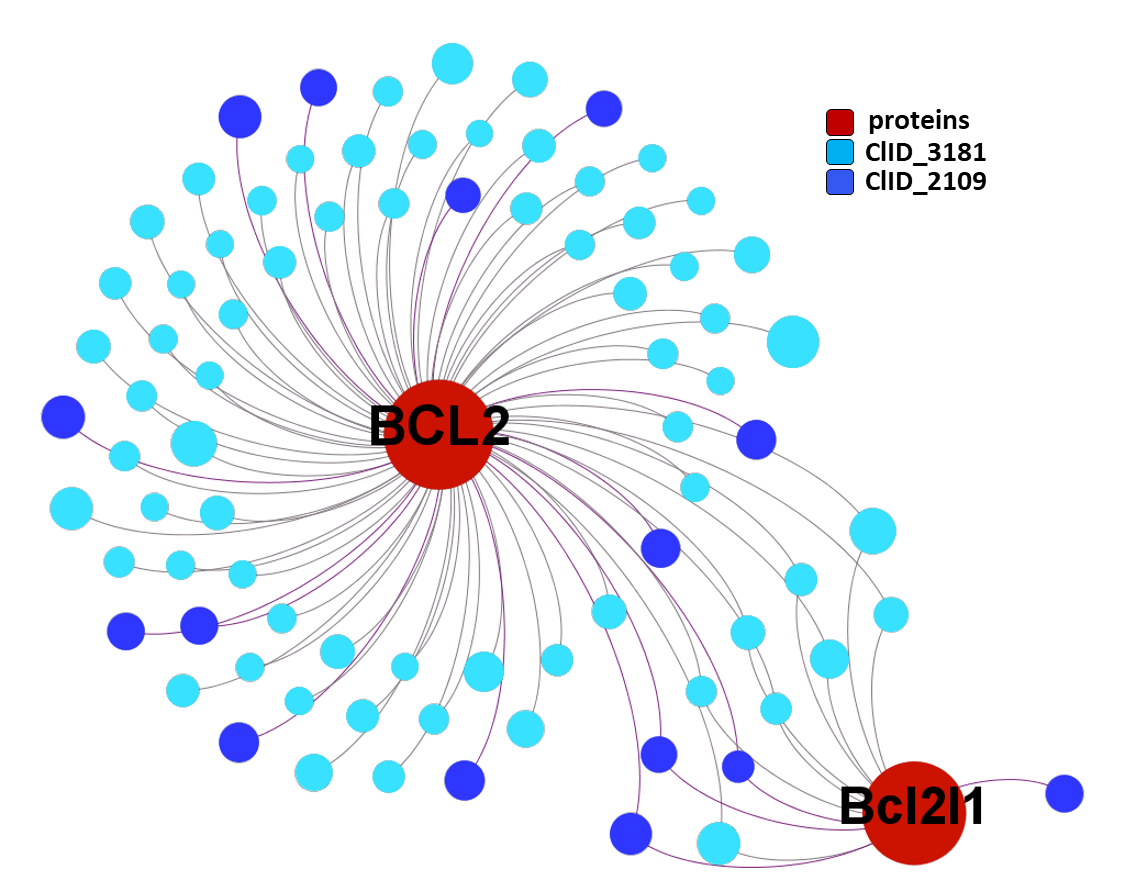
Now, we can select the compounds belonging to clusters that hit BCL2 proteins with higher specificity.
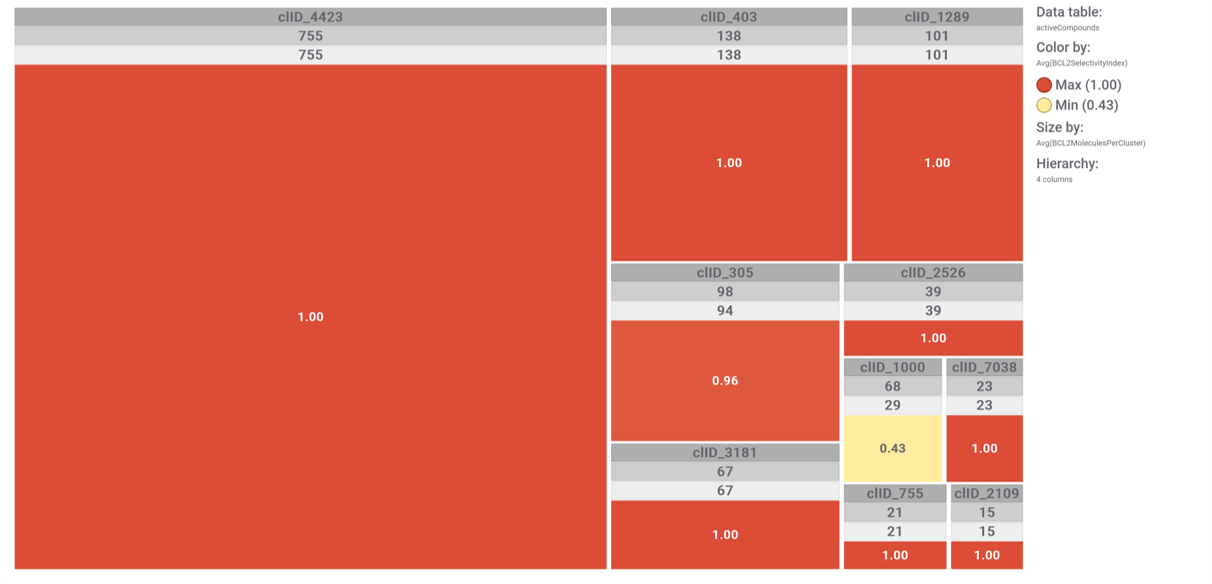
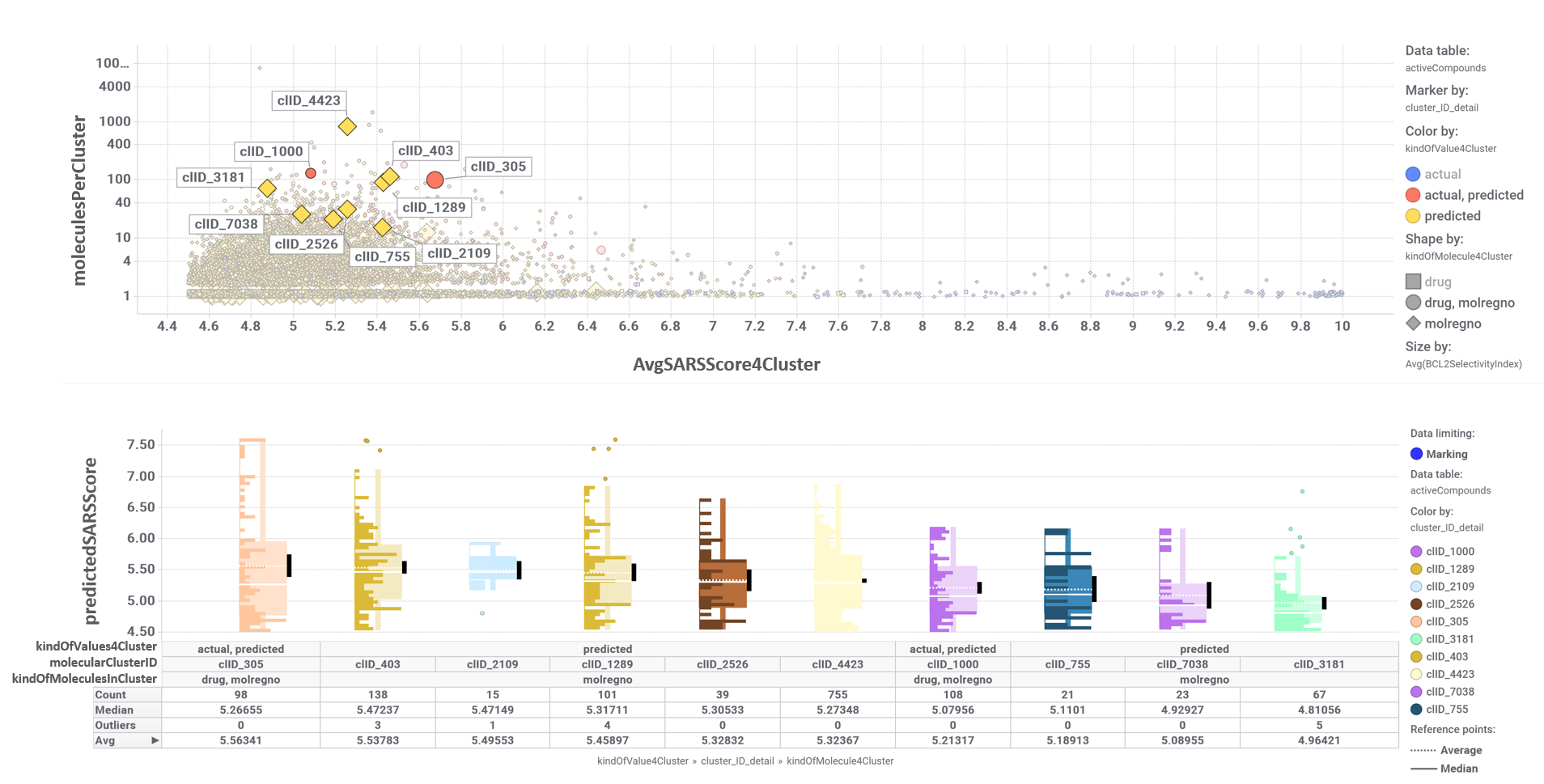
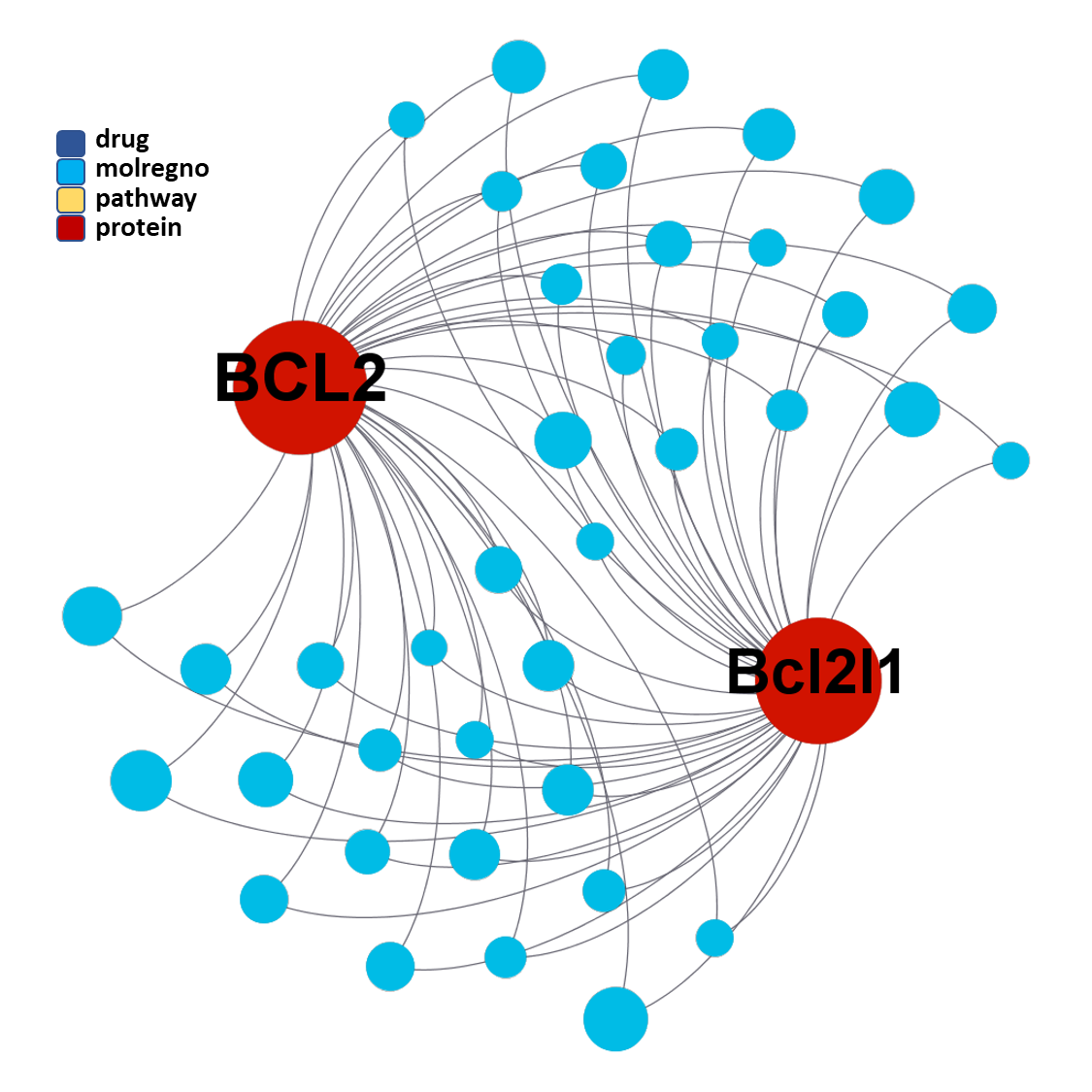
The tree map shows most specific molecular clusters, based on how many compounds hit on BCL2 and related proteins vs the total number of compounds that have molecular interaction records in ChEMBL. On the right, these BCL2 specific clusters are highlighted in the scatter plot representing SARS predicted or actual potency vs total number of molecules per cluster. The box plot represents the distribution of SARS score for each cluster.
Just click on each plot to get an expanded view.
