Kinesins are proteins belonging to a class of motor proteins found in eukaryotic cells. Kinesins move along microtubule (MT) filaments, and are powered by the hydrolysis of adenosine triphosphate (ATP) (thus kinesins are ATPases), a type of enzyme. The active movement of kinesins supports several cellular functions including mitosis, meiosis and transport of cellular cargo, such as in axonal transport. (taken from wikipedia).
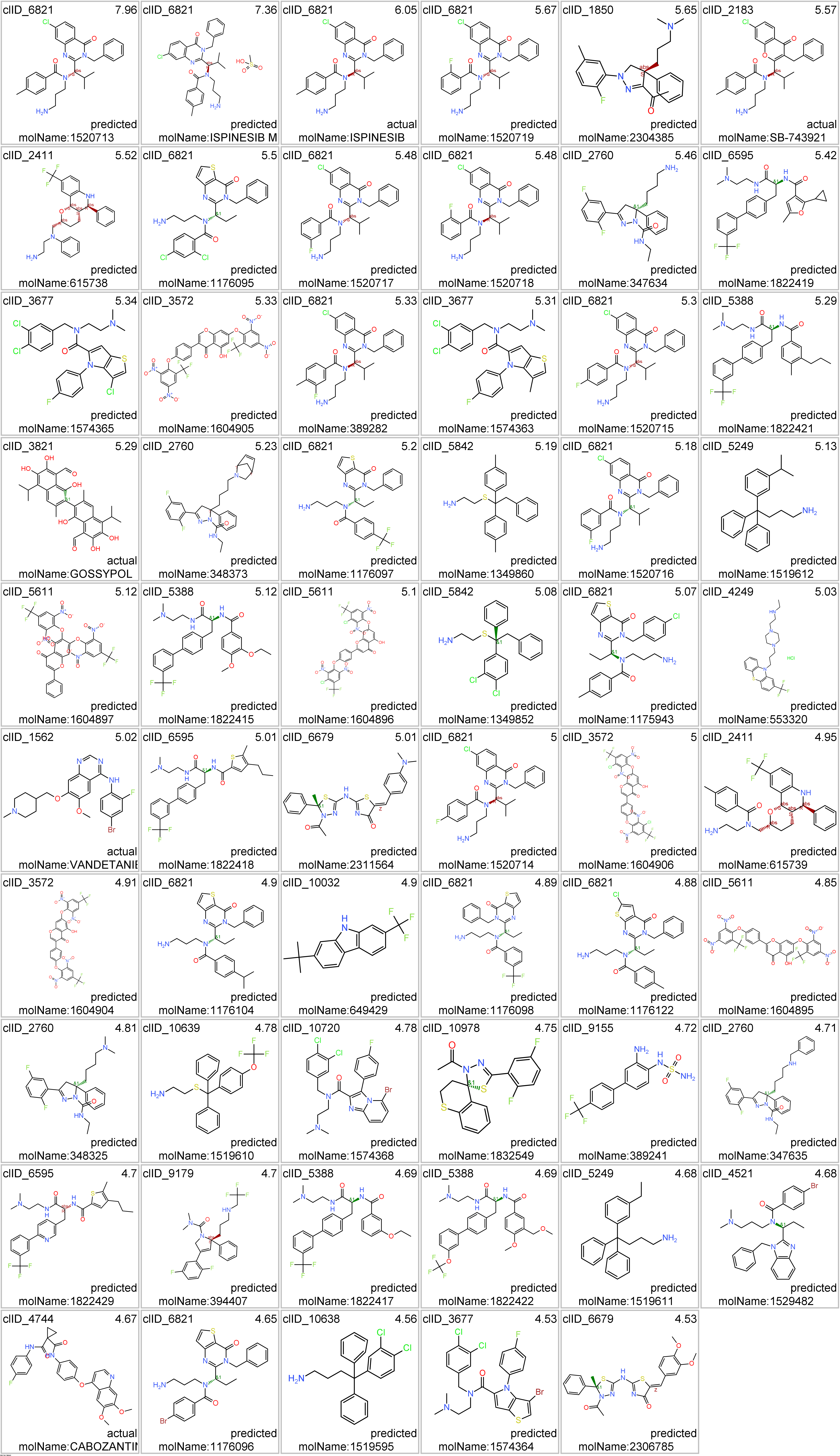
In this section we will focus on molecules predicted as active against coronaviruses by machine learning models based on SARS-CoV assays in ChEMBL DB that interact with different kinesins.
