SMN2
![By Emw (Own work) [CC BY-SA 3.0 (https://creativecommons.org/licenses/by-sa/3.0) or GFDL (http://www.gnu.org/copyleft/fdl.html)], via Wikimedia Commons](https://doctortarget.com/wp-content/uploads/2017/12/Protein_SMN1_PDB_1g5v-300x214.png)
By Emw (Own work) [CC BY-SA 3.0 (https://creativecommons.org/licenses/by-sa/3.0) or GFDL (http://www.gnu.org/copyleft/fdl.html)], via Wikimedia Commons
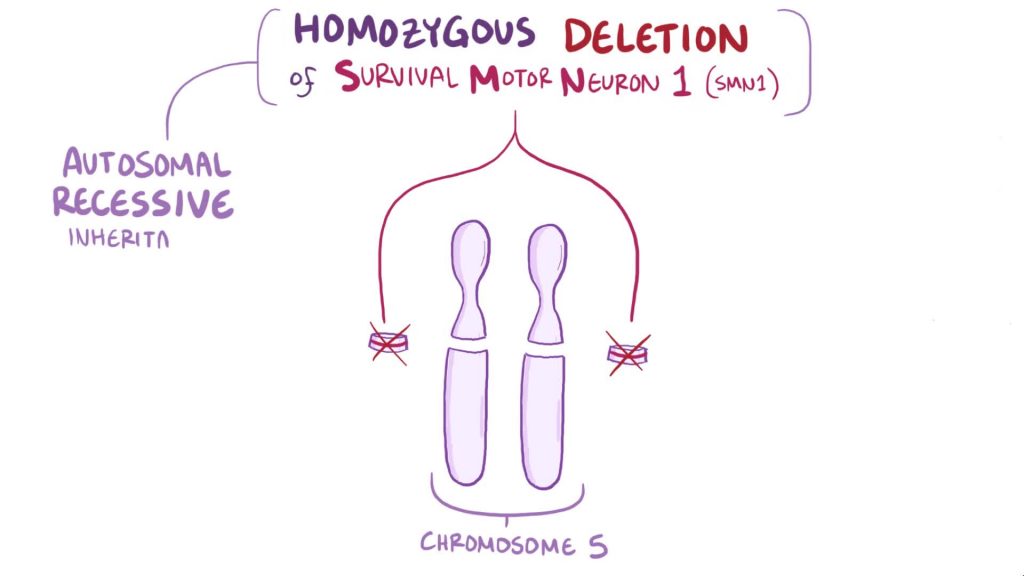
Note that the nine exons of both the telomeric and centromeric copies are designated historically as exon 1, 2a, 2b, and 3-8. It is thought that gene conversion events may involve the two genes, leading to varying copy numbers of each gene.[6]
Clinical significance
While mutations in the telomeric copy are associated with spinal muscular atrophy, mutations in this gene, the centromeric copy, do not lead to disease. This gene may be a modifier of disease caused by mutation in the telomeric copy. Description taken from Wikipedia.
- Spinal muscular atrophy (SMA) is caused by insufficient levels of the survival motor neuron protein SMN. The SMN locus on chromosome 5q13 contains two inverted copies of SMN called SMN1 and SMN2 which are 99% identical at the amino acid level. SMN1 is a fully functional protein and SMN2 skips axon 7 90% of the time. Skipping of exon 7 produces non-functional protein product. 10% of the SMN2 protein includes exon 7 and is fully functional. In the SMA disease state, mutations in the SMN1 locus are the cause of the disease state. Because only 10% of SMN2 is of the fully functional form, it is not sufficient to overcome the deficiency produced by the loss of the SMN1 product. A therapy that either increase the amount of SMN2 product made or to increase the inclusion of exon 7 has been proposed for the treatment of SMA.
- SMN2 assay has been designed to identify small molecules that can increase the amount of functional SMN2 product by appending a luciferase reporter gene after the native SMN2 gene, such that inclusion of exon 7 in the expressed product places the luciferase sequence in frame, thus generating functional luciferase enzyme.
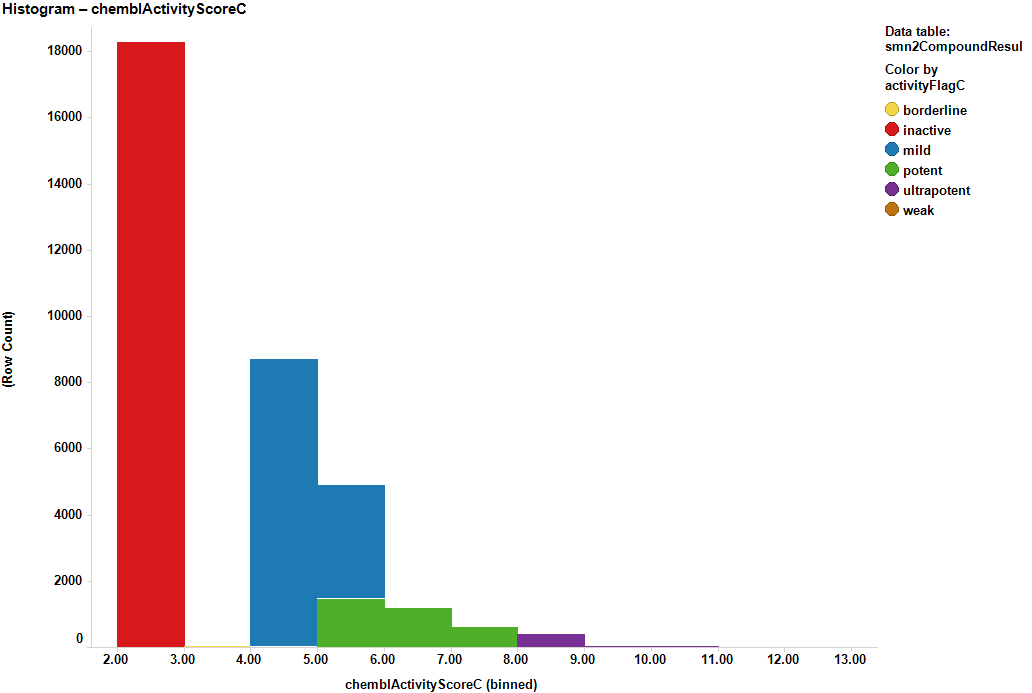
Table summary of the existing SMN2 assays in the ChEMBL DB. Among all, qHTS Assay for Enhancers of SMN2 Splice Variant Expression, with more than 30k records, has been used to annotate activity of molecules on SMN2. The rest of them have been excluded for annotation and construction of predictive models.
| ChEMBL Assay ID | Assay Source | Assay Type | Assay Organism | Activity Count | Description | Reference |
