Construction of the master dataset (SQL).
When an appropriate assay or set of assays is identified for study, the molecules that have been assayed against it are selected.
An sql statement is used to obtain the results of the selected molecules for all experiments in the database.
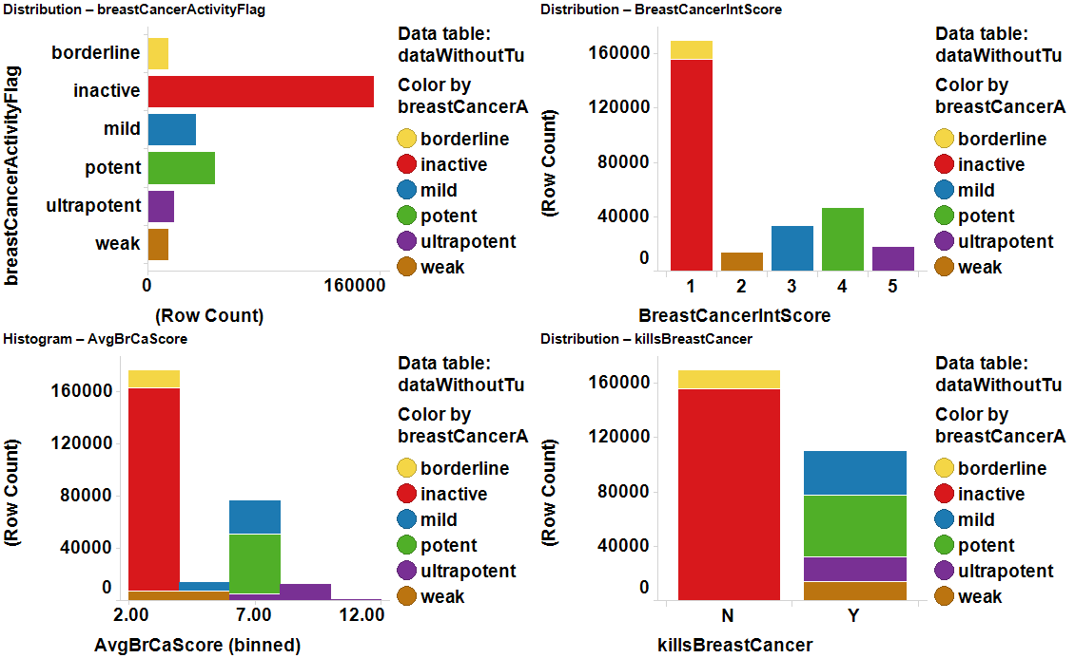
The rows with results of the selected molecules upon the assay of study are removed from the master dataset and placed in a separate one, pivoted by molecule, and the average potency for the assay calculated and added to the master dataset for regression. Activity flags and integer scores are subsequently added for further classification purposes. Now, the master table contains, apart from all the activityChemistryTarget columns, several columns that act as class descriptors. For Breast cancer it would be:
killsBreastCancer:’Y’, ‘N’; for binomial classification
BrCaActivesNNet:’0’, ‘1’ for binomial classification with NNet
breastCancerActivityFlag: ’inactive’, borderline, ‘weak’, ‘mild’, ‘potent’, ‘ultrapotent’ for multiclass,
BreastCancerIntScore: ‘1’, ‘2’, ‘3’, ‘4’, 5’, just in case
AvrBrCaScore: for regression
Names would be changed accordingly for other assays.
The columns with the activity values and flags are added to the master dataset to allow correlations, regressions and classifications.