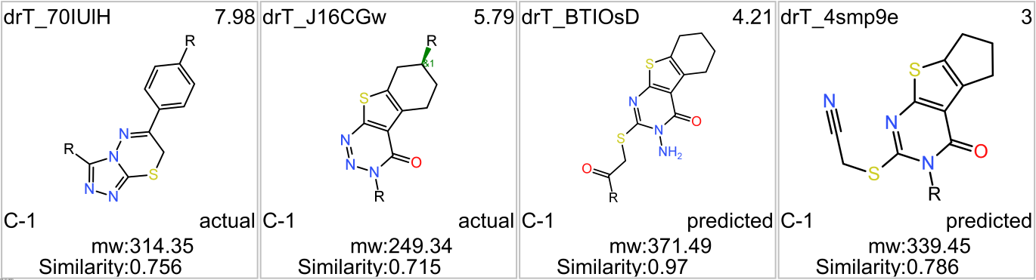
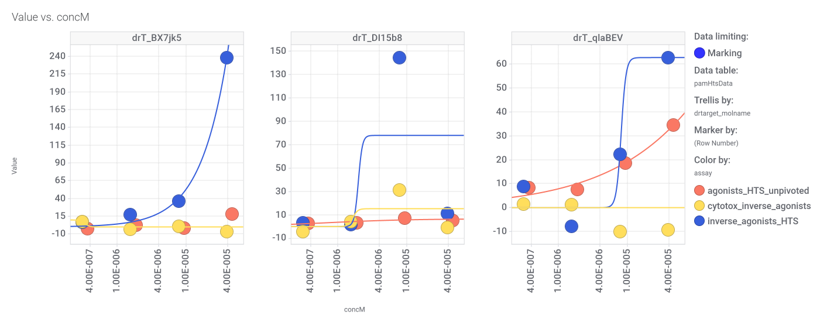
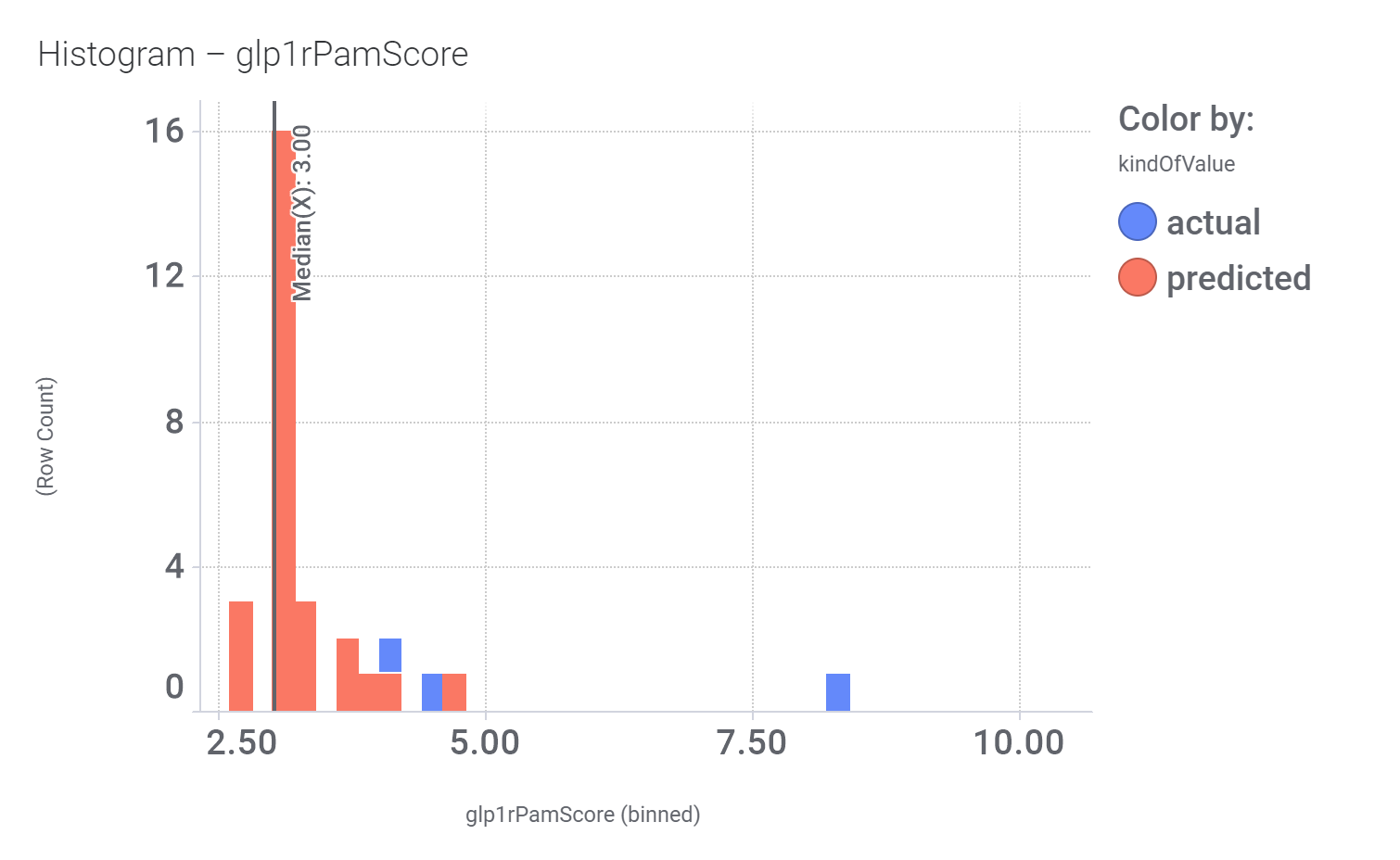