| CHEMBL1613842 | PubChem BioAssays | F | Homo sapiens | 33647 | PUBCHEM_BIOASSAY: qHTS Assay for Enhancers of SMN2 Splice Variant Expression. (Class of assay: confirmatory) | CHEMBL1201862 |
| CHEMBL1826381 | Scientific Literature | B | 133 | Activation of SMN expressed in HEK293 cells assessed as concentration required to reach 50% of maximum luciferase signal by SMN2-promotor driven lu… | J. Med. Chem., (2011) 54:18:6215 | |
| CHEMBL944092 | Scientific Literature | F | Mus musculus | 114 | Activation of SMN2 promoter in mouse NSC34 cells assessed as induction of beta-lactamase activity | J. Med. Chem., (2008) 51:3:449 |
| CHEMBL1738018 | PubChem BioAssays | F | Homo sapiens | 109 | PUBCHEM_BIOASSAY: Confirmation Concentration-Response Assay for Enhancers of SMN2 Splice Variant Expression for Further Probe SAR. (Class of assay:… | CHEMBL1201862 |
| CHEMBL1826380 | Scientific Literature | B | 98 | Activation of SMN expressed in HEK293 cells assessed as rate of induction by SMN2-promotor driven luciferase reporter gene assay relative to basal … | J. Med. Chem., (2011) 54:18:6215 | |
| CHEMBL1738105 | PubChem BioAssays | F | Photinus pyralis | 94 | PUBCHEM_BIOASSAY: Counterscreen Assay for Enhancers of SMN2 Splice Variant Expression: Interaction with Luciferase Reporter for Further Probe SAR. … | CHEMBL1201862 |
| CHEMBL944093 | Scientific Literature | F | Mus musculus | 90 | Activation of SMN2 promoter in mouse NSC34 cells assessed as induction of beta-lactamase activity relative to nonactivated cells | J. Med. Chem., (2008) 51:3:449 |
| CHEMBL1737944 | PubChem BioAssays | F | Homo sapiens | 61 | PUBCHEM_BIOASSAY: Counterscreen Assay for Enhancers of SMN2 Splice Variant Expression: Modulation of SMN1 Expression for Further Probe SAR. (Class … | CHEMBL1201862 |
| CHEMBL1614260 | PubChem BioAssays | F | Homo sapiens | 34 | PUBCHEM_BIOASSAY: Confirmation Concentration-Response Assay for Enhancers of SMN2 Splice Variant Expression. (Class of assay: confirmatory) [Relate… | CHEMBL1201862 |
| CHEMBL1614101 | PubChem BioAssays | B | Photinus pyralis | 26 | PUBCHEM_BIOASSAY: Counterscreen Assay for Enhancers of SMN2 Splice Variant Expression: Interaction with Luciferase Reporter. In this assay, we util… | CHEMBL1201862 |
| CHEMBL1613932 | PubChem BioAssays | F | Homo sapiens | 20 | PUBCHEM_BIOASSAY: Confirmation Concentration-Response Assay for Enhancers of SMN2 Splice Variant Expression for Probe SAR. (Class of assay: confirm… | CHEMBL1201862 |