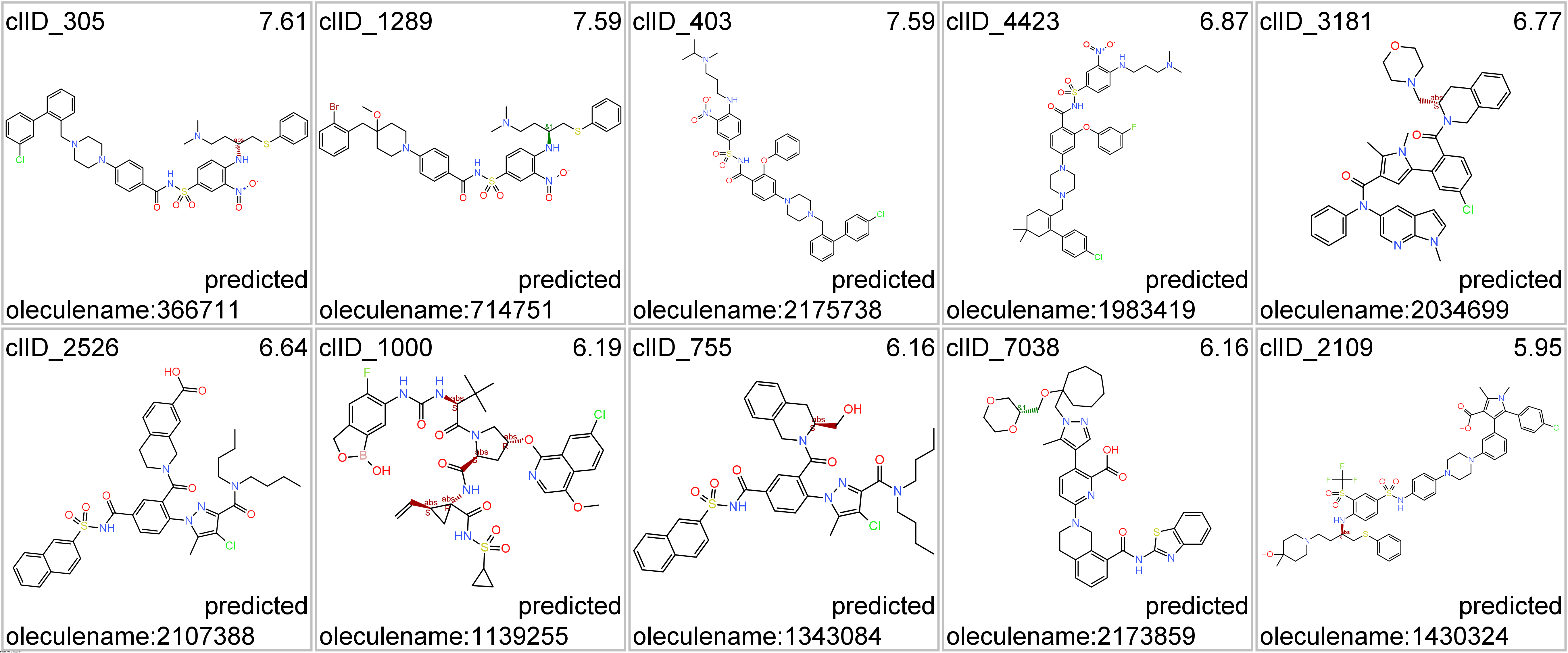
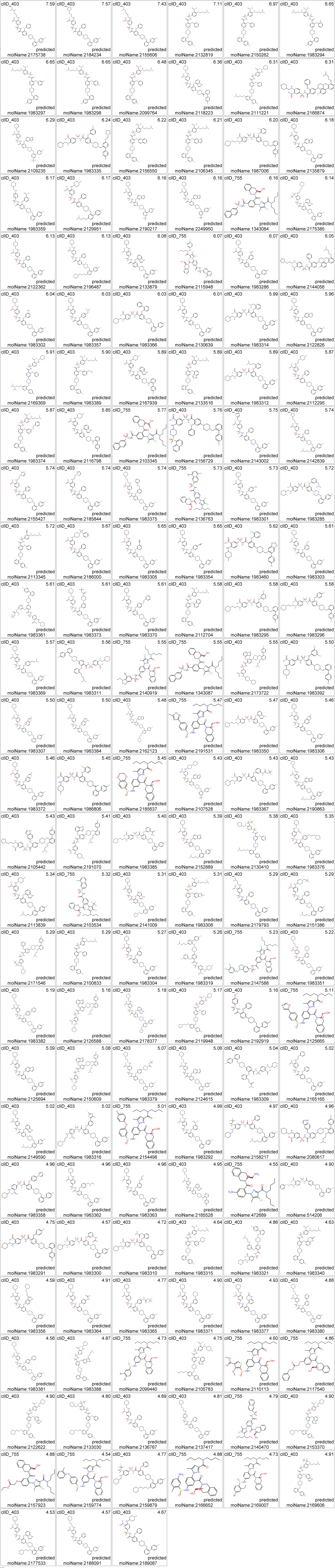
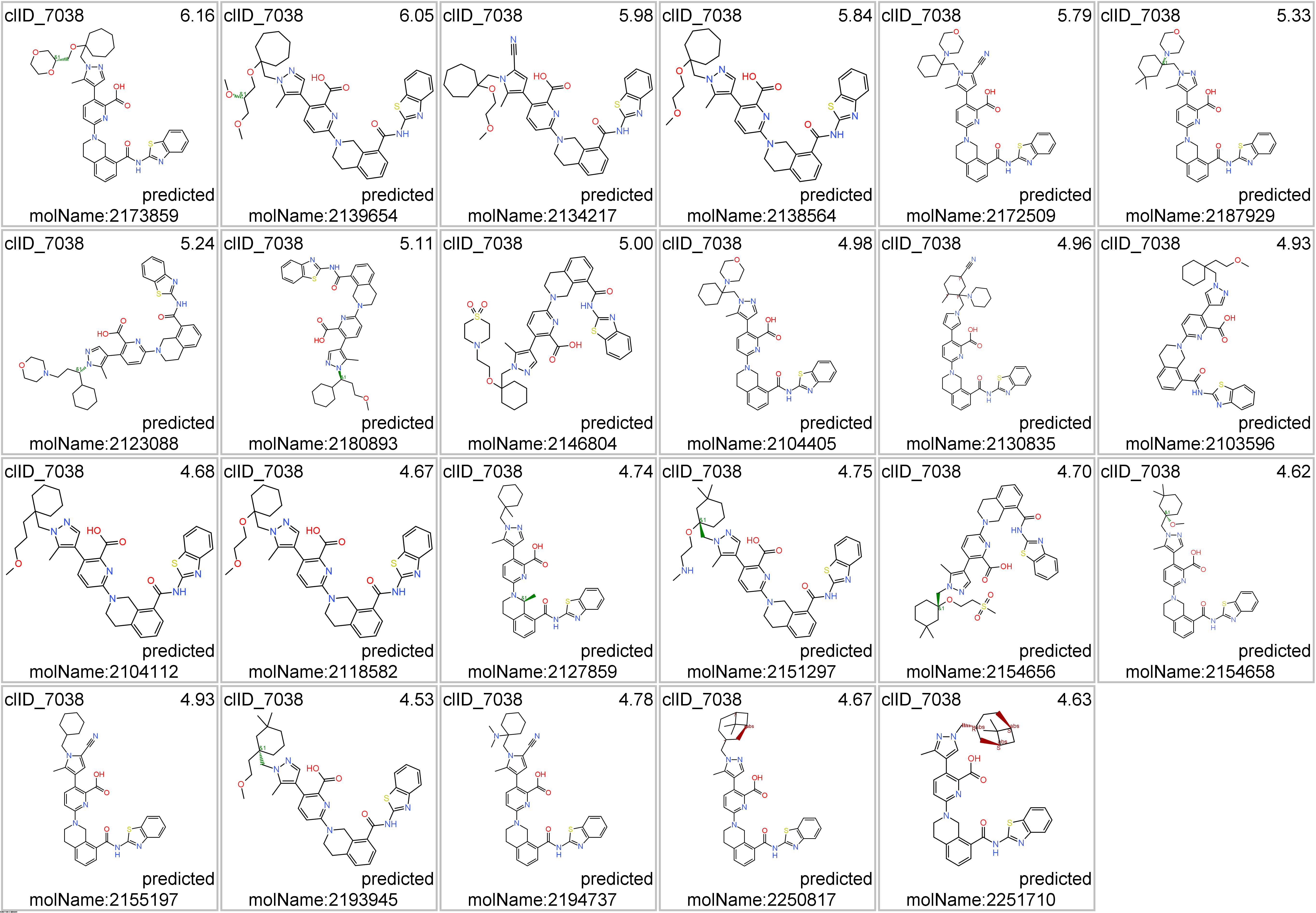
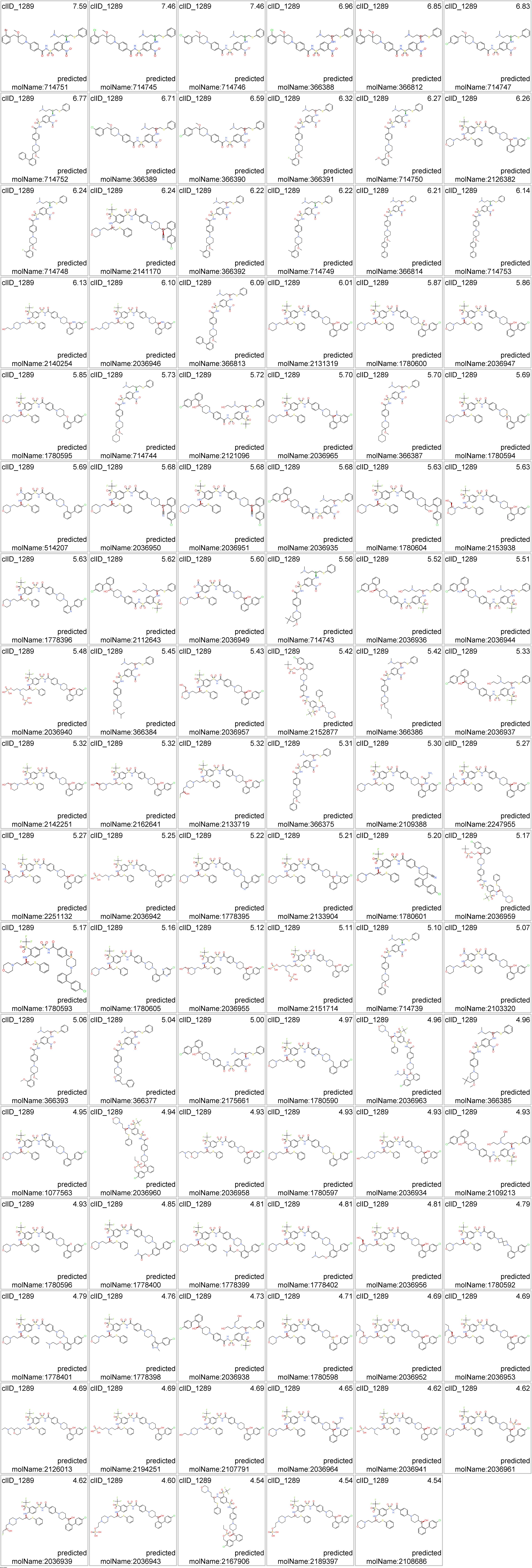
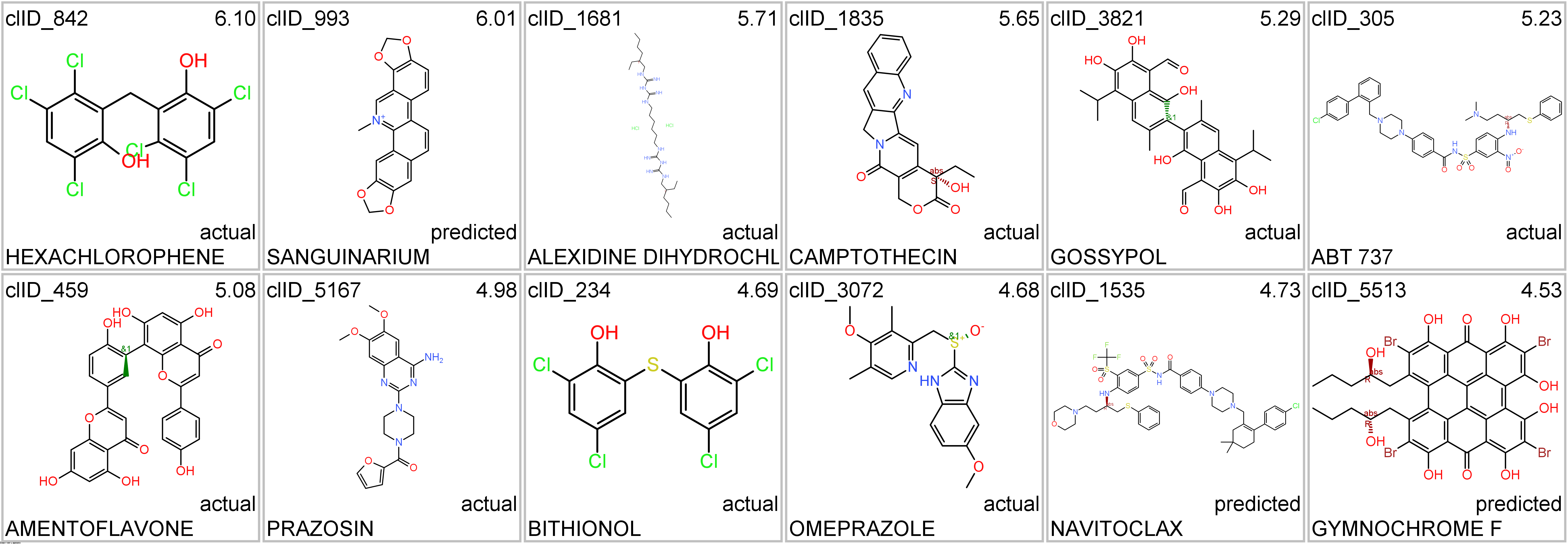
Representative structures of BCL2-specific clusters.
Click on the tabs below to visualize interactions and structures of molecules classified in most representative clusters.
