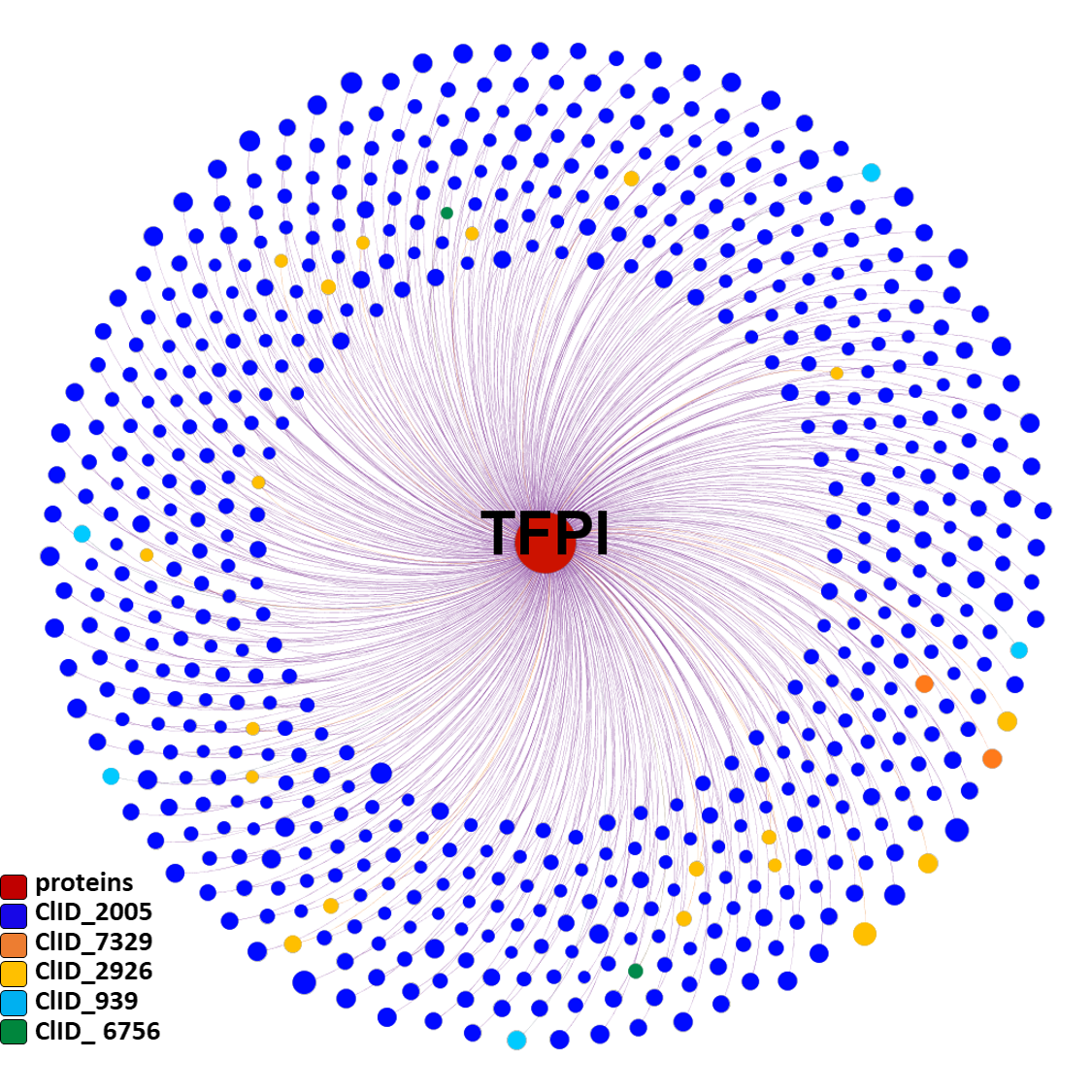
In ChEMBL there are about 3k experimental records for functional and binding assays carried out on the TFPI protein with 973 molecules.
