Prediction of activity
1.Data preparation: Construction of the master dataset for Machine Learning (SQL).
a)When an appropriate assay, or set of assays, is identified for study, the molecules that have been assayed are selected.
b)An SQL statement is used to obtain the results of the selected molecules for all experiments in the database.
c)The rows with results from the selected molecules from the assay of study are removed from the master dataset and placed in a separate one, pivoted by molecule, and the average potency for the assay calculated. Activity flags and integer scores are subsequently added for further classification purposes.
d)The columns with the activity values and flags are now added to the master dataset to allow correlations, regressions and classifications.
2.Model Evaluation: Prediction of activity (R).
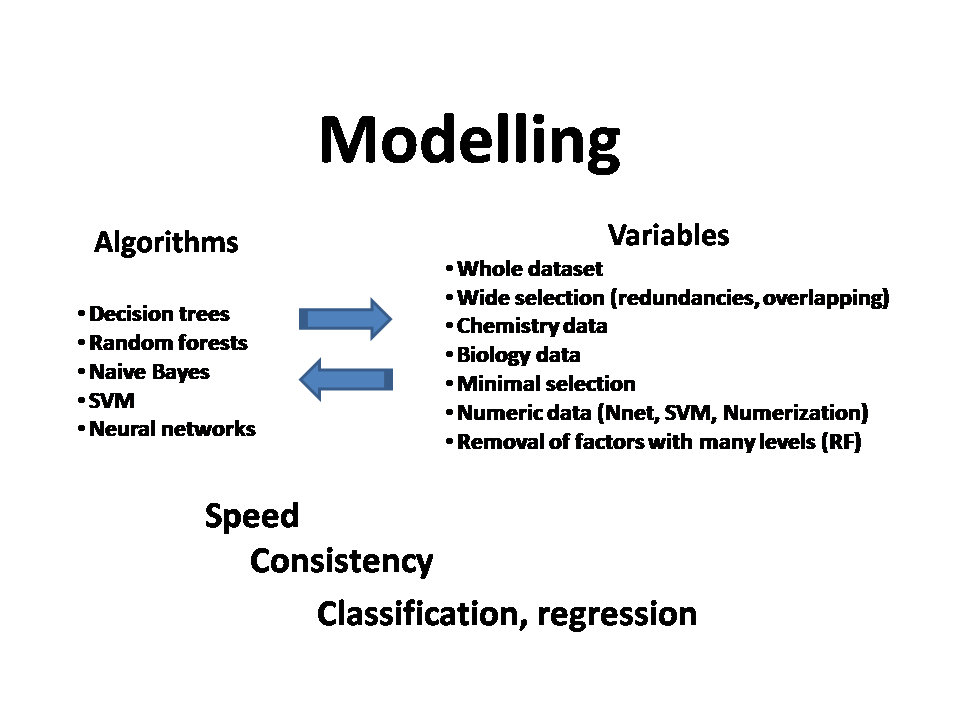
a) In R, a different combination of columns with different weights of chemical or biological variables are selected for regression or classification by decision trees, random forests Naive Bayes, support vector machines and neuronal networks (Caret, randomForest, e1071, naivebayes, neuralnet).
b)As there is an average activity value of the object assay, a class activity label identified for each molecule, all unique identifiers of molecules are excluded of the analysis (molregno, inchi key, smiles and formula).
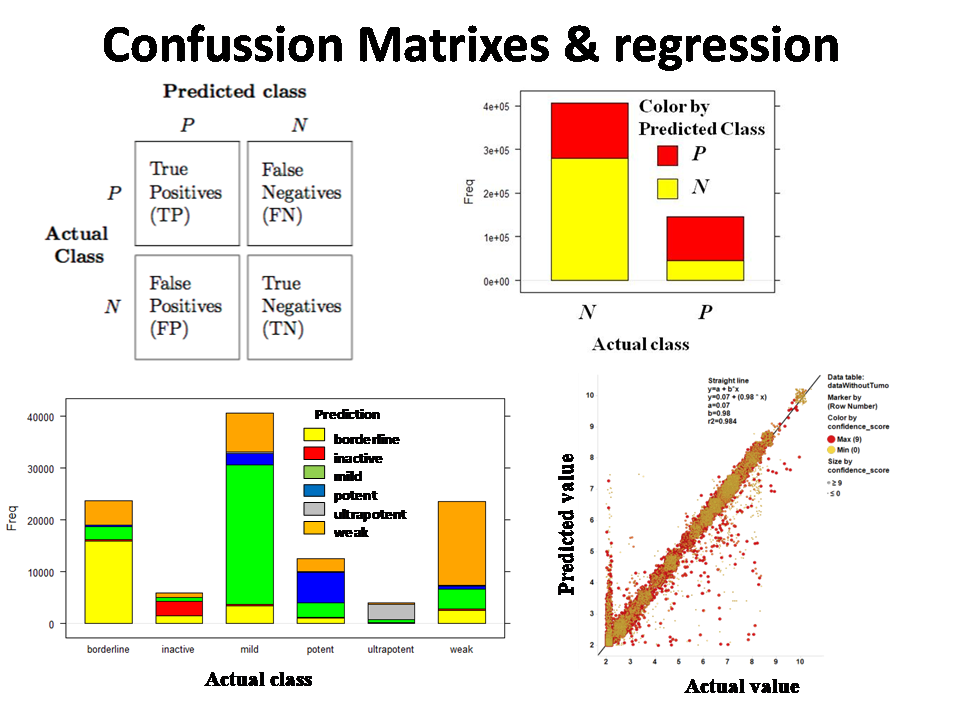
c)Confusion matrixes and statistics passed to R and SpotFire for visualization and comparison of several speed and performance parameters.
The ultimate evaluation of the predicted activities is made through statistics derived from confusion matrixes. Graphical comparisons of true and false positives and negatives, as below.