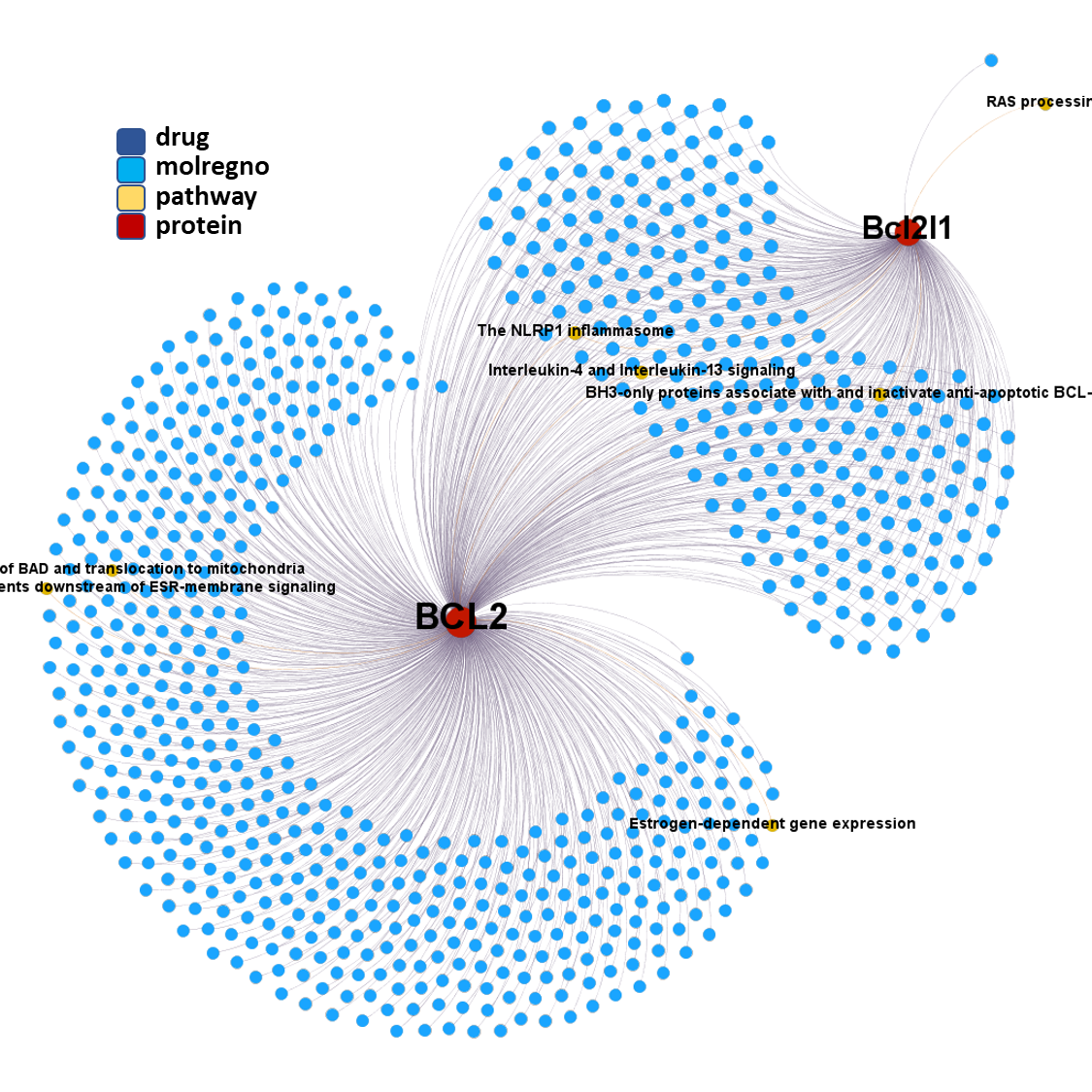
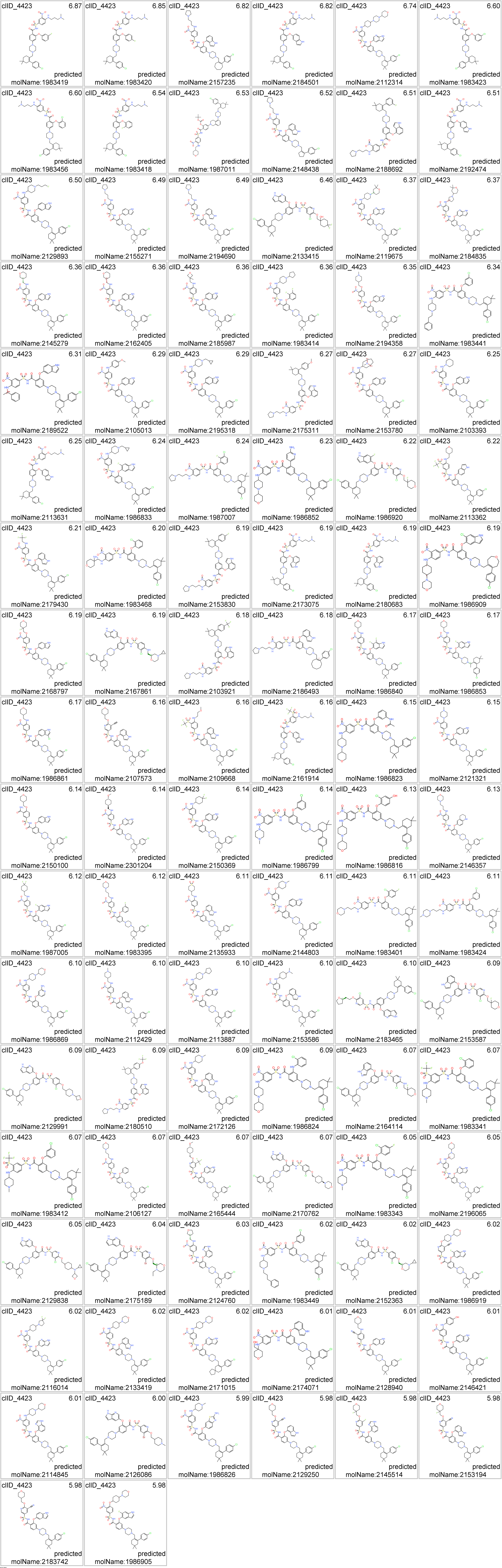
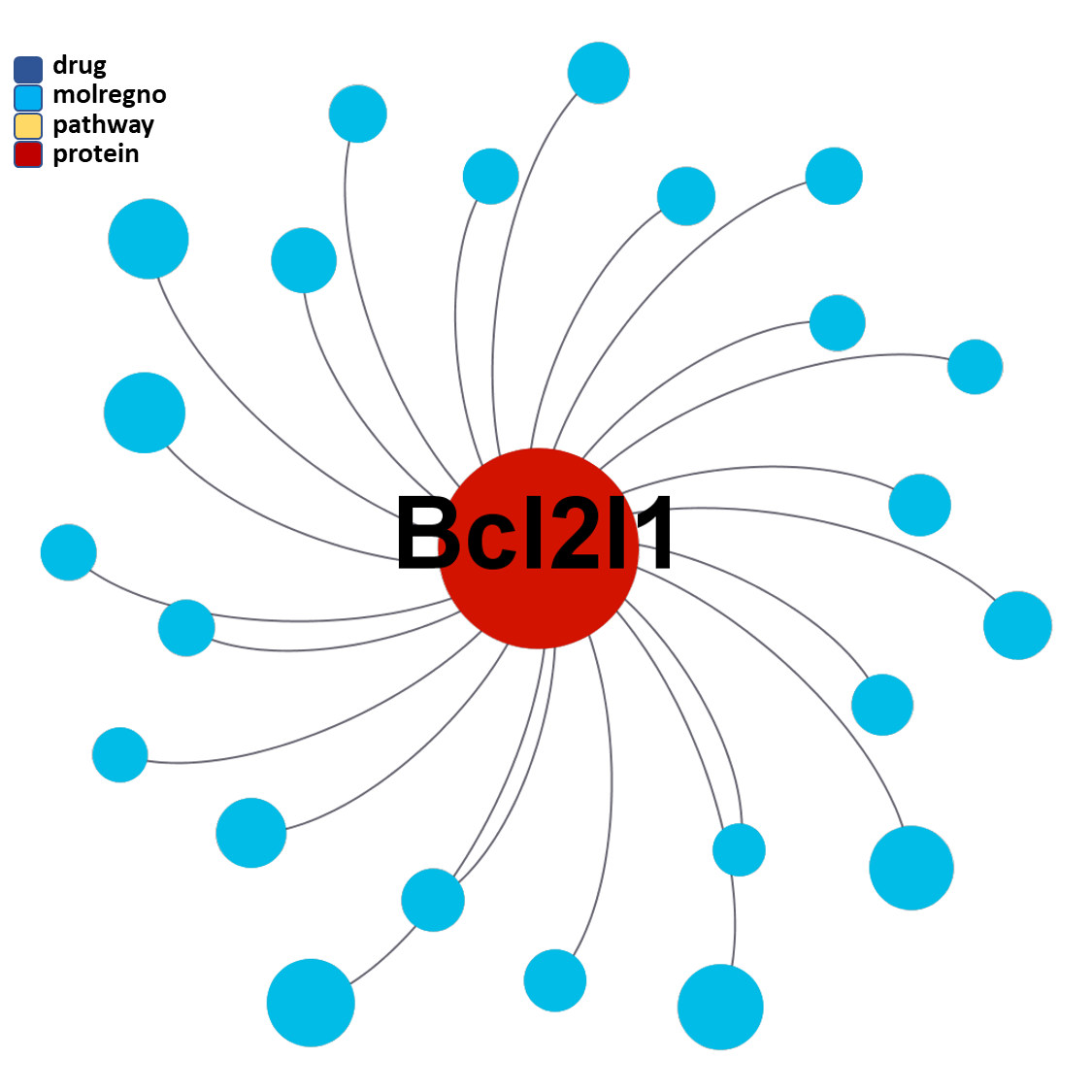
ClID_4423 compounds interact specifically with BCL2 and in combination with Bcl2l1 protein
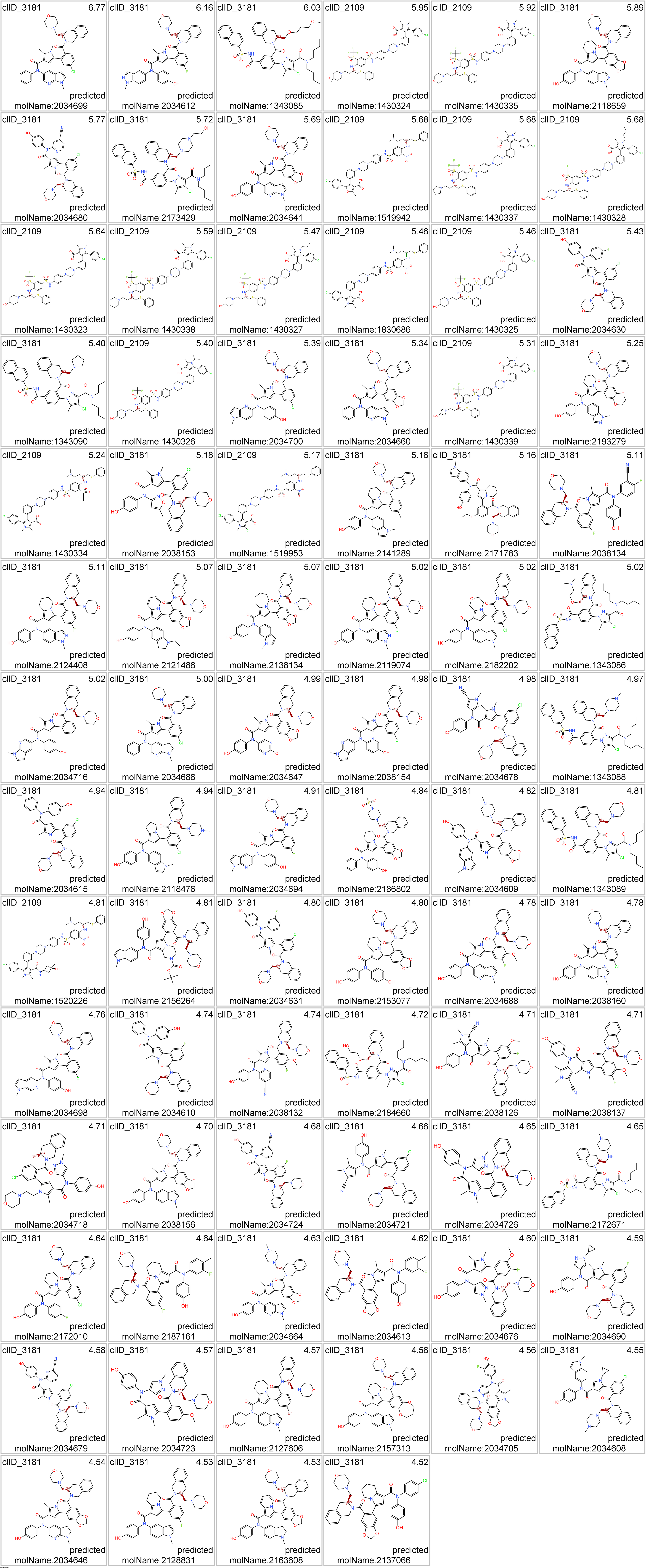
ClID_3181 & ClID_2109 compounds interact specifically with BCL2 and in combination with Bcl2l1 protein, similarly to ClID_4423