| CHEMBL1614136 | PubChem BioAssays | F | Photinus pyralis | 19 | PUBCHEM_BIOASSAY: Counterscreen Assay for Enhancers of SMN2 Splice Variant Expression: Interaction with Luciferase Reporter for Probe SAR. (Class o… | CHEMBL1201862 |
| CHEMBL1614348 | PubChem BioAssays | F | Homo sapiens | 7 | PUBCHEM_BIOASSAY: Counterscreen Assay for Enhancers of SMN2 Splice Variant Expression: Modulation of SMN1 Expression for Probe SAR. (Class of assay… | CHEMBL1201862 |
| CHEMBL945043 | Scientific Literature | F | Homo sapiens | 3 | Increase in number of cells with Cajal bodies containing SMN2 protein in human fibroblasts from spinal muscular atrophy patient at 100 nM after 5 d… | J. Med. Chem., (2008) 51:3:449 |
| CHEMBL945042 | Scientific Literature | F | Homo sapiens | 3 | Increase in number of Cajal bodies containing SMN2 protein in human fibroblasts from spinal muscular atrophy patient at 100 nM after 5 days relativ… | J. Med. Chem., (2008) 51:3:449 |
| CHEMBL945041 | Scientific Literature | F | Homo sapiens | 3 | Increase in SMN2 mRNA expression in human fibroblasts from spinal muscular atrophy patient | J. Med. Chem., (2008) 51:3:449 |
| CHEMBL1826384 | Scientific Literature | B | 2 | Activation of SMN expressed in HEK293 cells by SMN2-promotor driven luciferase reporter gene assay | J. Med. Chem., (2011) 54:18:6215 | |
| CHEMBL944099 | Scientific Literature | F | Mus musculus | 1 | Activation of SMN2 promoter in human NSC34 cells assessed as induction of beta-lactamase activity at 30 uM | J. Med. Chem., (2008) 51:3:449 |
| CHEMBL945040 | Scientific Literature | F | Mus musculus | 1 | Increase in SMN2 mRNA expression in mouse NSC34 cells at 500 nM relative to control | J. Med. Chem., (2008) 51:3:449 |
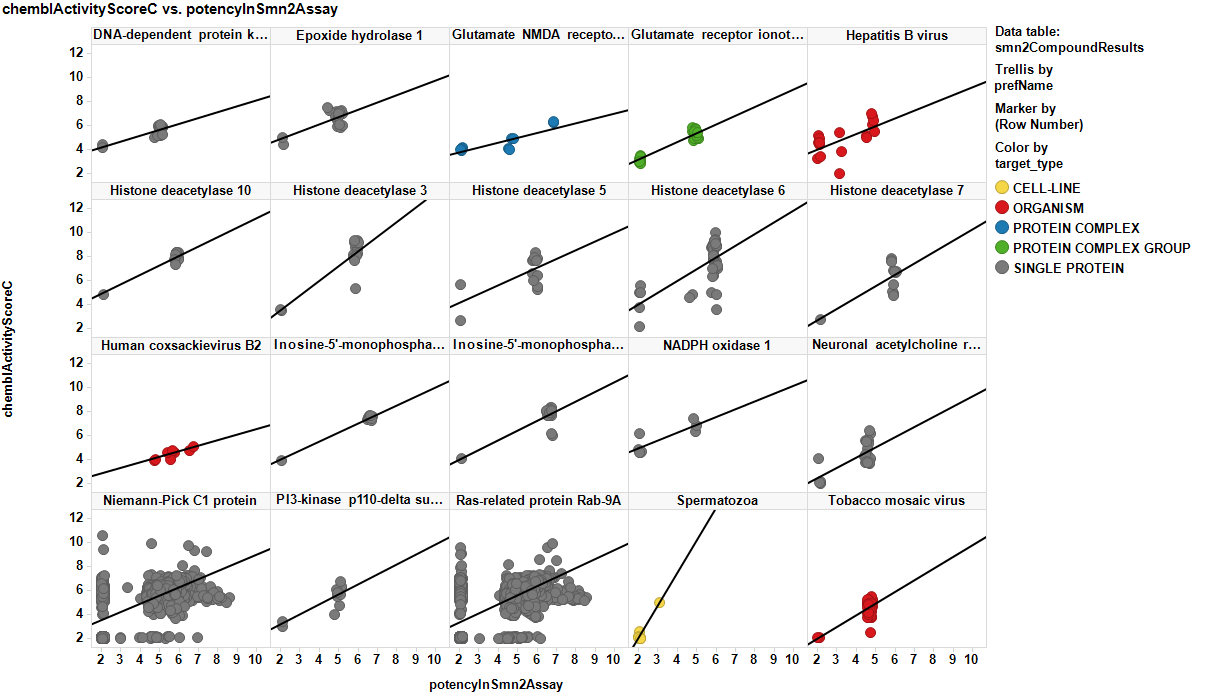
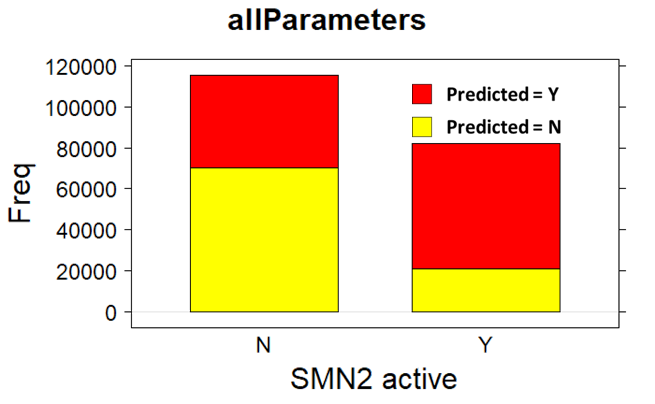
There are about 20 assays based on SMN2 constructs in the ChEMBL DB. Among all we have chosen the reporter assay described in the data description section. The assay contains 30k records, which is an interesting amount to apply ML methodology, but beyond that, it is a blackbox assay, which means that the patterns of expression of functional SMN2 protein can be elicited by a plethora of cellular mechanisms that can be further investigated to identify protein targets involved. Below, assay description of the most relevant SMN2 assays and distribution histogram of the activities recorded for the 30k molecules screen.
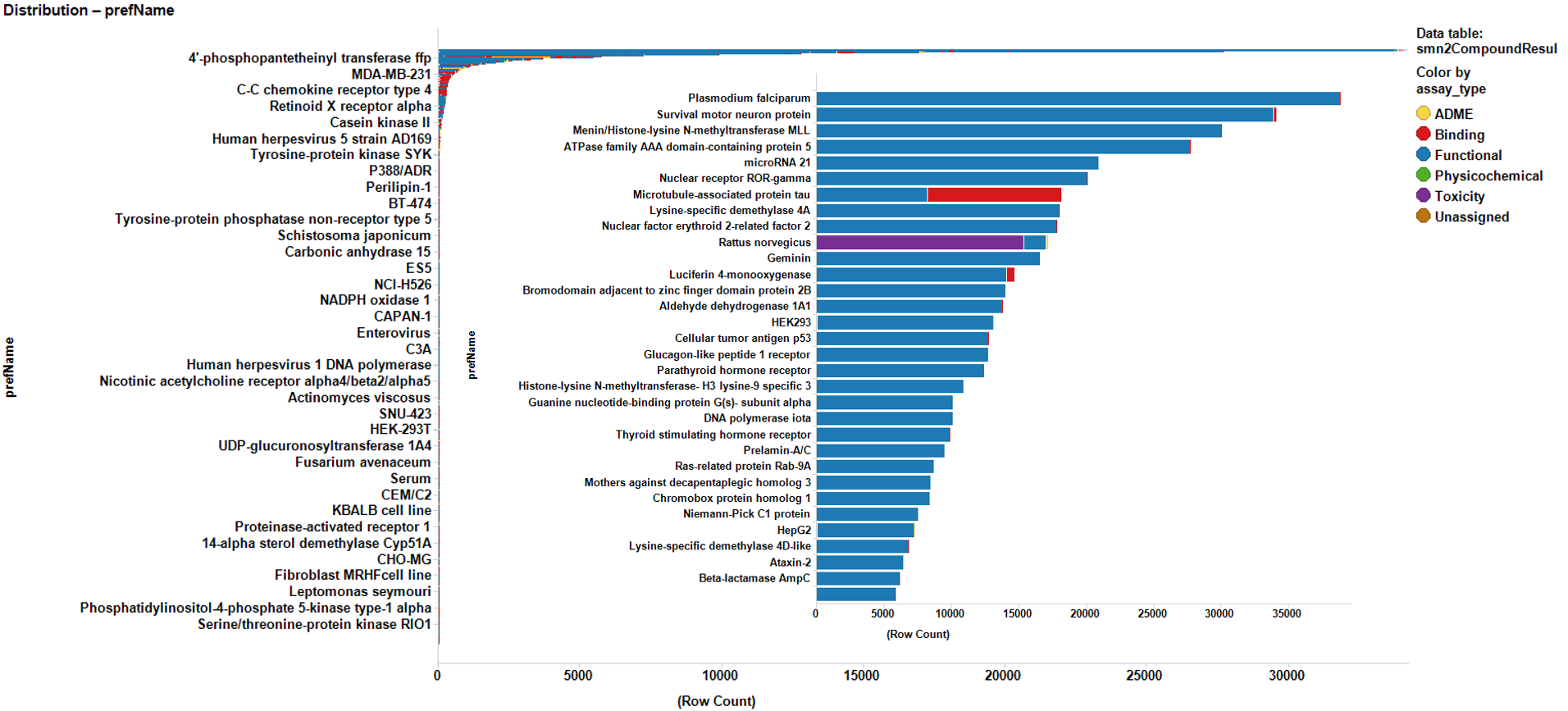
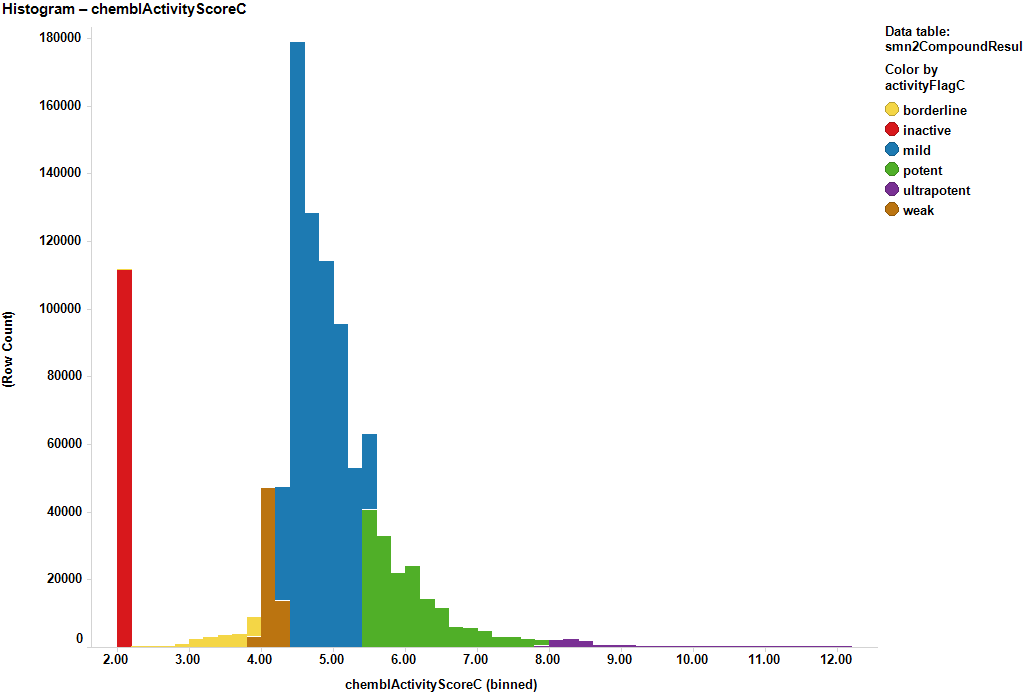