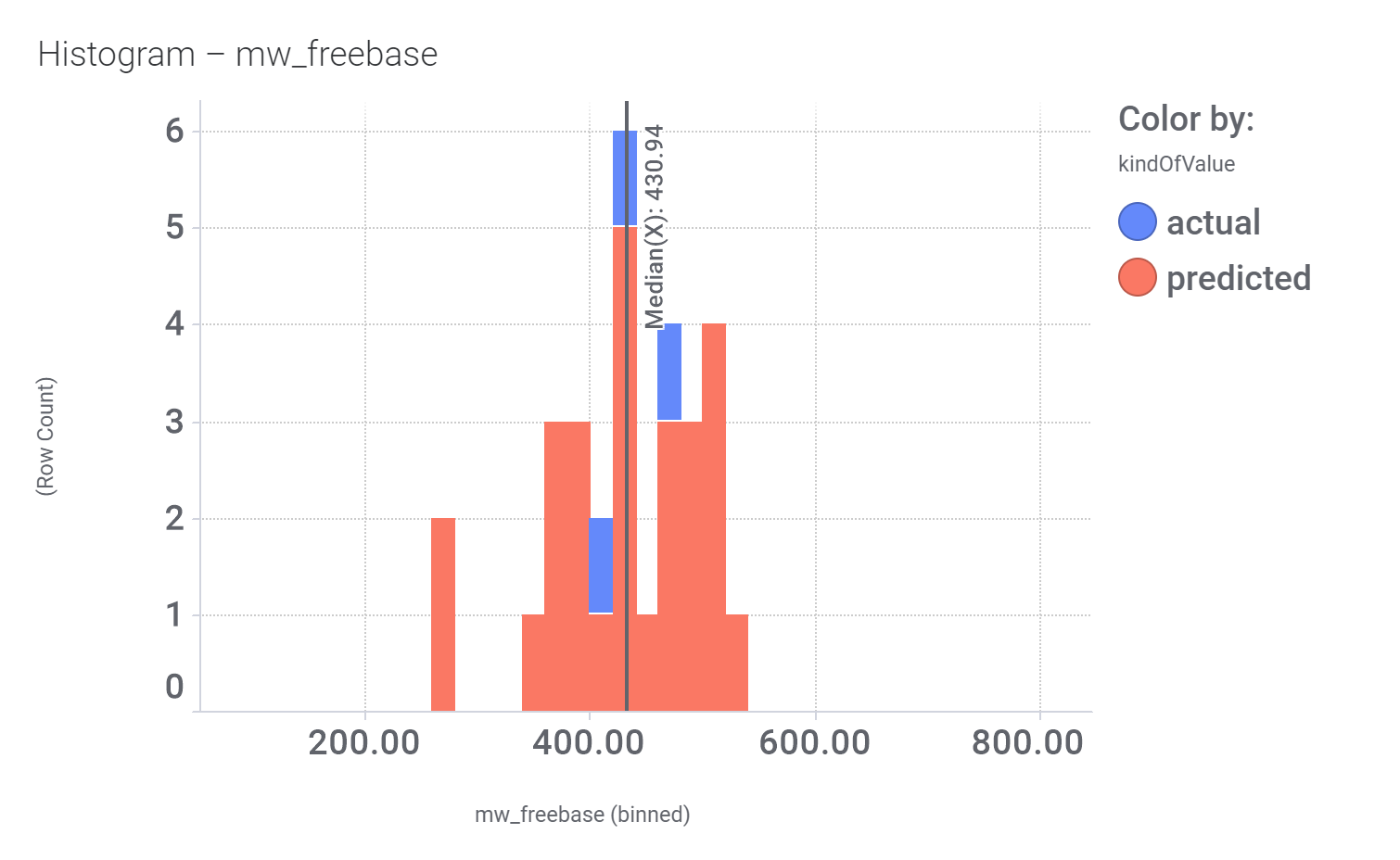
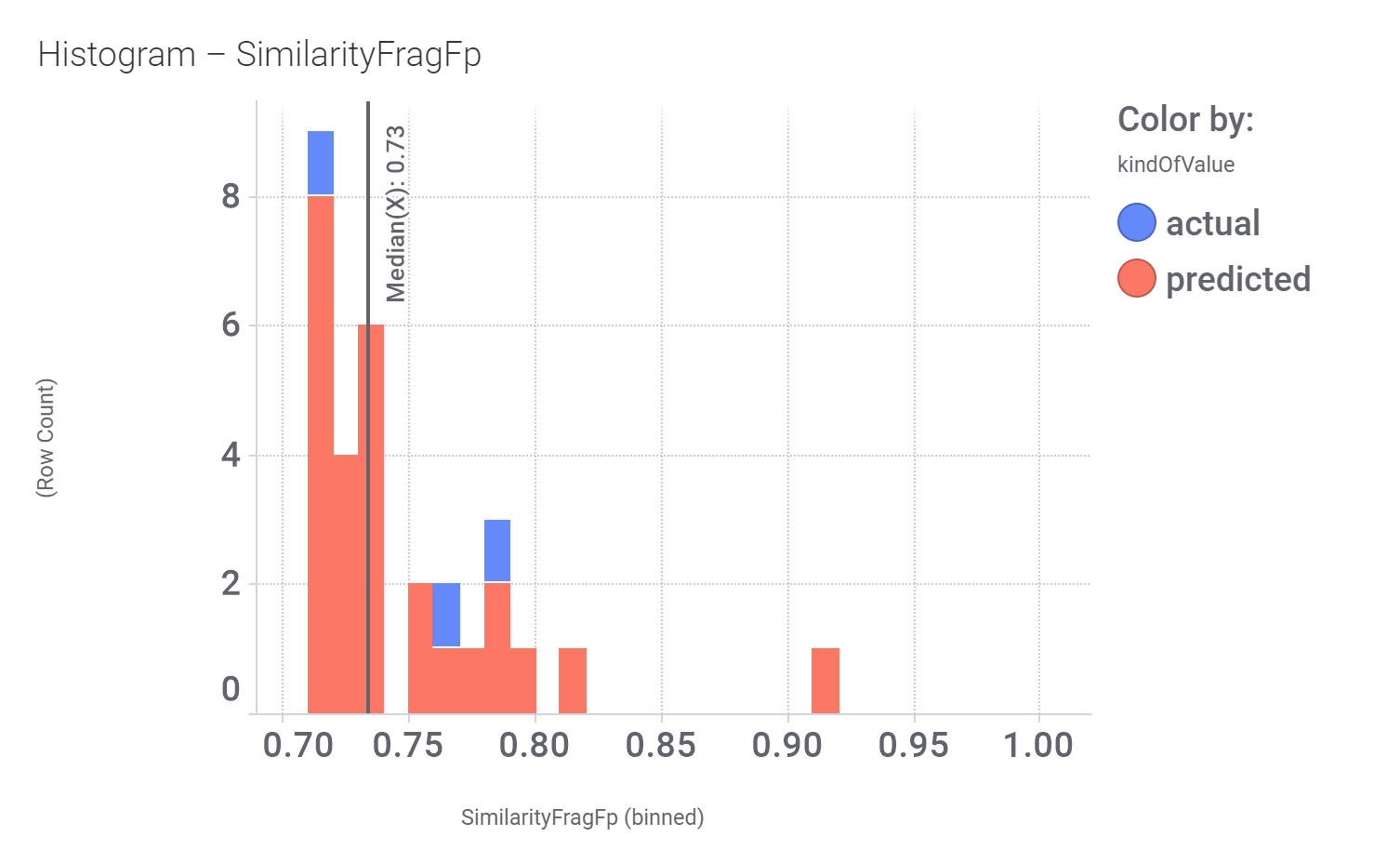
Among the predicted GLP1R PAM actives there are 30 compounds with > 70% similarity with Compound_20. Some exemplars here.