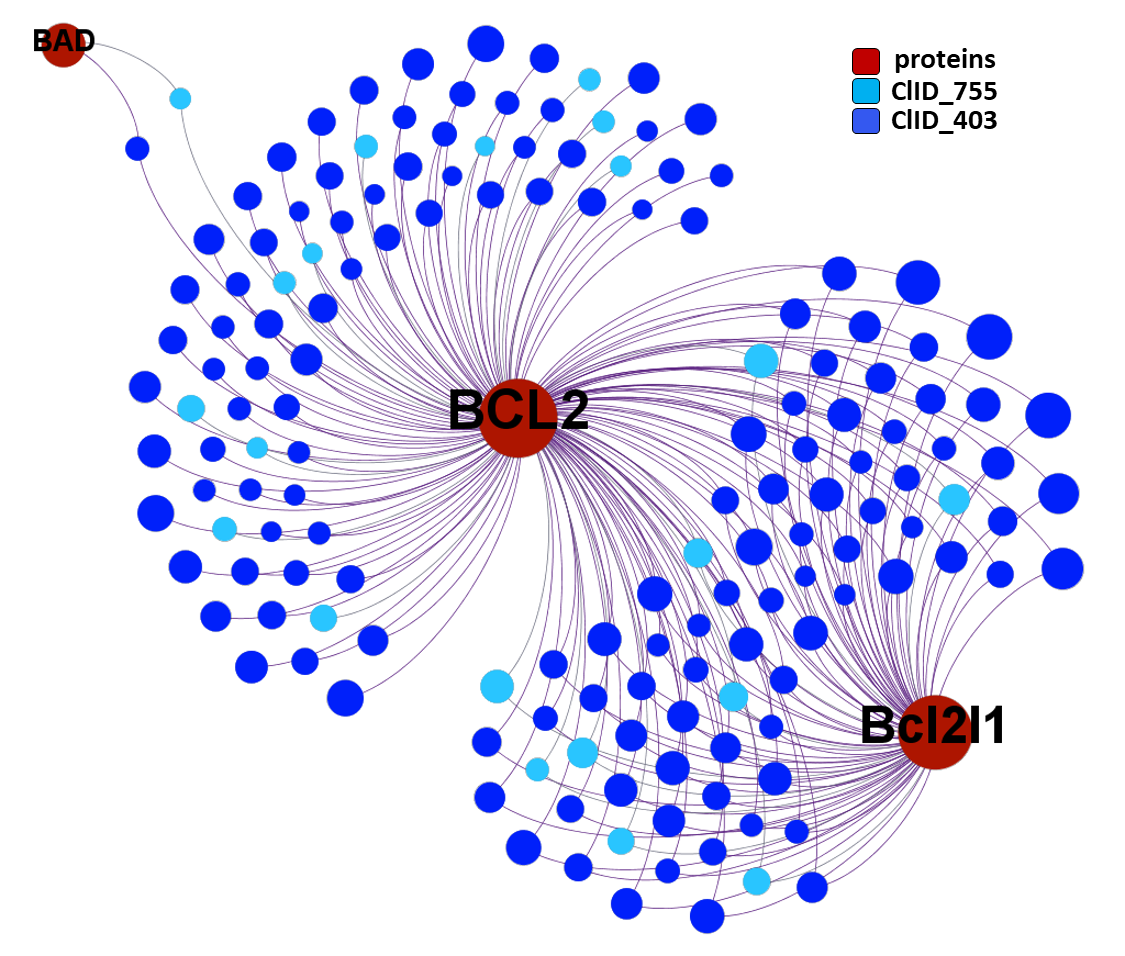
ClID_403 & ClID_755 compounds interact specifically with BCL2 and in combination with Bcl2l1 protein, similarly to ClID_4423, ClID_3181 and ClID_2109, with the inclusion of a small interaction rate with the Bcl2-associated agonist of cell death (BAD).