About 760 molecules can be considered TFPI actives, and most of them (>600 molecules) have been predicted as active against coronavirus infection.
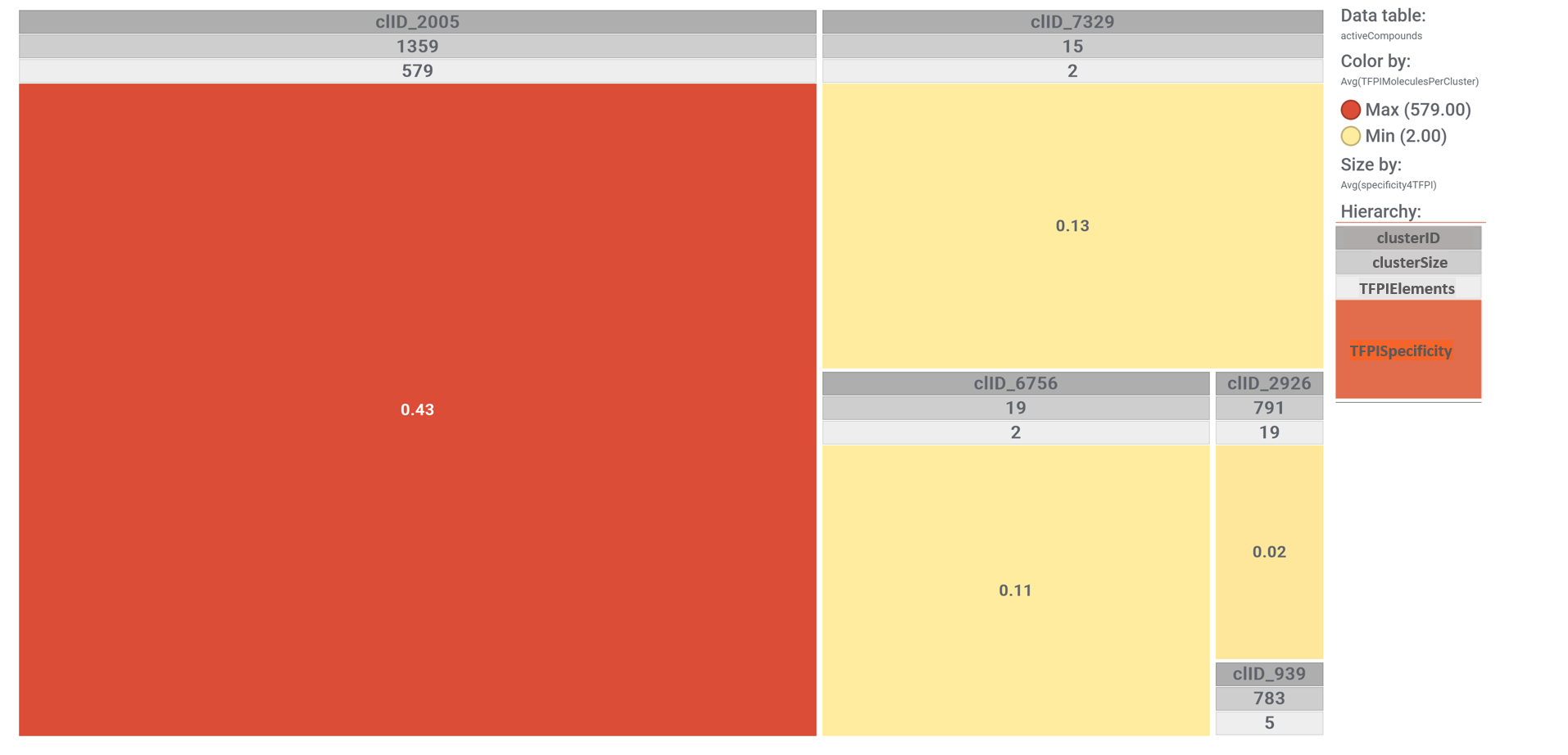