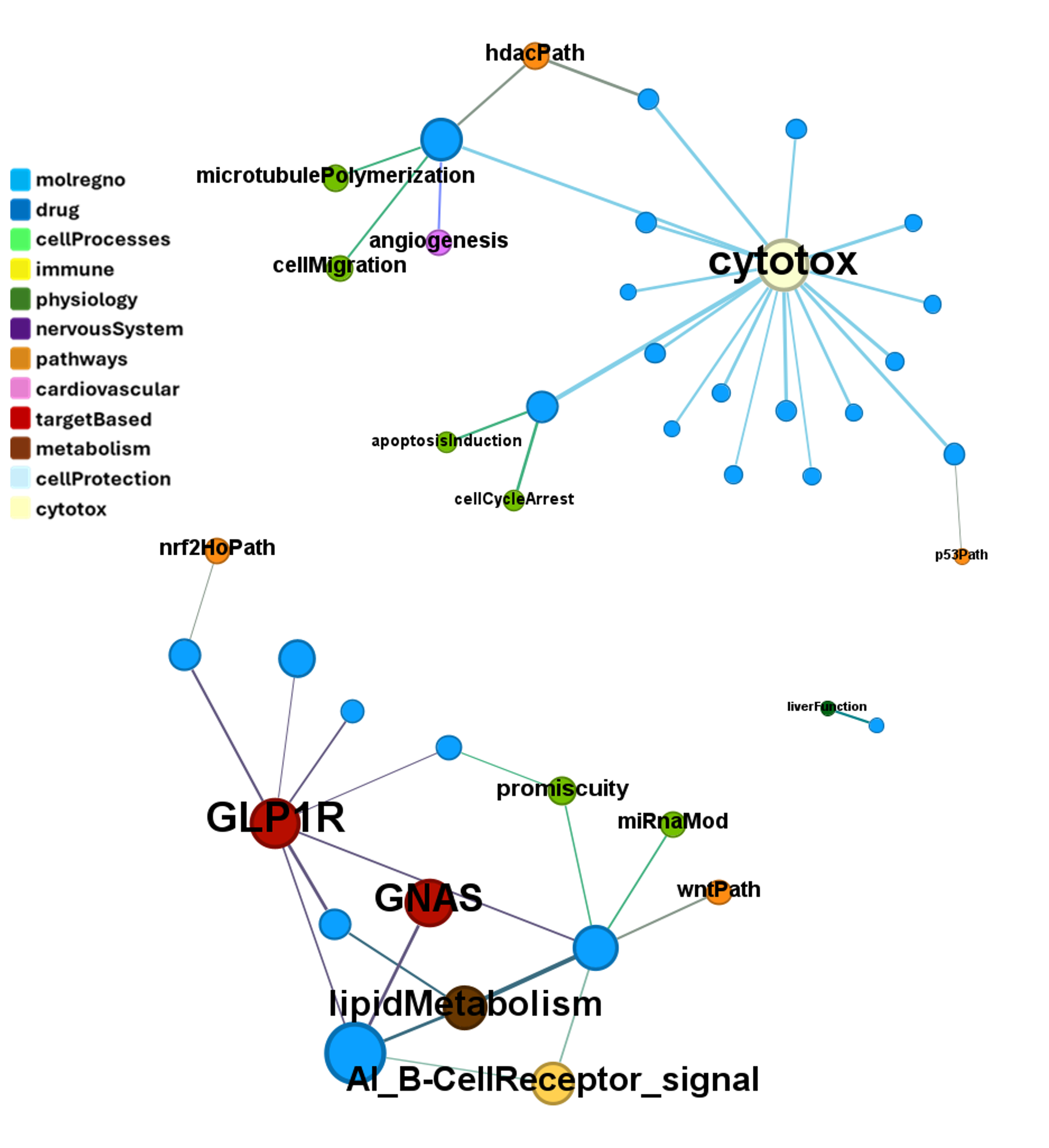
We can examine the effect of these molecules on relevant ChEMBL phenotypes using interaction graphs…