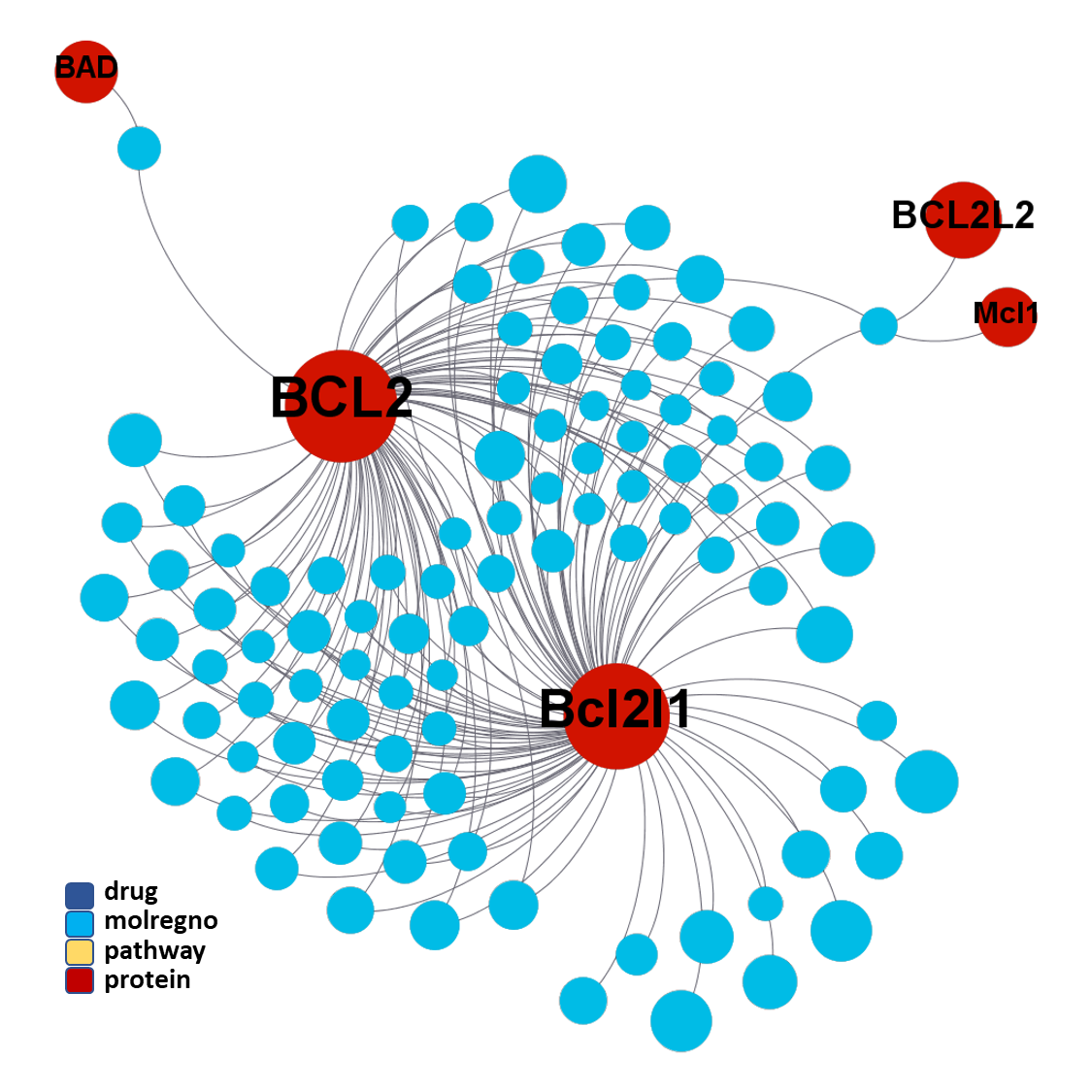
ClID_1289 compounds interact mainly with BCL2 & Bcl2l1 proteins, and eventually with the related Bcl2-associated agonist of cell death (BAD) & the Induced myeloid leukemia cell differentiation protein Mcl-1 – Induced myeloid leukemia cell differentiation protein Mcl-1 homolog (Mcl1).
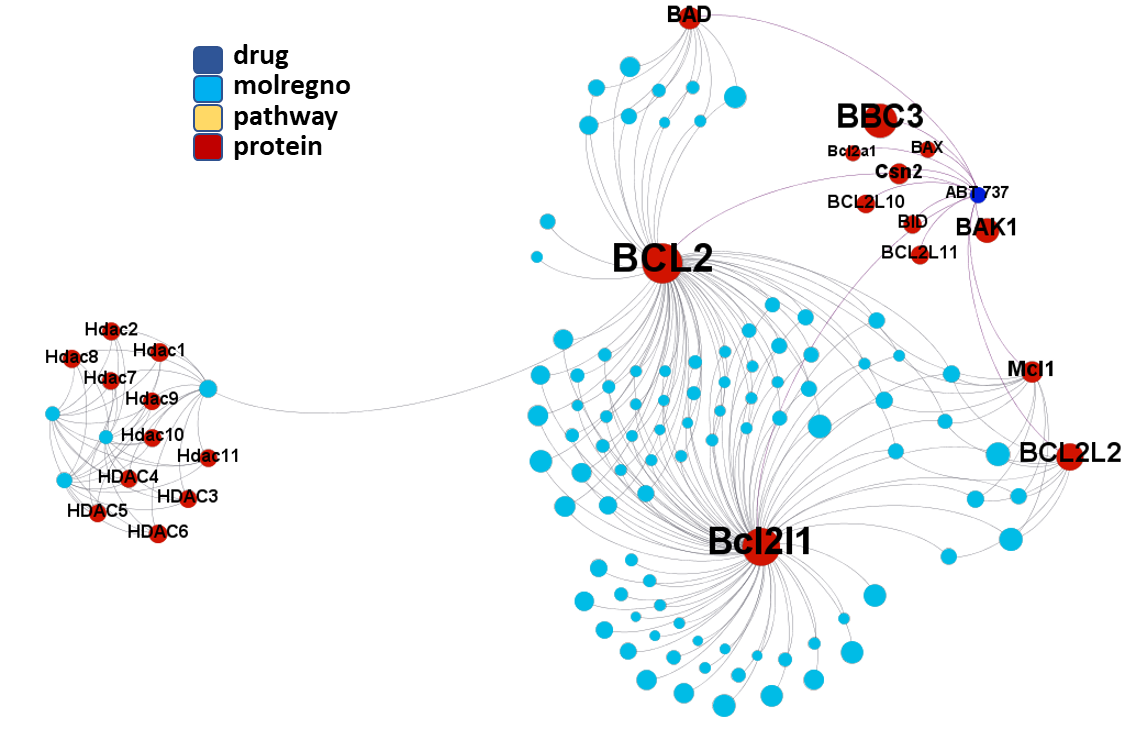
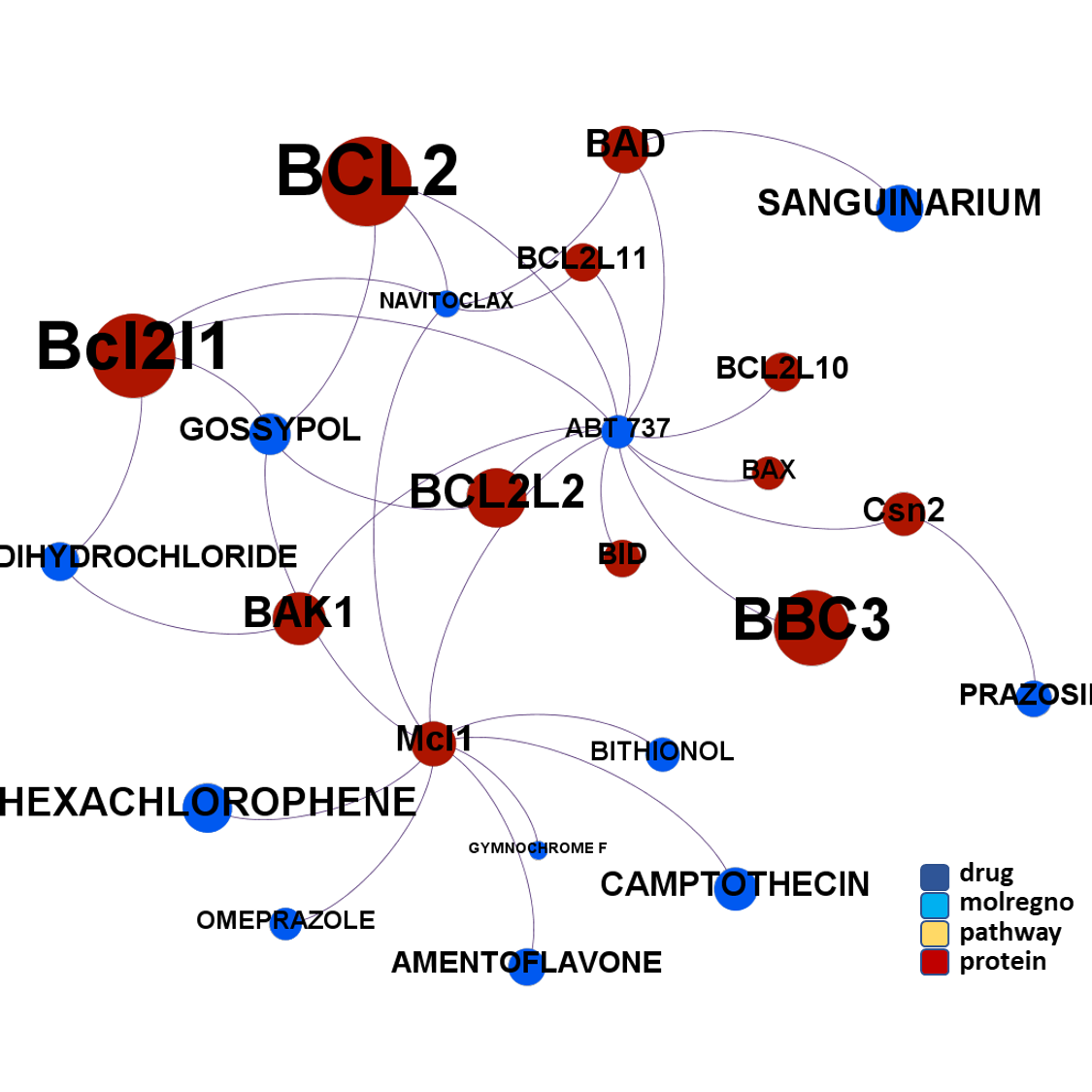
ClID_305 molecules includes ABT-737, the known BCL2 family proteins inhibitor and inductor of apoptosis. Besides the usual BCL2, Bcl2l1 and BAD, ABT-737 interacts with