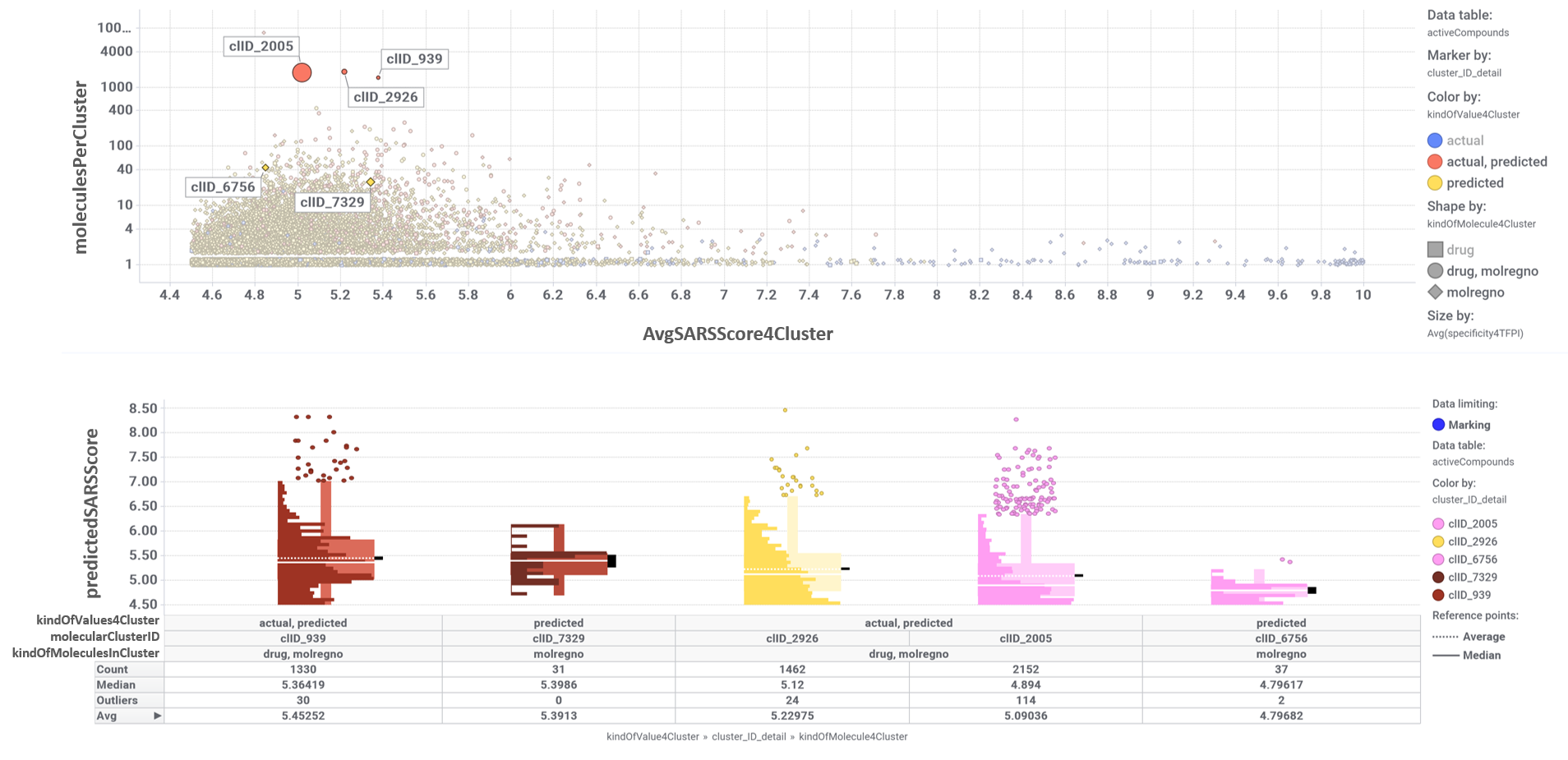
These SARS predicted active molecules belong to only 5 different molecular clusters
Just click on each plot below to get an expanded view.