The 30k molecules used in SMN2 reporter screen have also been assayed in many more experimental models in the ChEMBL DB producing almost 1M records in more than 50k assays. We can therefore use the activity of these compounds on all these assays to build ML models suitable for prediction of activity of molecules that have never faced to a SMN assay. Charts below display some examples of these assays and the global distribution of activities of their respective records.
And, of course, ahead of finding relations, or making predictions, why not to have a look upon the relationship between the activity in SMN2 and the rest of the assay these molecules participate in? The picture shows the existing correlation between SMN2 and other targets for the cases for R2 >0.5, slope>0.4 and N>10.
Evaluation of predictive models.
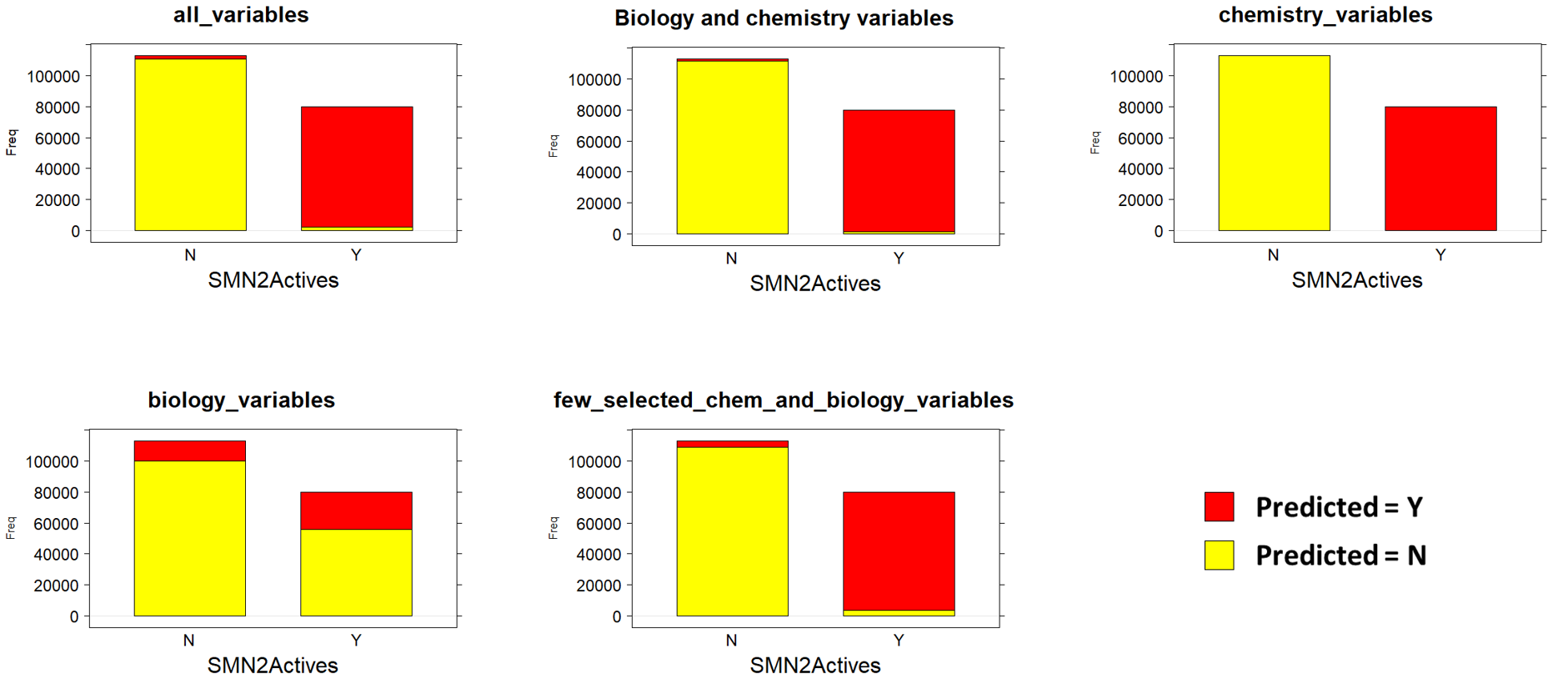
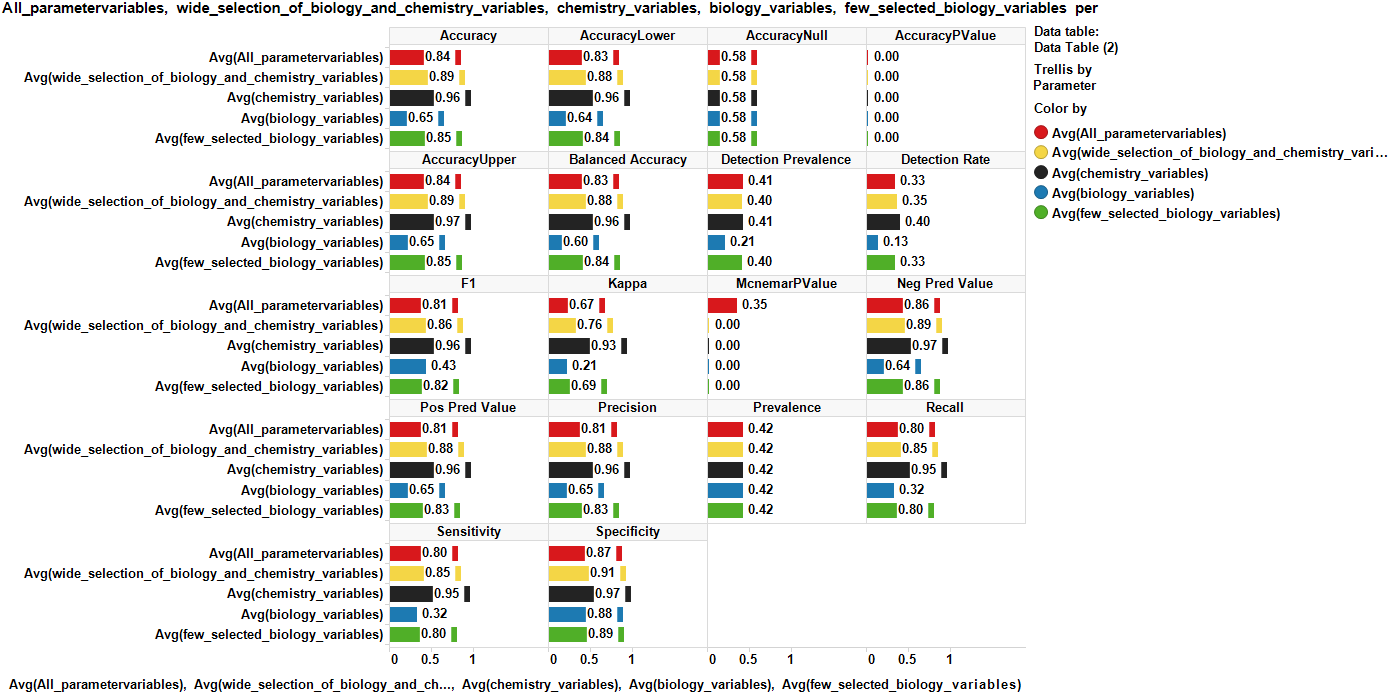
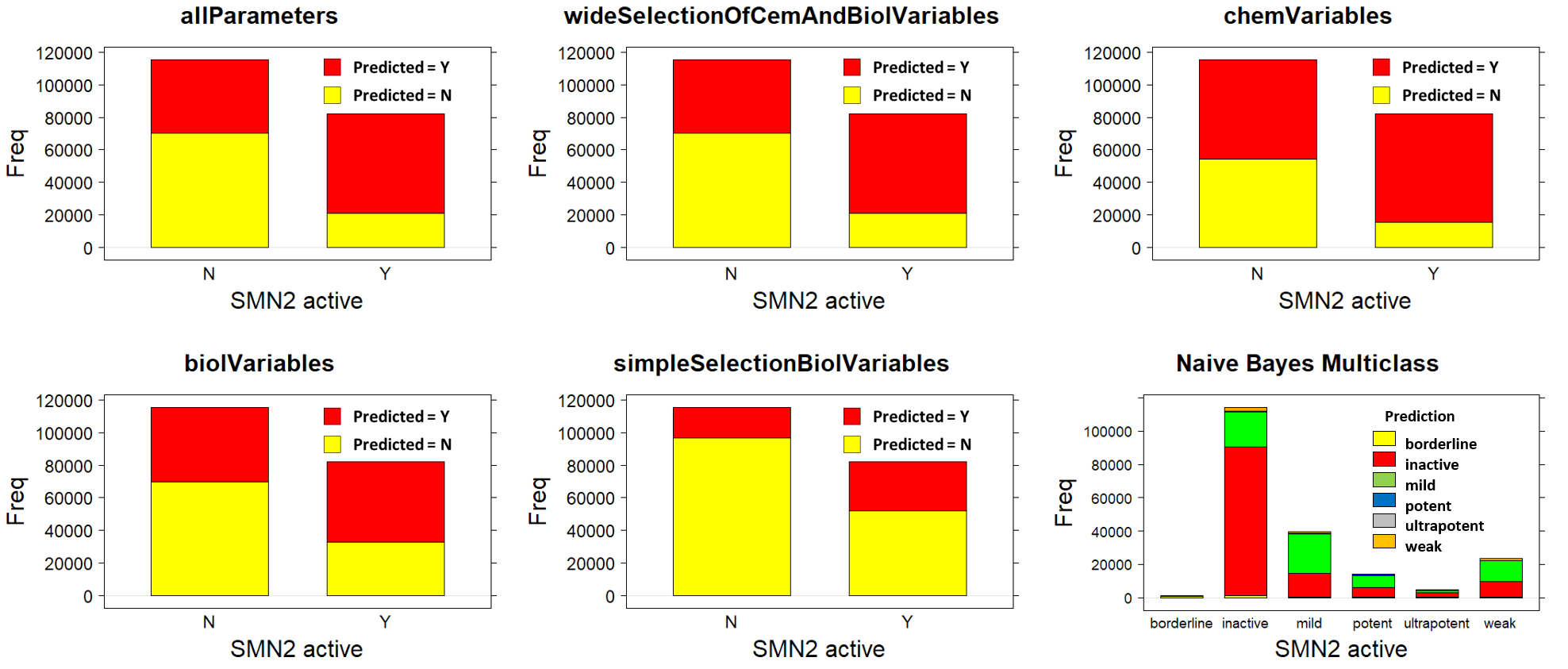
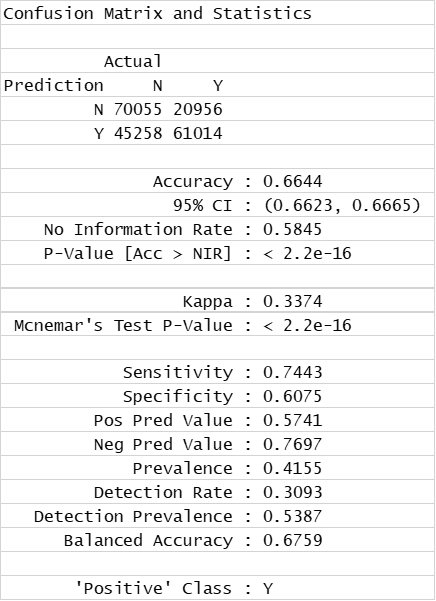
For model evaluation, data have been splitted in validation and test sets The validation set has been labelled for classification (Binary and multiple) and regression and used to generate the model. The test set is tested against the model and the predictions compared to the previously hidden activity columns. Accuracy of predictions is then assessed by the classical machine learning evaluation parameters in the charts below.
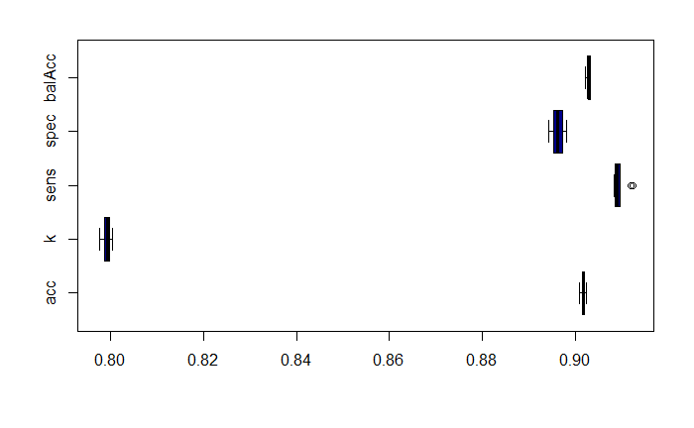
Validations set are passed through decision trees (Rpart: classification and regression) random forests (classification and regression), and Naive Bayes (binary and multiple classification) algorithms. Each algorithm is tested against different combinations of variables: All possible variables, a wide combination of biological and chemical variables, just biological, just chemical, and a minimal combination of both. For each combination of classification algorithm and variable set, a confusion matrix and a bar diagram with most meaningful statistical quality descriptors is charted below. If regression, there is a correlation plot instead.
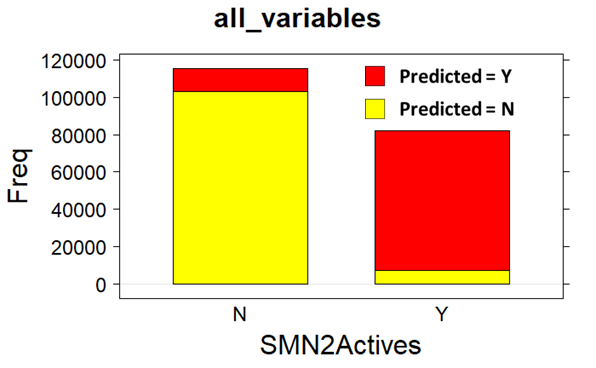
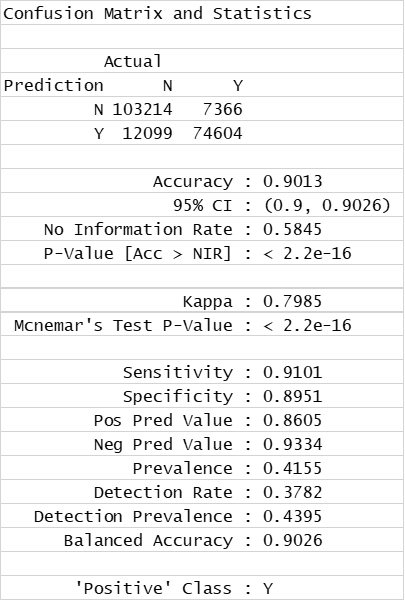
Decision Trees Classification (confusion matrix)
Decision Trees (QC descriptors)
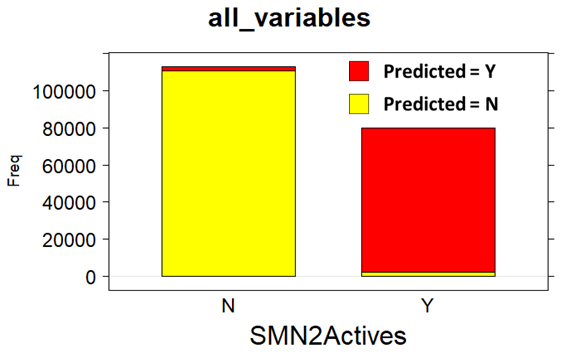
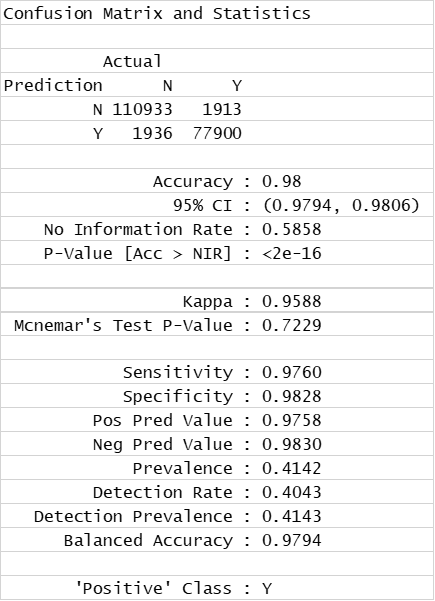
Random Forests Classification (confusion matrix)
Random Forests Classification (QC descriptors)
Naive Bayes Classification (ConfusionMatrix)
Naive Bayes (QC descriptors)
All algorithms compared in their respective best condition tested.