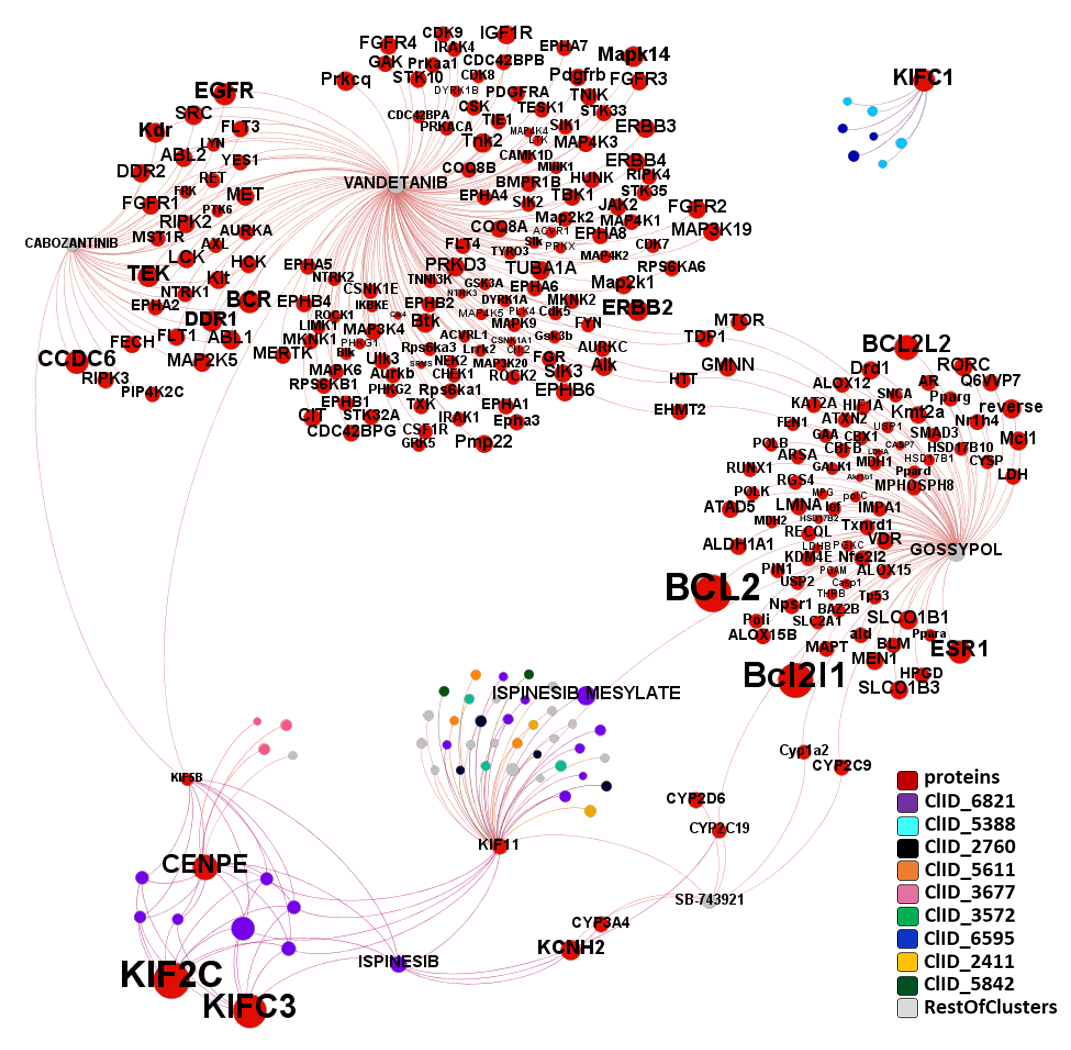
The network graph below displays all molecular interactions of the 59 kinesin interacting compounds with all proteins recorded in ChEMBL. Compounds have been classified in clusters according to their structural similarity. A specific color code has been assigned to most relevant clusters. ClIDs 5388 & 6595 seem KIFC1 specific. Main molecular cluster, ClID_6821 molecules share their activity among KIF11, KIF2c, KIFC3 and KIF5B. ChEMBL keeps records of VEGFR inhibitors vandetanib and cabozantinib interactions with kinesins alongside many other proteins, simmilarly to the pleiotropic gossypol.