several additional BCL-related proteins and casein. A subgroup of elements in this cluster also shows activity in histone deacetylases.
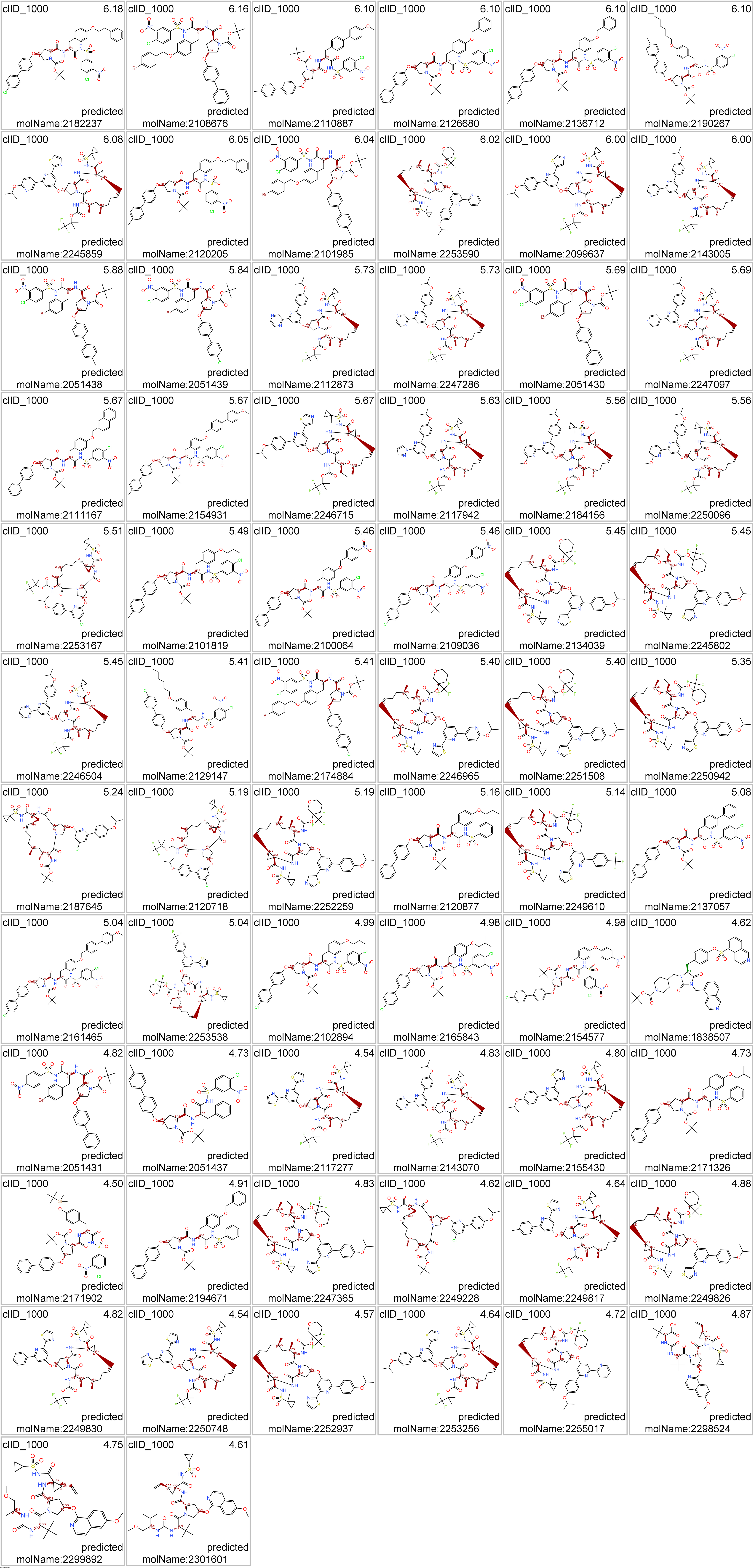
A fraction of ClID_1000 molecules shows some specificity for the Induced myeloid leukemia cell differentiation protein Mcl-1 – Induced myeloid leukemia cell differentiation protein Mcl-1 homolog (Mcl1), while some others display crossed interactions between BCL2, Bcl1l1 and Mcl1. An additional group of compounds presents annotations of activity upon the NS3 and NS4A proteins of the HCV protease complex.