And the three binary classification algorithms visual confusion matrixes compared in their best conditions…
When classification algorithms result in acceptable quality they are submitted to 10 iterations cross-validations to assess predictions robustness and stability of QC descriptors
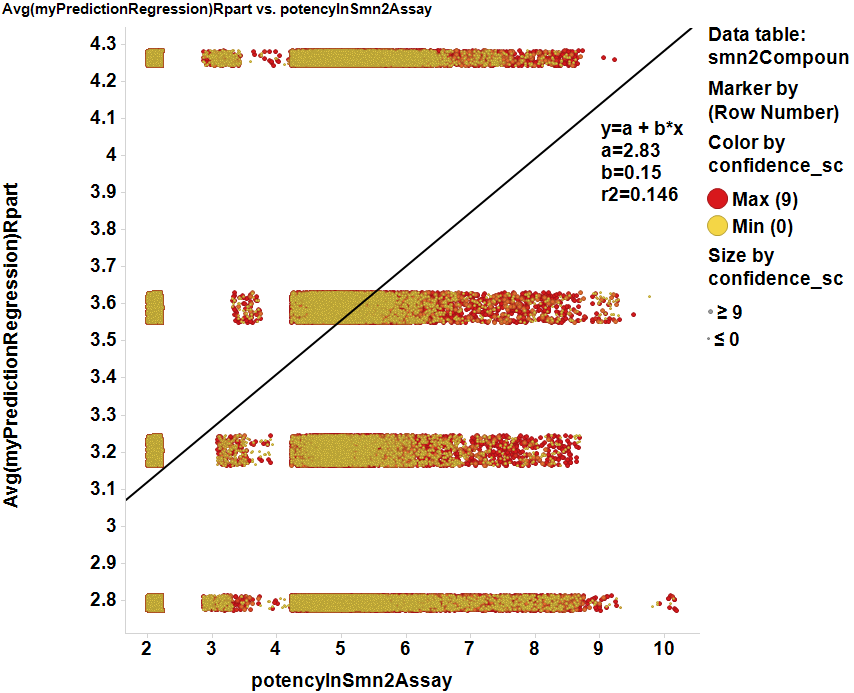
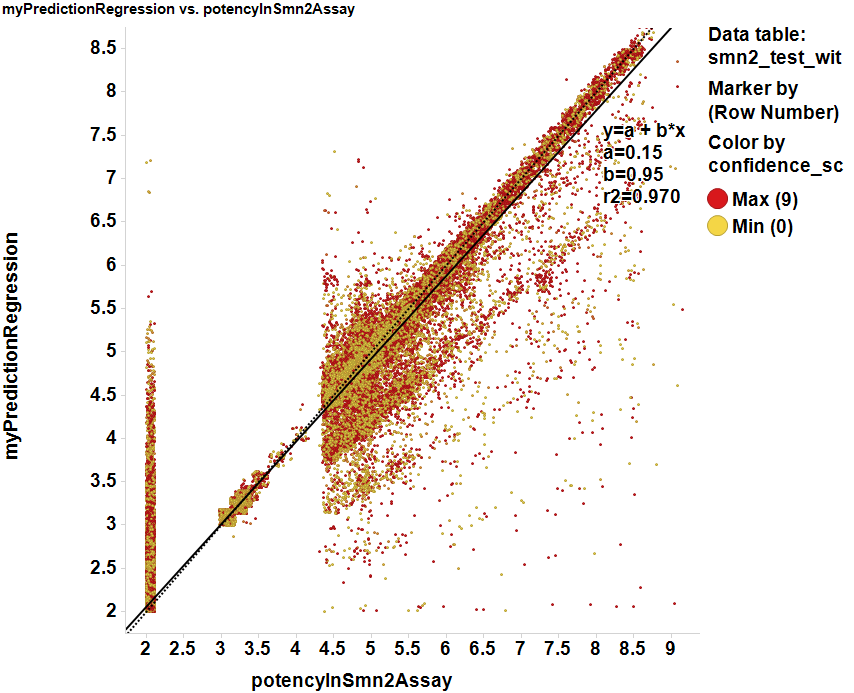
Scores prediction by regression. Although prediction by decision tree results in a poor estimation of potencies, Random forest regression shows a high correlation with actual ChemblScore data.
Based on these quality indicators, compound selection will be carried output using a dashboard that contains classification predictions from Rpart and random forests so as forests regression.. Naive Bayes output to be included in the dashboard just as an optional end filter.
Compound selection. Description of dashboard uses.
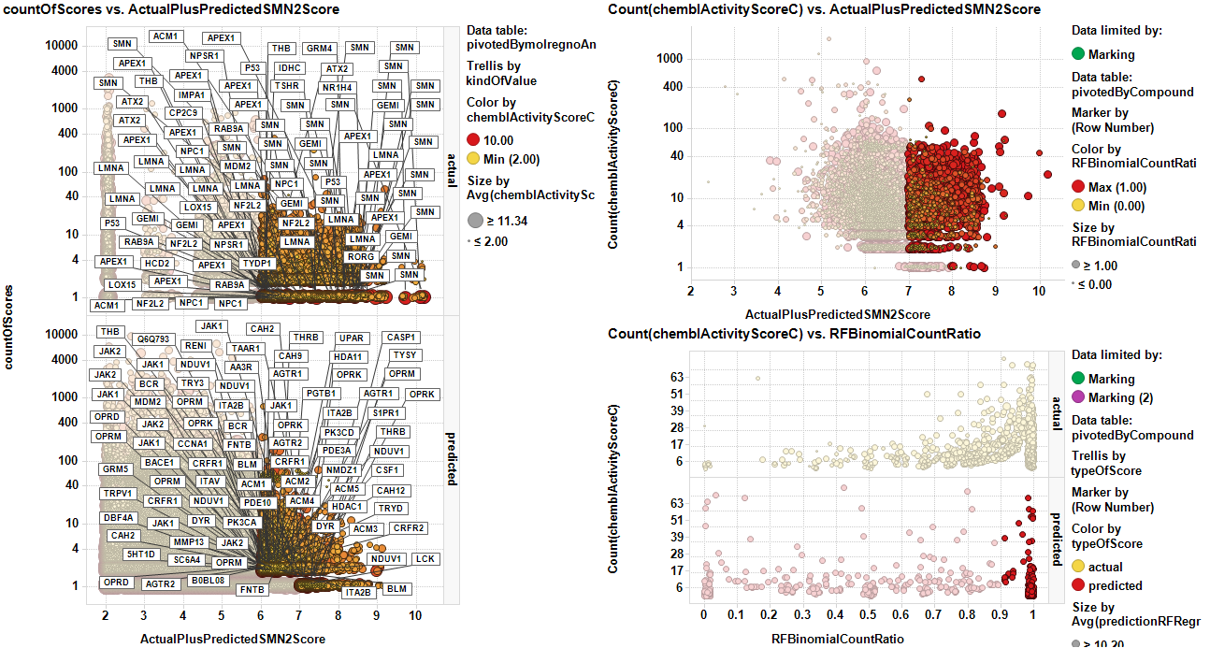
Let’s look at a typical multimethod compound selection dashboard. It is a panel where predictions through different models are interactively compared to allow quality control of the selection. As most compounds include multiple experimental events recorded in the database, and there is a prediction for each event, we need to calculate an index of activity for each compound. For regressions, this is the average SMN2 score (ActualPlusPredictedSMN2Score), which uses actual values when they exist. Classification will use the ratio between the number of times a compound has been predicted active vs the total number of events (countRatio). In this example we will use binomial random forests classification.
Left chart in the dashboard below visualizes the ActualPlusPredictedSMN2Score for each unique molecule-target pair vs the number of occasions that this pair has a result in the DB. Marking the most active (either actual or predicted) in a convenient number of occasions in this chart causes the marked compounds to appear in the top right plot that compares the average SMN2 potency (actual and predicted. ) with the number of records for each molecule. Marking the desired compounds in this plot raises the down right chart with the random forests classification countRatio.
