| WNT ligand secretion is abrogated by the PORCN inhibitor LGK974 | 3.432191895 | 1.255272505 | 8.119656295 | 0.91125719 | 0.721649166 | 0.136213546 |
| TRIF(TICAM1)-mediated TLR4 signaling | 0.375641506 | 2.932213515 | 0.275365275 | |||
| Defective ABCC2 causes Dubin-Johnson syndrome | 2.928223009 | 1.707570176 | 7.564016194 | 0.542246917 | ||
| Platelet degranulation | 2.637047185 | 2.949390007 | 8.223484378 | 0.266118007 | 0.462136716 | 0.050580791 |
| HHAT G278V abrogates palmitoylation of Hh-Np | 2.579502725 | 1.146128036 | 6.305133485 | 0.59545952 | 0.350781567 | 0.044154982 |
| Misspliced GSK3beta mutants stabilize beta-catenin | 3.063502619 | 2.416640507 | 8.543645745 | 0.46828187 | 0.791988351 | 0.109967218 |
| Transcriptional activation of cell cycle inhibitor p21 | 2.134380652 | 2.687528961 | 6.956290266 | 0.612655269 | 0.411479407 | 0.056135515 |
| Misspliced LRP5 mutants have enhanced beta-catenin-dependent signaling | 1.994489694 | 0 | 3.988979387 | 0.22775185 | ||
| Activation of BAD and translocation to mitochondria | 3.434566515 | 3.515476441 | 10.38460947 | 0.607060859 | 2.877830509 | 0.517837845 |
| MyD88-independent TLR4 cascade | 3.657116572 | 1.851258349 | 9.165491493 | 0.375641506 | 2.932213515 | 0.396698047 |
| Hh mutants that don't undergo autocatalytic processing are degraded by ERAD | 3.338979272 | 2.830588669 | 9.508547212 | 0.810828406 | 4 | 0.77644988 |
| XAV939 inhibits tankyrase, stabilizing AXIN | 2.324402569 | 1.361727836 | 6.010532973 | 0.646849315 | 0.534012074 | 0.068620648 |
| SMAC(DIABLO)-mediated dissociation of IAP:caspase complexes | 2.455607959 | 2.507855872 | 7.41907179 | 0.471749863 | 0.61901022 | 0.077239527 |
| Cleavage of the damaged pyrimidine | 4.182684107 | 1.397940009 | 9.763308222 | 0.549246385 | 0.843447774 | 0.138916223 |
| Activation of BIM and translocation to mitochondria | 2.52679244 | 2.506505032 | 7.560089912 | 0.687123288 | 0.661356881 | 0.101531185 |
| Defective ABCC8 can cause hypoglycemias and hyperglycemias | 2.240951409 | 0.698970004 | 5.180872823 | 0.332602409 | ||
| Defective ABCC9 causes dilated cardiomyopathy 10, familial atrial fibrillation 12 and hypertrichotic osteochondrodysplasia | 2.328003179 | 0.698970004 | 5.354976361 | 0.347918252 | ||
| Toxicity of botulinum toxin type B (BoNT/B) | 1.499401263 | 0 | 2.998802526 | 0.140646246 | ||
| Defective CSF2RA causes pulmonary surfactant metabolism dysfunction 4 (SMDP4) | 3.108908384 | 0 | 6.217816768 | 0.4238221 |