The sources
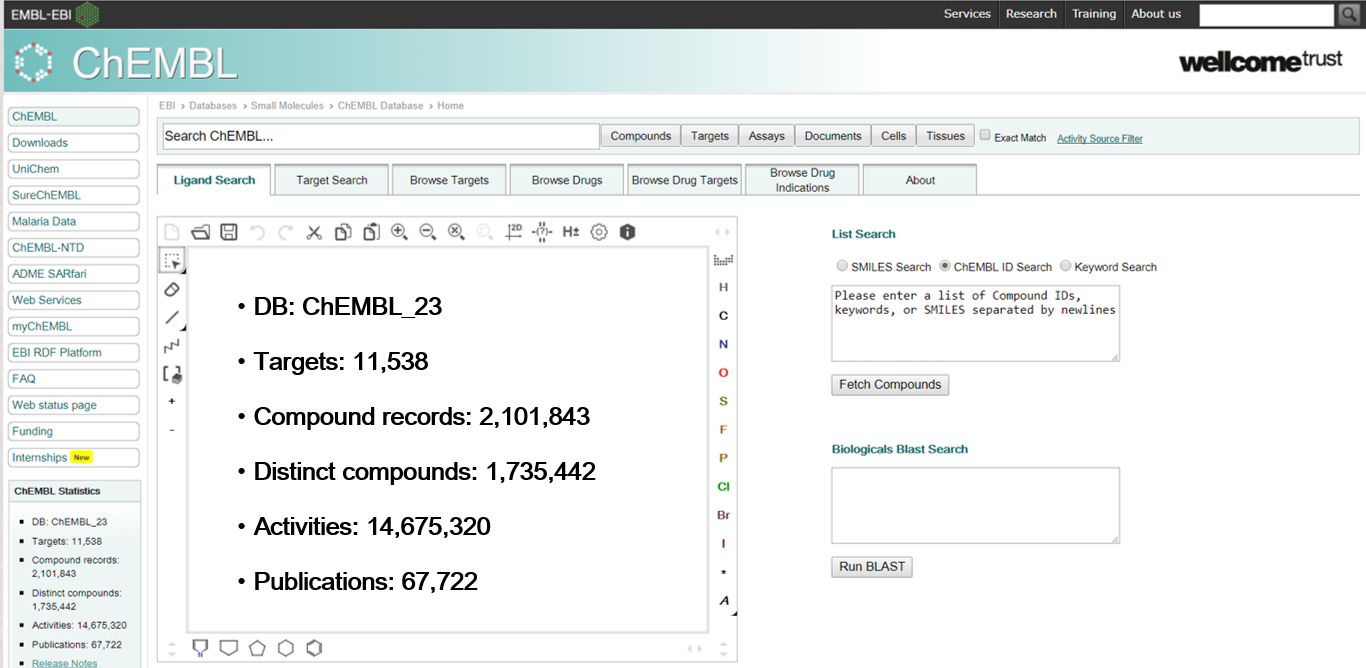
ChemBl database
ChemBl or ChEMBLdb is a manually curated chemical database of bioactive molecules with drug-like properties. It is maintained by the European Bioinformatics Institute (EBI), of the European Molecular Biology Laboratory (EMBL), based at the Wellcome Trust Genome Campus, Hinxton, UK
The database, originally known as StARlite, was developed by a biotechnology company called Inpharmatica Ltd. later acquired by Galapagos NV. The data was acquired for EMBL in 2008 with an award from The Wellcome Trust, resulting in the creation of the ChemBl chemogenomics group at EMBL-EBI, led by John Overington
The ChemBl database contains compound bioactivity data against drug targets. Bioactivity is reported in Ki, Kd, IC50, and EC50 Data can be filtered and analyzed to develop compound screening libraries for lead identification during drug discovery
ChemBl version 2 (ChEMBL_02) was launched in January 2010, including 2.4 million bioassay measurements covering 622,824 compounds, including 24,000 natural products. This was obtained from curating over 34,000 publications across twelve medicinal chemistry journals. ChemBl’s coverage of available bioactivity data has grown to become “the most comprehensive ever seen in a public database.”In October 2010 ChemBl version 8 (ChEMBL_08) was launched, with over 2.97 million bioassay measurements covering 636,269 compounds.
ChEMBL_10 saw the addition of the PubChem confirmatory assays, in order to integrate data that is comparable to the type and class of data contained within ChemBl.
ChEMBLdb can be accessed via a web interface or downloaded by File Transfer Protocol. It is formatted in a manner amenable to computerized data mining, and attempts to standardize activities between different publications, to enable comparative analysis. ChemBl is also integrated into other large-scale chemistry resources, including PubChem and the ChemSpider system of the Royal Society of Chemistry.
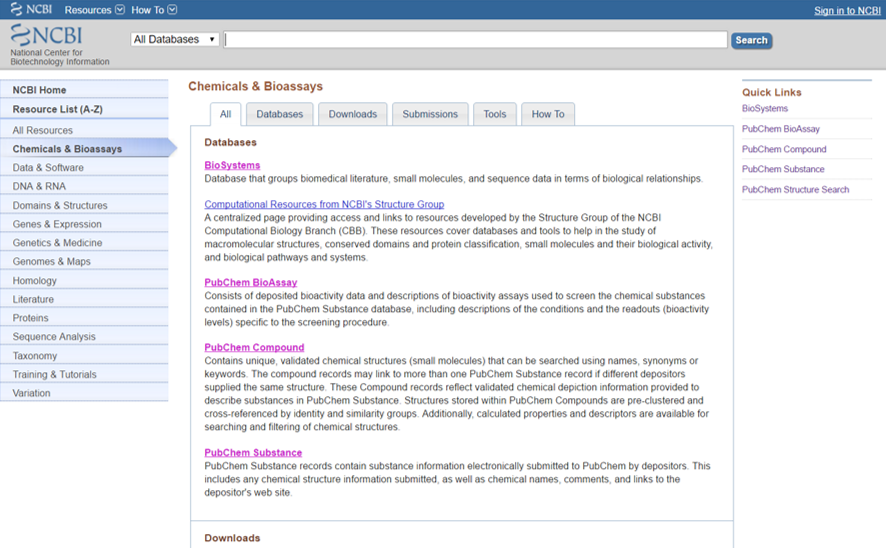
The NCBI database
The National Center for Biotechnology Information (NCBI) is part of the United States National Library of Medicine (NLM), a branch of the National Institutes of Health (NIH). The NCBI is located in Bethesda, Maryland and was founded in 1988 through legislation sponsored by Senator Claude Pepper.
The NCBI houses a series of databases relevant to biotechnology and biomedicine and is an important resource for bioinformatics tools and services. Major databases include GenBank for DNA sequences and PubMed, a bibliographic database for the biomedical literature. Other databases include the NCBI Epigenomics database. All these databases are available online through the Entrez search engine.
NCBI was directed by David Lipman, one of the original authors of the BLAST sequence alignment program and a widely respected figure in bioinformatics. He also leads an intramural research program, including groups led by Stephen Altschul (another BLAST co-author), David Landsman, Eugene Koonin (a prolific author on comparative genomics), John Wilbur, Teresa Przytycka, and Zhiyong Lu. David Lipman stood down from his post in May 2017.
NCBI is listed in the Registry of Research Data Repositories re3data.org.