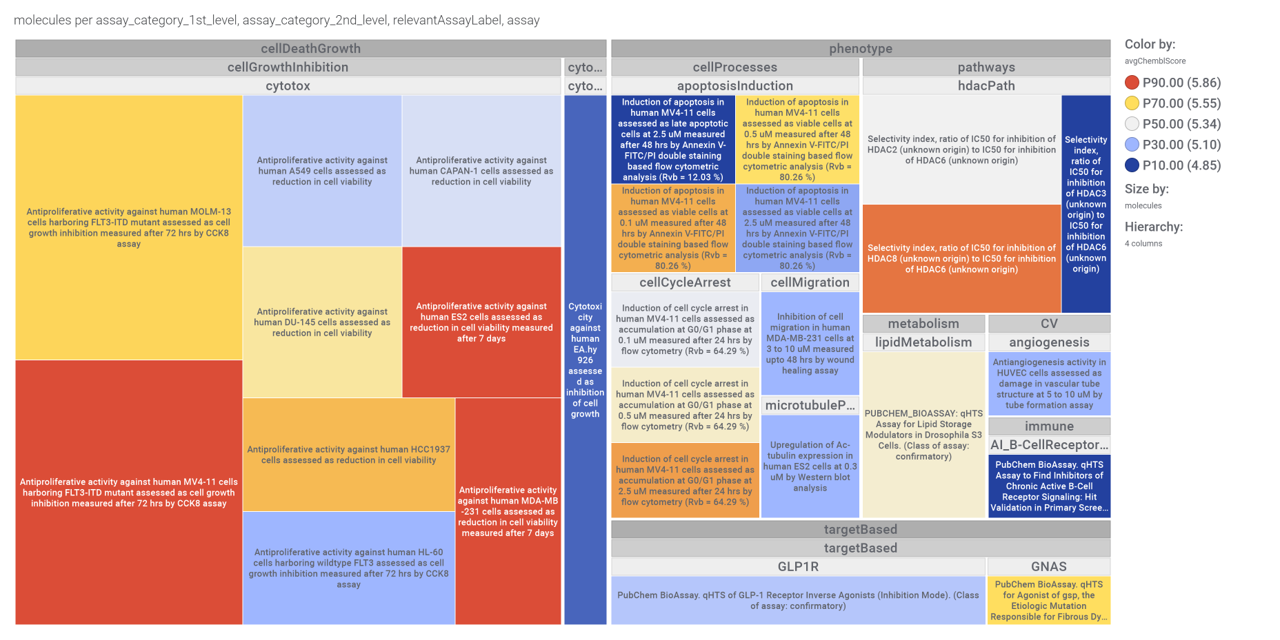
…and inspect specific assays involved in such phenotypes with treeMaps.
3 compounds have also full curve data from PubChem GLP1R screens.