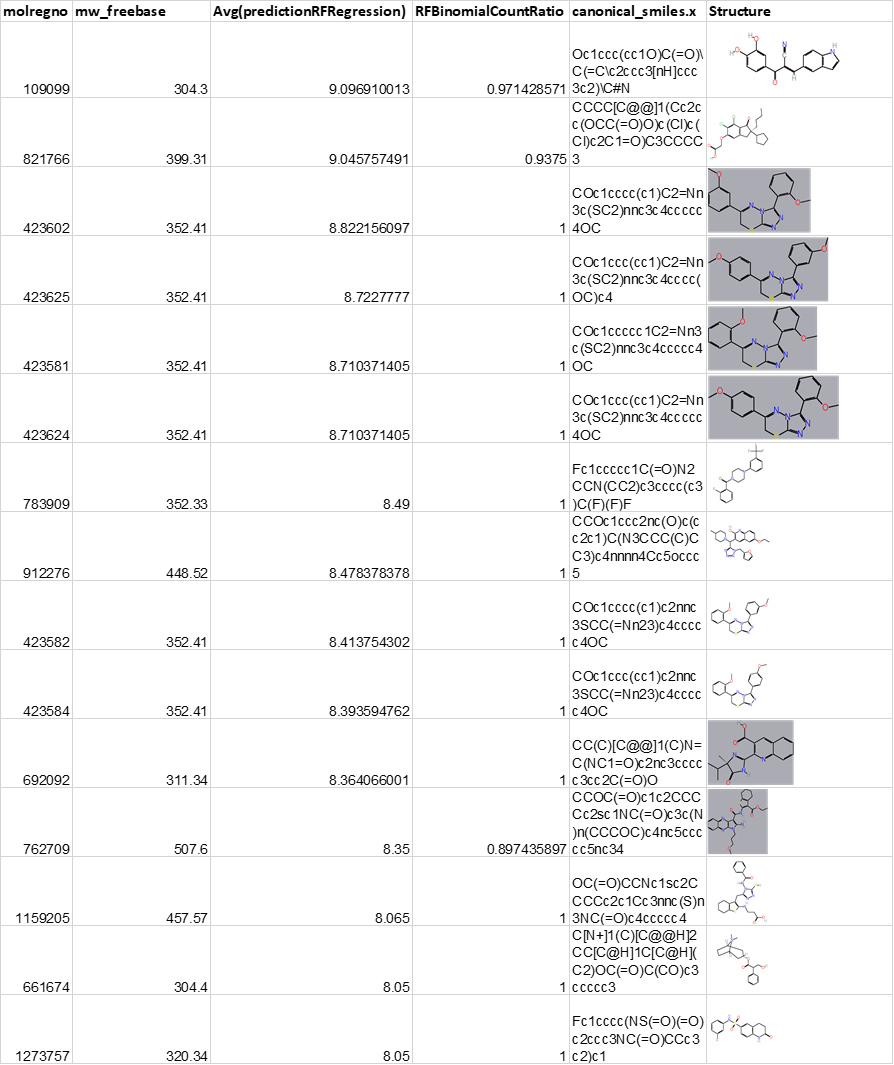
Now, it’s just a matter of making the selection at your convenience. The table here contains the 143 most potent predicted SMN2 actives with the highest count ratios, as indicated in more intense dot colors in the dashboard.
| molregno | mw_freebase | Count(chemblActivityScoreC) | Avg(predictionRFRegression) | RFBinomialCountRatio | canonical_smiles.x |
|---|---|---|---|---|---|
| 109099 | 304.3 | 70 | 9.0969100130081 | 0.97142857142857 | Oc1ccc(cc1O)C(=O)\C(=C\c2ccc3[nH]ccc3c2)\C#N |
| 821766 | 399.31 | 16 | 9.0457574905607 | 0.9375 | CCCC[C@@]1(Cc2cc(OCC(=O)O)c(Cl)c(Cl)c2C1=O)C3CCCC3 |
| 423602 | 352.41 | 13 | 8.8221560968745 | 1 | COc1cccc(c1)C2=Nn3c(SC2)nnc3c4ccccc4OC |
| 423625 | 352.41 | 3 | 8.7227776996806 | 1 | COc1ccc(cc1)C2=Nn3c(SC2)nnc3c4cccc(OC)c4 |
| 423624 | 352.41 | 1 | 8.7103714054693 | 1 | COc1ccc(cc1)C2=Nn3c(SC2)nnc3c4ccccc4OC |
| 423581 | 352.41 | 1 | 8.7103714054693 | 1 | COc1ccccc1C2=Nn3c(SC2)nnc3c4ccccc4OC |
| 783909 | 352.33 | 19 | 8.49 | 1 | Fc1ccccc1C(=O)N2CCN(CC2)c3cccc(c3)C(F)(F)F |
| 912276 | 448.52 | 37 | 8.4783783783784 | 1 | CCOc1ccc2nc(O)c(cc2c1)C(N3CCC(C)CC3)c4nnnn4Cc5occc5 |
| 423582 | 352.41 | 7 | 8.413754301856 | 1 | COc1cccc(c1)c2nnc3SCC(=Nn23)c4ccccc4OC |
| 423584 | 352.41 | 2 | 8.3935947616949 | 1 | COc1ccc(cc1)c2nnc3SCC(=Nn23)c4ccccc4OC |
| 692092 | 311.34 | 1 | 8.3640660013008 | 1 | CC(C)[C@@]1(C)N=C(NC1=O)c2nc3ccccc3cc2C(=O)O |
| 762709 | 507.6 | 39 | 8.35 | 0.8974358974359 | CCOC(=O)c1c2CCCCc2sc1NC(=O)c3c(N)n(CCCOC)c4nc5ccccc5nc34 |
| 1159205 | 457.57 | 2 | 8.065 | 1 | OC(=O)CCNc1sc2CCCCc2c1Cc3nnc(S)n3NC(=O)c4ccccc4 |
| 1418094 | 304.4 | 10 | 8.05 | 0.9 | [Br-].C[N+]1(C)[C@@H]2CC[C@H]1C[C@H](C2)OC(=O)C(CO)c3ccccc3 |
| 661674 | 304.4 | 6 | 8.05 | 1 | C[N+]1(C)[C@@H]2CC[C@H]1C[C@H](C2)OC(=O)C(CO)c3ccccc3 |
| 1273757 | 320.34 | 4 | 8.05 | 1 | Fc1cccc(NS(=O)(=O)c2ccc3NC(=O)CCc3c2)c1 |
| 984482 | 207.25 | 23 | 8.0260869565217 | 0.95652173913044 | Sc1nnc(c2cocc2)n1CC=C |
| 807723 | 493.53 | 11 | 8.0159393939394 | 0.90909090909091 | CCCN1C(=O)NC(=O)C(=C1N)C(=O)CSC2=Nc3ccccc3C(=O)N2c4ccc(OC)cc4 |
| 807129 | 430.46 | 16 | 8.005 | 0.9375 | N=C1N(CC2CCCO2)C3=C(C=C1C(=O)NCc4cccnc4)C(=O)N5C=CC=CC5=N3 |
| 1218332 | 302.32 | 10 | 8 | 1 | CCOC(=O)COc1ccc2C3=C(CCCC3)C(=O)Oc2c1 |
| 359668 | 417.48 | 1 | 7.9775 | 1 | Cc1ccc(C(=O)NC(CCS)C(=O)N[C@H](Cc2ccccc2)C(=O)O)c(O)n1 |
| 992556 | 403.47 | 21 | 7.9505952380952 | 1 | CC1CCN(CC1)C(C(=O)NC2CCCCC2)c3cc4OCOc4cc3[N+](=O)[O-] |
| 150696 | 233.19 | 4 | 7.94625 | 1 | C[C@@](O)(C(=O)Nc1ccccc1)C(F)(F)F |
| 116405 | 233.19 | 4 | 7.94625 | 1 | CC(O)(C(=O)Nc1ccccc1)C(F)(F)F |
| 1160938 | 233.31 | 1 | 7.94625 | 1 | CC(C)(C)NC(=O)C(O)\C=C\c1ccccc1 |
| 252654 | 192.24 | 4 | 7.91 | 1 | Cc1nc(cs1)c2c[nH]c(C=O)c2 |
| 1702332 | 211.65 | 5 | 7.9 | 1 | C\C(=N/NC(=O)N)\c1ccccc1Cl |
| 834050 | 492.57 | 2 | 7.8644166666667 | 1 | CCN(C(=O)CN1CCN(CC1)c2ccccc2OC)C3=C(N)N(Cc4ccccc4)C(=O)NC3=O |
| 1104771 | 368.45 | 3 | 7.8375 | 1 | Cc1ccc(NC(=O)C(=O)NCCN2CCN(CC2)S(=O)(=O)C)cc1 |
| 938752 | 320.34 | 7 | 7.8339285714286 | 1 | Fc1ccccc1NS(=O)(=O)c2ccc3NC(=O)CCc3c2 |
| 255016 | 285.26 | 58 | 7.7918103448276 | 1 | OC(=O)Cn1cnc2c(O)nc(Nc3ccccc3)nc12 |
| 984429 | 305.36 | 7 | 7.7571428571429 | 1 | CC(=O)N(CC1CCCO1)c2nnc(s2)c3cnccn3 |
| 1009982 | 473.52 | 29 | 7.7499137931034 | 1 | COc1ccc(cc1)C2=CN(C(=O)N2CC(=O)Nc3ccc(OC)cc3OC)c4ccc(C)cc4 |
| 888665 | 366.26 | 12 | 7.7463541666667 | 1 | Cc1ccc2SCC(NC(=O)c3ccc(Cl)c(Cl)c3)C(=O)c2c1 |
| 1312048 | 201.25 | 37 | 7.7375 | 1 | c1ccc(cc1)c2nc3sccn3n2 |
| 1538251 | 201.25 | 2 | 7.7375 | 1 | c1csc(c1)c2ccc3nncn3c2 |
| 1839634 | 252.31 | 5 | 7.715602503252 | 1 | Cl.N[C@H]1C[C@H]1c2cccc(NC(=O)c3ccccc3)c2 |
| 1672353 | 488.66 | 15 | 7.6875 | 1 | SCCC(=O)OCC(COC(=O)CCS)(COC(=O)CCS)COC(=O)CCS |
| 869149 | 389.47 | 3 | 7.6708333333333 | 1 | COc1ccc(CN(C2CCS(=O)(=O)C2)C(=O)c3cccc(OC)c3)cc1 |
| 1302993 | 246.26 | 54 | 7.6517100527401 | 1 | COc1cc2ccc(CC(=O)O)cc2cc1OC |
| 1698463 | 333.39 | 4 | 7.65 | 1 | Cc1nn(Cc2ccccc2)c(C)c1\C=N\NC(=O)c3ccncc3 |
| 405235 | 304.37 | 20 | 7.6425 | 1 | CCCNc1nnc(CN2C(=O)Oc3ccc(C)cc23)s1 |
| 548008 | 267.12 | 3 | 7.635625 | 1 | Cl.Brc1ccc2OC(Cc2c1)C3=NCCN3 |
| 1110569 | 277.7 | 2 | 7.63 | 1 | COc1ccc(Nc2cc(Cl)ccc2C(=O)O)cc1 |