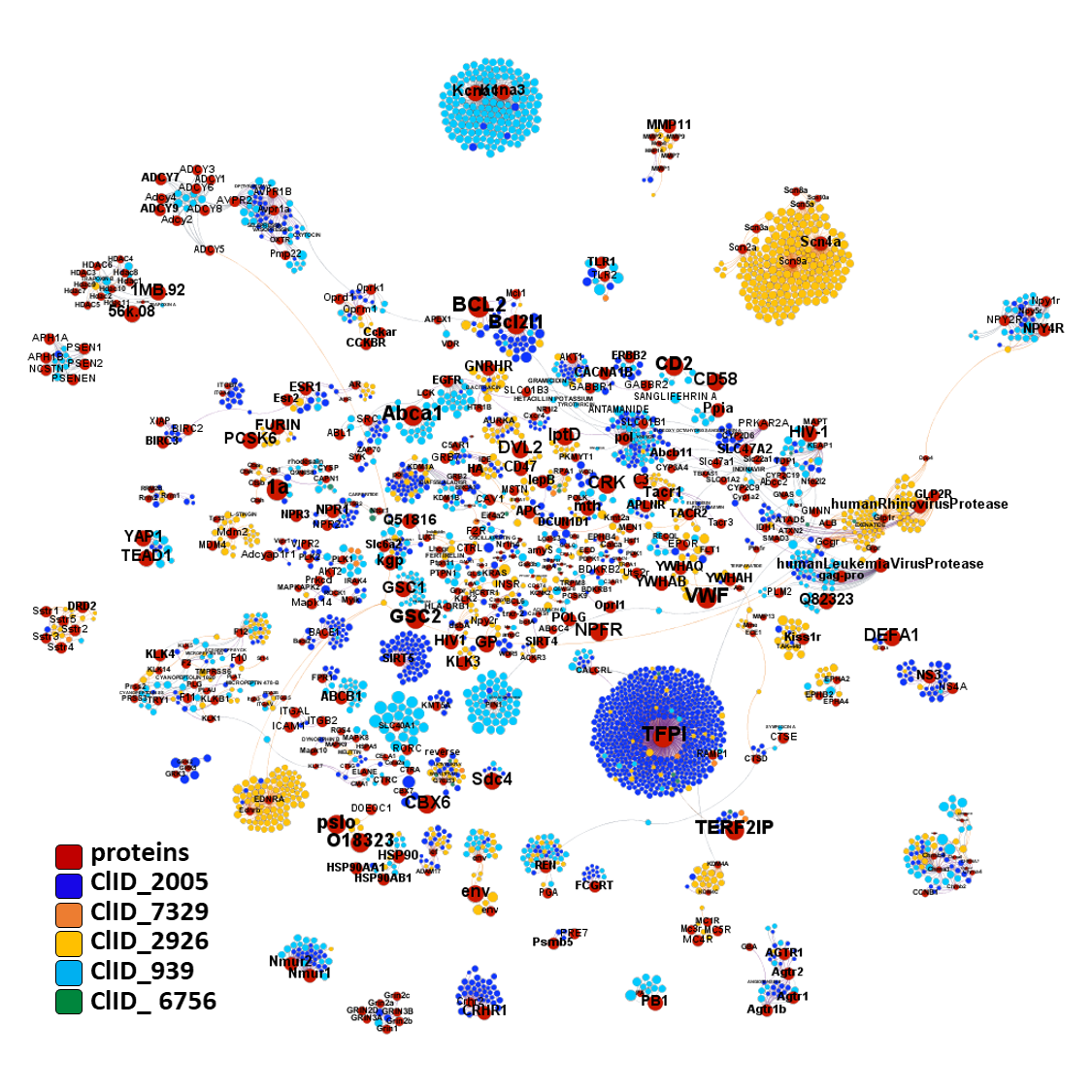
A graph displaying interactions of all components of molecular clusters with protein targets in ChEMBL DB, shows again little specificity for TFPI for most of them. Only ClID_2005 keeps a significant number of molecules hitting TFPI.