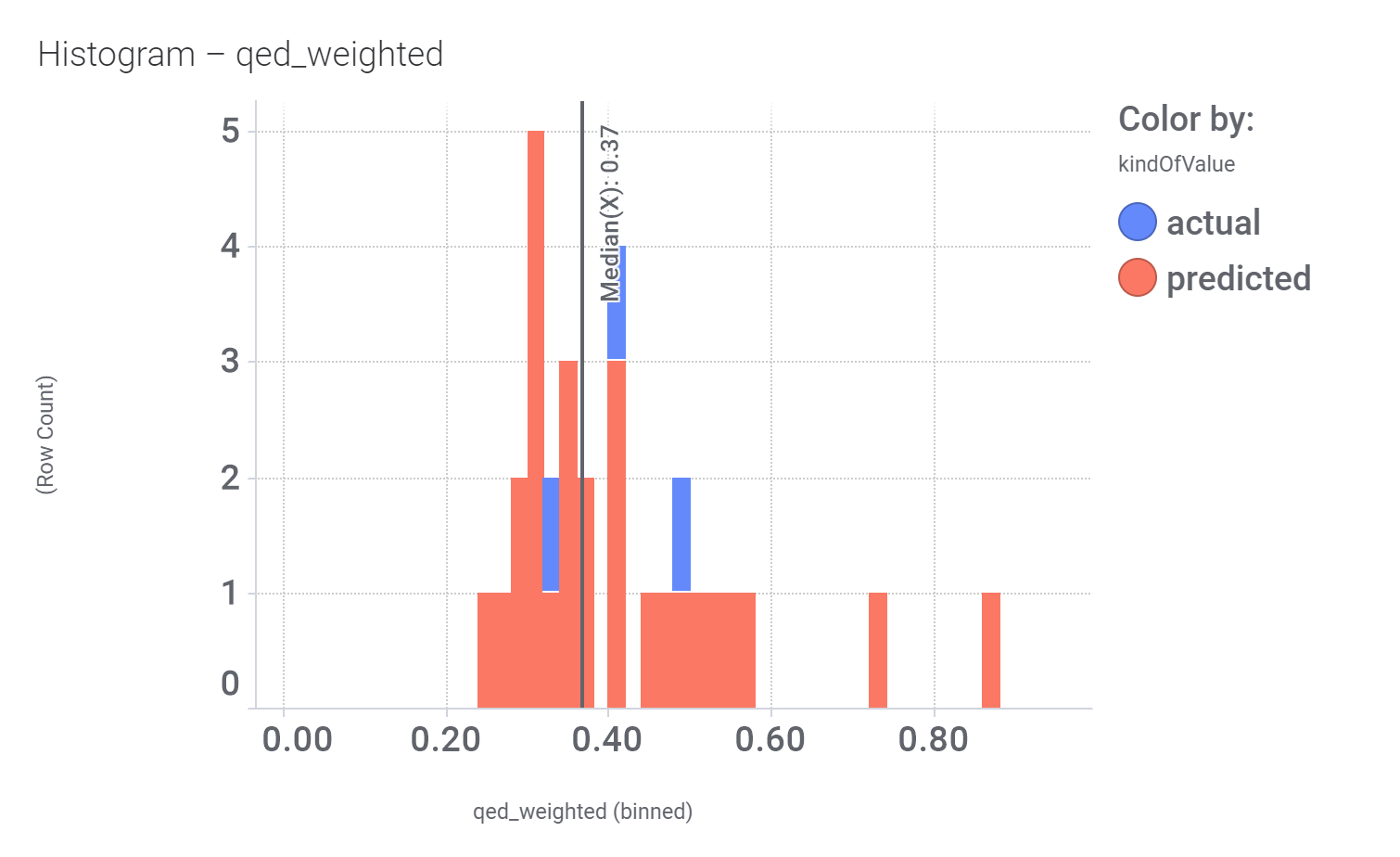
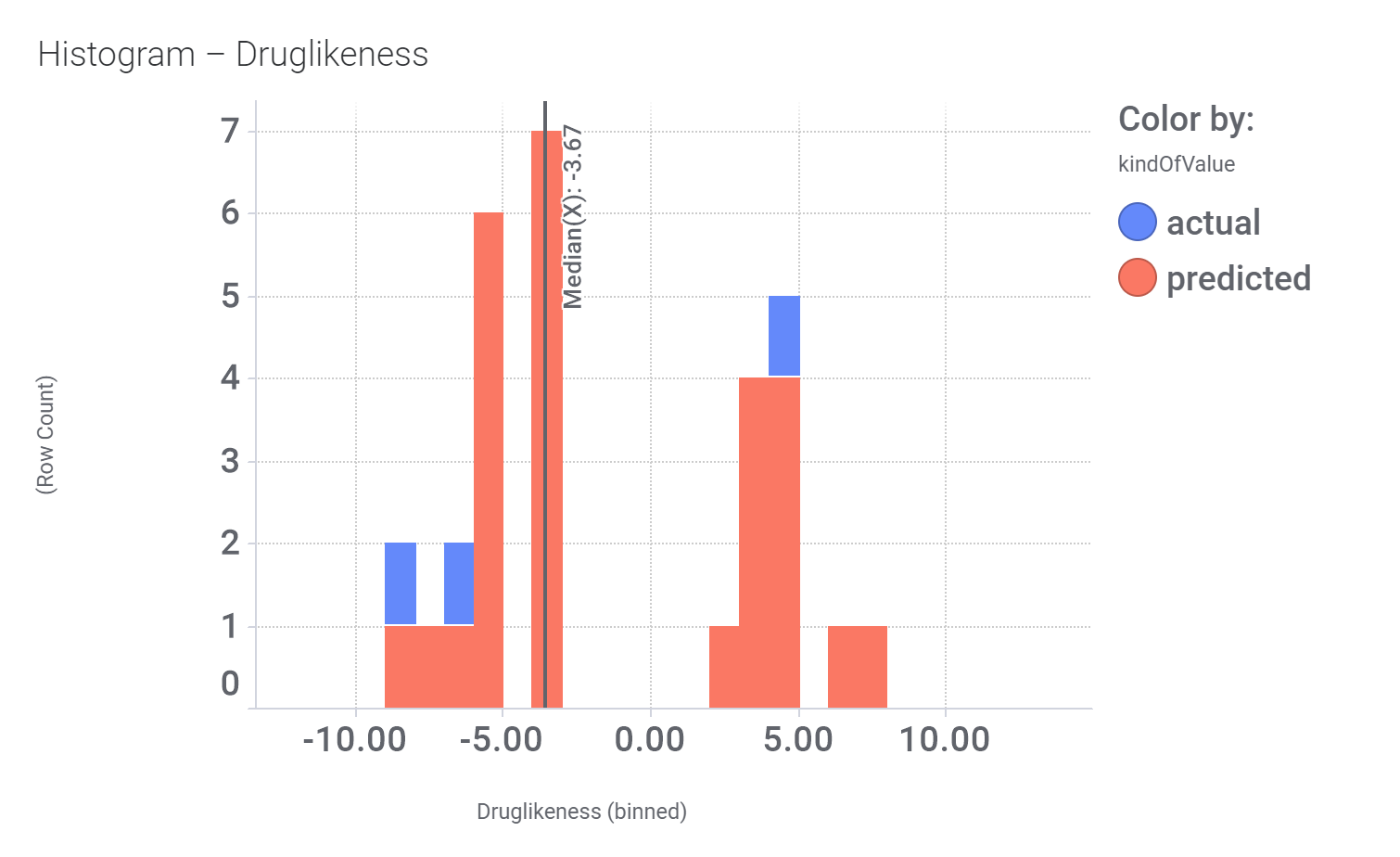
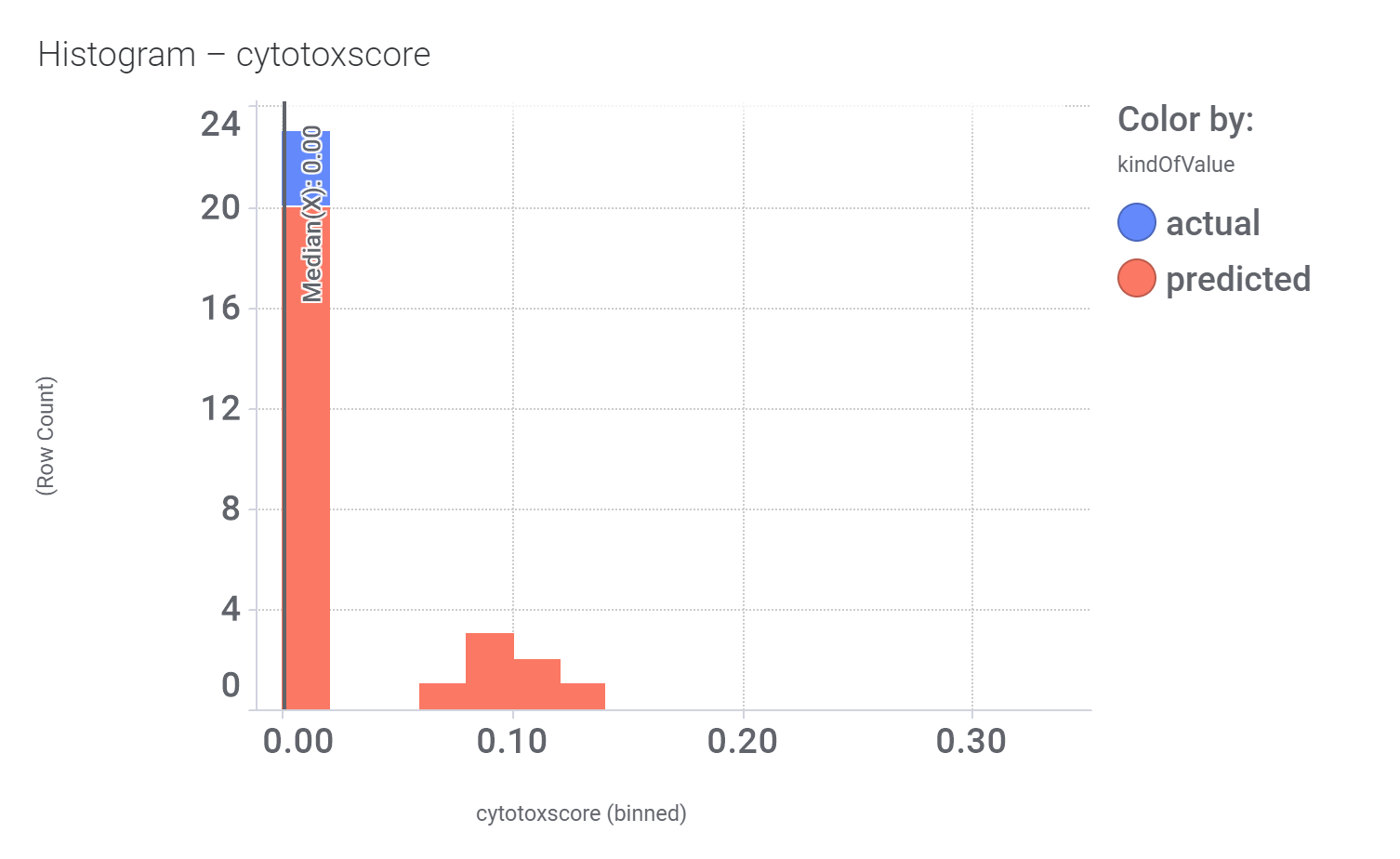
Let’s have a look on some of their bio-physicochemical and developability properties.
Druglikeness estimated by QedWeighted.
Druglikeness estimated by DataWarrior.
Some references on compound_20 series.
Br J Pharmacol. 2022 Feb; 179(4): 511–525. doi: 10.1111/bph.15446
Non‐peptide agonists and positive allosteric modulators of glucagon‐like peptide‐1 receptors: Alternative approaches for treatment of Type 2 diabetes
Faisal Malik 1 and Zhijun Li 1
Abstract
Glucagon‐like peptide‐1 (GLP‐1) receptors belong to the pharmaceutically important Class B family of GPCRs and are involved in many biologically significant signalling pathways. Its incretin peptide ligand GLP‐1 analogues are effective treatments for Type 2 diabetes. Although developing non‐peptide low MW drugs targeting GLP‐1 receptors remains elusive, considerable progress has