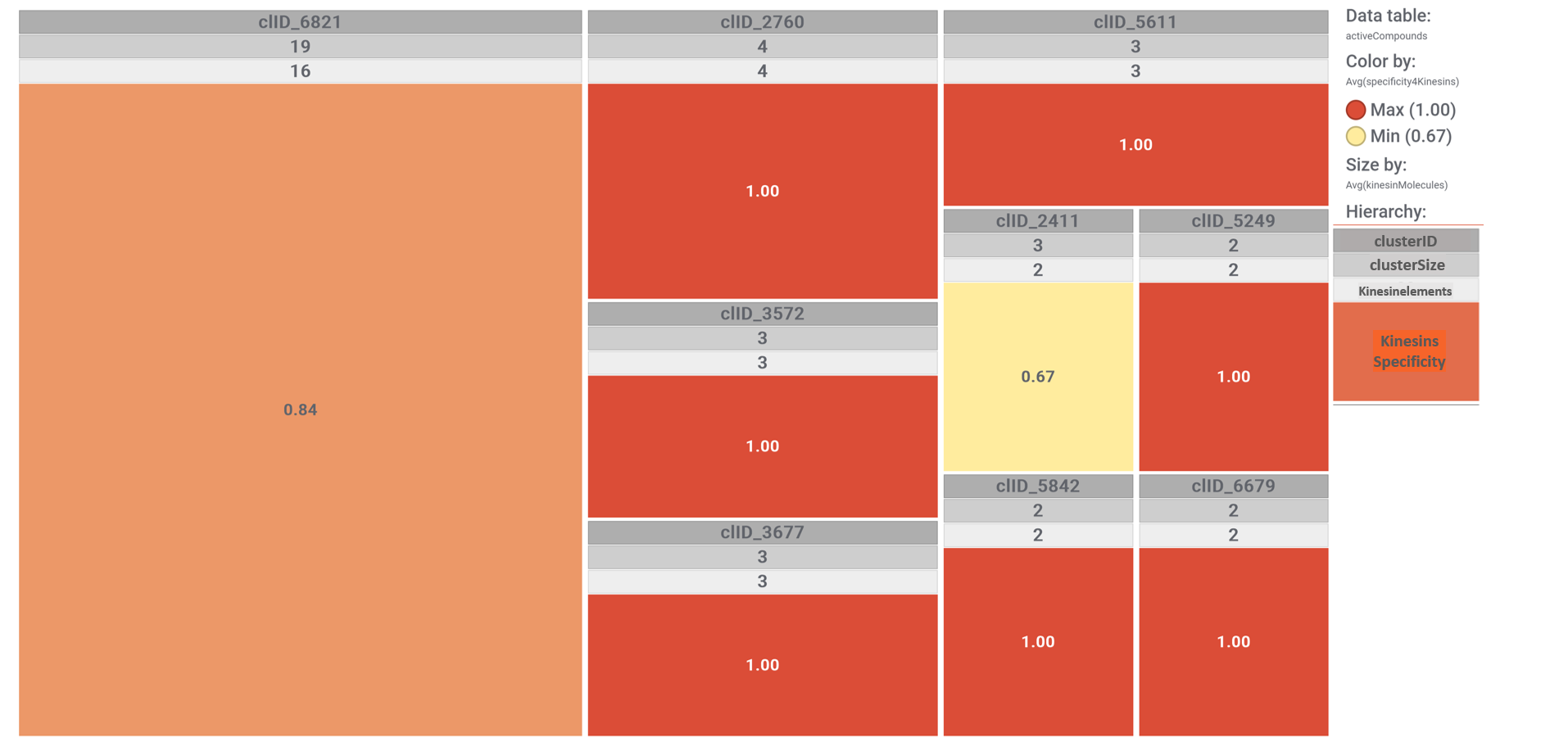
Each molecular cluster specificity for this set of molecules can be visualized with a simple tree map, where each cluster is sized according to its number of elements and its color graded by the ratio between the number of cluster elements hitting on kinesins and the cluster size. See legend.