| 865534 | 312.43 | 26 | 7.5925 | 1 | Cc1cccc(C)c1NC(=S)NC(=O)CCc2ccccc2 |
| 1962360 | 402.4 | 2 | 7.575 | 1 | OC(=O)[C@H](Cc1ccc(O)cc1)NC(=O)c2cccc3C(=O)c4ccccc4Nc23 |
| 183470 | 284.28 | 2 | 7.56375 | 1 | [Na+].[O-]S(=O)(=O)C(F)(F)c1cccc(c1)c2ccccc2 |
| 1273631 | 323.35 | 2 | 7.56 | 1 | Cn1ccnc1NC(=O)c2ccc3cc4C(=O)NCCCn4c3c2 |
| 922240 | 288.34 | 13 | 7.5115384615385 | 1 | CCC(=O)c1cn(Cc2ccccc2C#N)c3ccccc13 |
| 69225 | 178.21 | 4 | 7.5 | 1 | Nc1nc(ns1)c2cccnc2 |
| 69259 | 178.21 | 4 | 7.5 | 1 | Nc1nc(ns1)c2ccncc2 |
| 985902 | 516.63 | 16 | 7.5 | 1 | COc1ccc(CNC(=O)[C@H](C)[C@H]2C[C@]2(C)[C@H](NC(=O)OCc3ccccc3)c4ccccc4)cc1OC |
| 130479 | 239.29 | 3 | 7.4925 | 1 | CCCCN1C(=O)c2ccccc2S1(=O)=O |
| 1208689 | 355.41 | 3 | 7.4781103203779 | 1 | COc1ccccc1c2nnc(SC(C)C(=O)O)n2c3ccccc3 |
| 746541 | 302.35 | 42 | 7.4660714285714 | 0.95238095238095 | COc1ccccc1NC(=S)NC(=O)\C=C\c2occc2 |
| 23297 | 444.54 | 2 | 7.4625 | 1 | CC[C@H](C)[C@H](NC(=O)[C@@H](S)c1ccccc1)C(=O)N[C@@H](Cc2ccc(O)cc2)C(=O)O |
| 23927 | 444.54 | 2 | 7.4625 | 1 | CC(C)[C@H](NC(=O)[C@@H](S)Cc1ccccc1)C(=O)N[C@@H](Cc2ccc(O)cc2)C(=O)O |
| 748748 | 399.46 | 7 | 7.4375 | 1 | CCc1ccc(NC(=O)c2ccc(NS(=O)(=O)c3c(C)onc3C)cc2)cc1 |
| 807870 | 498.55 | 19 | 7.4276315789474 | 1 | CCOc1cc(ccc1OS(=O)(=O)c2ccc(C)cc2)C(c3c(C)n[nH]c3O)c4c(C)n[nH]c4O |
| 1207664 | 269.26 | 2 | 7.425 | 1 | Cc1ccc(Nc2ncc3c(O)nnc(O)c3n2)cc1 |
| 31016 | 255.24 | 2 | 7.424227503252 | 1 | NCCc1c[nH]c(n1)c2ccccc2C(F)(F)F |
| 886994 | 274.36 | 13 | 7.4134615384615 | 1 | N#Cc1ccccc1OCCOCCNC2CCCC2 |
| 1402809 | 238.24 | 2 | 7.4 | 1 | O.NC(=O)OCC(COC(=O)N)c1ccccc1 |
| 57376 | 262.27 | 3 | 7.38125 | 1 | CC(C)(C)C(=O)OCn1nnnc1c2cnccn2 |
| 623319 | 339.37 | 2 | 7.365625 | 1 | COc1ccc(cc1)N2C=Nc3c(sc4nc(C)nc(N)c34)C2=O |
| 1109330 | 285.29 | 2 | 7.3625 | 1 | COc1ccc(cc1)C(=O)NC(C(=O)O)c2ccccc2 |
| 744335 | 498.53 | 5 | 7.3625 | 1 | COc1ccc(cc1)c2nc3NC(=C(C(c4ccc(OC)c(OC)c4)n3n2)C(=O)Nc5cccnc5)C |
| 503123 | 218.33 | 5 | 7.3625 | 1 | CC(=CCCC(C)(O)c1ccc(C)cc1)C |
| 775141 | 305.36 | 15 | 7.3625 | 1 | CN(C(=O)C1CCCO1)c2nnc(s2)c3cnc(C)cn3 |
| 969153 | 250.28 | 4 | 7.359375 | 1 | COC(=O)CSc1nnnn1c2ccccc2 |
| 1320560 | 235.06 | 55 | 7.3575 | 1 | OCC1COc2cc(Cl)c(Cl)cc2O1 |
| 1073814 | 292.33 | 3 | 7.355 | 1 | COc1c2CCC(C)(C)Oc2c(C(=O)C)c(OC)c1C=O |
| 796832 | 339.43 | 11 | 7.352803030303 | 0.90909090909091 | CC(C)c1nnc(NC(=O)CS(=O)(=O)Cc2ccccc2)s1 |
| 769821 | 322.29 | 5 | 7.35 | 1 | Fc1ccc(cc1F)C(=O)CSc2oc(nn2)c3occc3 |
| 898090 | 339.39 | 7 | 7.3414285714286 | 1 | COc1ccccc1NC(=O)N2C[C@@H]3C[C@@H](C2)C4=CC=CC(=O)N4C3 |
| 359615 | 497.52 | 2 | 7.34 | 1 | OC(=O)[C@@H](Cc1ccccc1)NC(=O)C(CCS)NC(=O)c2oc(cc2)c3ccccc3[N+](=O)[O-] |
| 360050 | 430.52 | 2 | 7.3375 | 1 | Cc1cc(cc(C)c1O)C(=O)NC(CCS)C(=O)N[C@H](Cc2ccccc2)C(=O)O |
| 868579 | 311.34 | 4 | 7.3275 | 1 | Cc1c2C=NN(CC(=O)O)C(=O)c2c(C)n1Cc3ccccc3 |
| 1238753 | 255.34 | 4 | 7.315625 | 1 | CN1CCN(CC1)S(=O)(=O)c2ccc(N)cc2 |
| 1244843 | 347.23 | 2 | 7.3125 | 1 | CN(CC(=O)NC1CC1)S(=O)(=O)c2ccc(Br)cc2 |
| 825043 | 394.42 | 13 | 7.2961538461539 | 0.92307692307692 | COCCN1C(C(=C(O)C1=O)C(=O)c2ccc(OCC=C)cc2)c3cccnc3 |
| 851047 | 349.43 | 12 | 7.28625 | 0.91666666666667 | O=C1N(CCCN2CCOCC2)C(=Nc3ccccc13)c4ccccc4 |
| 827328 | 249.35 | 7 | 7.2707142857143 | 1 | Cc1cc(C)c(C)c(OCCN2CCOCC2)c1 |
| 297984 | 423.39 | 2 | 7.265 | 1 | COc1cc(cc(\C=C\C(=O)O)c1OC)S(=O)(=O)Nc2ccc(C(=O)O)c(O)c2 |
| 736479 | 255.74 | 18 | 7.2548611111111 | 1 | Cc1cc(OCCN2CCOCC2)ccc1Cl |
| 280445 | 243.22 | 2 | 7.25 | 1 | Oc1nc(O)c(C#N)c(NCc2cccnc2)n1 |
| 887078 | 483.56 | 2 | 7.25 | 1 | CCN(CC)CCN1C(C(=C(O)C1=O)C(=O)c2c(C)[nH]c(C(=O)OC)c2C)c3ccc(OC)cc3 |
| 688354 | 229.67 | 2 | 7.25 | 1 | Clc1cccc(c1)c2cnc3ncccn23 |
| 122441 | 496.53 | 6 | 7.2125 | 1 | OC(=O)CCNC(=O)[C@H](CC1CCCCC1)NC(=O)[C@H](CCCc2ccccc2)CP(=O)(O)O |
| 552739 | 257.12 | 3 | 7.210625 | 1 | Cl.Clc1ccc2CC(Oc2c1Cl)C3=NCCN3 |
| 737321 | 340.46 | 13 | 7.2048076923077 | 1 | CCC(=O)N(Cc1cc2c(C)ccc(C)c2nc1O)C3CCCCC3 |
| 1099633 | 334.42 | 7 | 7.2010714285714 | 1 | Cc1csc(NC(=O)CSc2nccn2Cc3occc3)n1 |
| 187199 | 444.57 | 4 | 7.2 | 1 | OC1(CNCc2ccccc2)CCN(CCCc3c[nH]c4ccc(cc34)n5cnnc5)CC1 |
| 14961 | 370.47 | 3 | 7.1958333333333 | 1 | CCN1CCCC1CNC(=O)c2cc(ccc2OC)N(C)S(=O)(=O)N |
| 132747 | 227.69 | 4 | 7.19375 | 1 | CCC(C)(O)C(=O)Nc1ccccc1Cl |