been made in discovering non‐peptide agonists and positive allosteric modulators (PAMs) of GLP‐1 receptors with demonstrated efficacy. Many of these compounds induce biased signalling in GLP‐1 receptor‐mediated functional pathways. High‐quality structures of GLP‐1 receptors in both inactive and active states have been reported, revealing detailed molecular interactions between GLP‐1 receptors and non‐peptide agonists or PAMs. These progresses raise the exciting possibility of developing non‐peptide drugs of GLP‐1 receptors as alternative treatments for Type 2 diabetes. The insight into the interactions between the receptor and the non‐peptide ligand is also useful for developing non‐peptide ligands targeting other Class B GPCRs.
3.2.1. Compound 20
The discovery of GLP‐1 receptor PAMs, such as compound 2 and BETP, inspired screening efforts, applying computer‐aided molecular design techniques. The first report employing structure‐based virtual screening adopted the X‐ray crystal structure of Class A rhodopsin receptors to construct 3D models of two Class B GPCRs, the corticotropin‐releasing hormone CRF1 receptor and the glucagon receptor (de Graaf et al., 2011). The model of the glucagon receptor was subsequently used to screen a database of 1.9 million commercially available drug‐like compounds in an effort to identify non‐competitive antagonists. Twenty‐three compounds were selected and evaluated in vitro in a whole cell‐based functional assay for binding to glucagon receptors and human GLP‐1 receptors and for modulation of receptor‐induced cAMP accumulation. Two compounds were confirmed to inhibit glucagon receptor activity in a dose‐dependent manner, and one was characterized as a PAM of GLP‐1 receptors, compound 16 (compound 20) (Figure 3, [16]). Although the potency of compound 20 is modest, this work demonstrates the usefulness of the computational approach in discovering PAMs for Class B GPCRs.