| 290126 | 349.38 | 3 | 7.1875 | 1 | NC(=O)CNC(=O)COCC1CCCN1C(=O)OCc2ccccc2 |
| 153953 | 584.66 | 2 | 7.175 | 1 | Cc1cc(C)nc(O[C@H](C(=O)O)[C@]2(NCC(=O)N(Cc3ccc(cc3)c4ccccc4)c5ccccc25)c6ccccc6)n1 |
| 861885 | 368.45 | 11 | 7.1727272727273 | 1 | CCOC(=O)[C@H]1CCC[C@H](C1)NS(=O)(=O)c2ccc(NC(=O)C)cc2 |
| 360264 | 355.47 | 4 | 7.1725 | 1 | O=C(C1CN(C2CCCCC2)C(=O)C1)N3CCN(CC3)c4ccccc4 |
| 804660 | 352.41 | 14 | 7.1660714285714 | 0.92857142857143 | COc1ccc(cc1)N(C(C)C(=O)NCc2occc2)S(=O)(=O)C |
| 580465 | 454.95 | 2 | 7.1625 | 1 | Cn1nccc1c2cc(NC(=O)Nc3ccc(Cl)cc3)ccc2OCCN4CCNCC4 |
| 1345337 | 462.89 | 2 | 7.15 | 1 | Clc1cc(Nc2ncncc2c3occ(CNC(=O)C=C)n3)ccc1OCc4ccccn4 |
| 1698917 | 289.31 | 14 | 7.15 | 1 | COC(=O)\C=C\1/SC(=NC1=O)N\N=C\c2ccccc2 |
| 881856 | 500.55 | 4 | 7.15 | 1 | CCc1nnc(NC(=O)CSc2nnc(Cc3cc(O)nc(O)n3)n2c4ccc(OC)cc4)s1 |
| 751364 | 500.55 | 5 | 7.15 | 1 | CCOc1ccc(cc1)n2c(Cc3cc(O)nc(O)n3)nnc2SCC(=O)Nc4nnc(C)s4 |
| 466426 | 408.41 | 11 | 7.1409090909091 | 1 | COc1ccc(cc1)N2N=C(c3nc4ccccc4[nH]3)c5nc6ccccc6n5C2=O |
| 1786409 | 412.37 | 2 | 7.135 | 1 | CN(C(=O)c1cn2c(cnc2cn1)c3ccc(cc3)C(F)(F)F)c4ccc(C)nn4 |
| 789115 | 288.3 | 18 | 7.1331481481482 | 1 | CC(=O)OC(C)(C)C1Cc2cc3C=CC(=O)Oc3cc2O1 |
| 261971 | 311.38 | 2 | 7.125 | 1 | Oc1c(cnc2ccccc12)C(=O)NC3CCN4CCCC3C4 |
| 895463 | 311.36 | 10 | 7.1215 | 1 | CN(C)S(=O)(=O)c1ccc2c(c1)nc(CCC(=O)O)n2C |
| 396730 | 352.75 | 19 | 7.1160446357981 | 1 | Fc1ccc(Cl)cc1c2nc(Nc3ccncc3)c4nccnc4n2 |
| 1017627 | 997.39 | 13 | 7.1153846153846 | 0.92307692307692 | CC(C)(C)CC(C)(C)c1ccc(OCOCCOCCO)c(Cc2cc(cc(Cc3cc(ccc3OCOCCOCCO)C(C)(C)CC(C)(C)C)c2OCOCCOCCO)C(C)(C)CC(C)(C)C)c1 |
| 1003526 | 309.14 | 5 | 7.113875 | 1 | Cc1onc(NC(=O)CSCC(=O)O)c1Br |
| 763492 | 320.38 | 2 | 7.109 | 1 | COc1ccc(cc1OC)C(=O)NCC(=O)N2CCC(C)CC2 |
| 2003920 | 443.46 | 2 | 7.1025 | 1 | CC1(C)OCC(=N[C@](C)(c2cc(N[C@H]3CCc4cc(cnc34)C#N)ccc2F)C1(F)F)N |
| 1534460 | 416.51 | 2 | 7.1 | 1 | CCCCNC(=O)C[C@@H]1C[C@]2(CCCCC=C2N(Cc3occc3)C1=O)C(=O)OC |
| 160137 | 150.13 | 3 | 7.0975 | 1 | Cc1ccc2nc(O)oc2n1 |
| 1225532 | 355.41 | 5 | 7.094985320378 | 1 | COc1ccccc1c2nnc(SCC(=O)O)n2c3ccc(C)cc3 |
| 867806 | 307.26 | 6 | 7.09 | 1 | COC(=O)c1cnc2N(C)C(=O)N(C)C(=O)c2c1C(=O)OC |
| 379867 | 336.47 | 2 | 7.07625 | 1 | C[C@@H]1C[C@@H](O)[C@H]2[C@@](C)(CO)CCC[C@]2(C)[C@@]1(O)CCc3cocc3 |
| 449995 | 468.59 | 2 | 7.0725 | 1 | COc1cc2nc(nc(NC3CCCCNC3=O)c2cc1OC)N4CCC(CC4)N5CCCC5 |
| 2009201 | 405.4 | 2 | 7.0725 | 1 | OC(=O)C[C@@H](Cc1ccc(cc1)c2ccccc2)NC(=O)c3cc(ncn3)C(=O)O |
| 951444 | 230.26 | 5 | 7.055 | 1 | Cc1nn(Cc2ccccc2)c(C)c1C(=O)O |
| 145945 | 312.32 | 3 | 7.0541666666667 | 1 | OC(=O)C1CCN(CC1)c2ccc3[nH]c4nc(O)nc4cc3c2 |
| 815015 | 357.35 | 5 | 7.0525 | 1 | CS(=O)(=O)C1=C(O)C(=O)N(CCCC(=O)O)C1c2ccccc2F |
| 830234 | 396.48 | 49 | 7.0510204081633 | 0.93877551020408 | C\C(=C/c1ccccc1)\C=C\2/SC(=S)N(NC(=O)c3cccc(O)c3)C2=O |
| 97880 | 531.69 | 7 | 7.05 | 1 | COc1ccc(CNC(=O)C(N2CCN(CC(O)COc3c(C)cccc3C)CC2)c4ccc(C)cc4)cc1 |
| 206152 | 260.25 | 53 | 7.05 | 1 | CC(=O)Nc1ccc(Nc2cc(O)nc(O)n2)cc1 |
| 854753 | 213.3 | 4 | 7.05 | 1 | CCCC(C)C(=O)Nc1nnc(C)s1 |
| 915974 | 236.65 | 6 | 7.049613751626 | 1 | OC(=O)c1ccc(Cn2cc(Cl)cn2)cc1 |
| 1224007 | 311.38 | 4 | 7.0475 | 1 | CCOC(=O)CC1N(CCNC1=O)C(=O)NC2CCCCC2 |
| 917463 | 246.3 | 5 | 7.0425 | 1 | C[C@@H](C(=O)O)c1ccc(C[C@@H]2CCCC2=O)cc1 |
| 959645 | 434.38 | 7 | 7.0342142857143 | 1 | Fc1cccc(c1)N2C(=O)[C@H]3NN=C([C@H]3C2=O)C(=O)CCN4C(=O)c5ccccc5C4=O |
| 912837 | 301.34 | 6 | 7.0208333333333 | 1 | Cc1ccc(OCC(=O)NCC(=O)n2nc(C)cc2C)cc1 |
| 950421 | 263.4 | 7 | 7.0155952380952 | 1 | Cc1ccccc1CSCC(=O)NC2CCCC2 |
| 903207 | 291.34 | 11 | 7.015 | 0.90909090909091 | CC1CCCC(C)N1C(=O)COC(=O)\C=C\c2occc2 |
| 1563165 | 253.25 | 2 | 7.0125 | 1 | Nc1ccc(cc1)C2=CC(=O)c3c(O)cccc3O2 |
| 1328495 | 281.31 | 57 | 7.011727503252 | 1 | OC[C@@]1(OC[C@@H]1c2ccccc2)n3nc4ccccc4n3 |
| 1209849 | 311.34 | 3 | 7 | 1 | Cn1nccc1c2coc3c(cccc23)C(=O)N4CCOCC4 |
| 942098 | 349.4 | 4 | 6.9625 | 1 | Cc1ccc(cc1C(=O)c2nccn2C)S(=O)(=O)N3CCOCC3 |
| 1834654 | 252.63 | 2 | 6.961727503252 | 1 | OC(=O)c1cc(ncn1)c2ccc(F)c(Cl)c2 |
| 832689 | 283.35 | 8 | 6.9271875 | 1 | CC(=O)Nc1nnc(SCc2cccc(F)c2)s1 |
| 410704 | 483.54 | 4 | 6.905 | 1 | CNC(=O)Cc1ccccc1CNC(=O)c2nc(N3CCCCS3(=O)=O)c4cccnc4c2O |
And here, some of the most potent structures