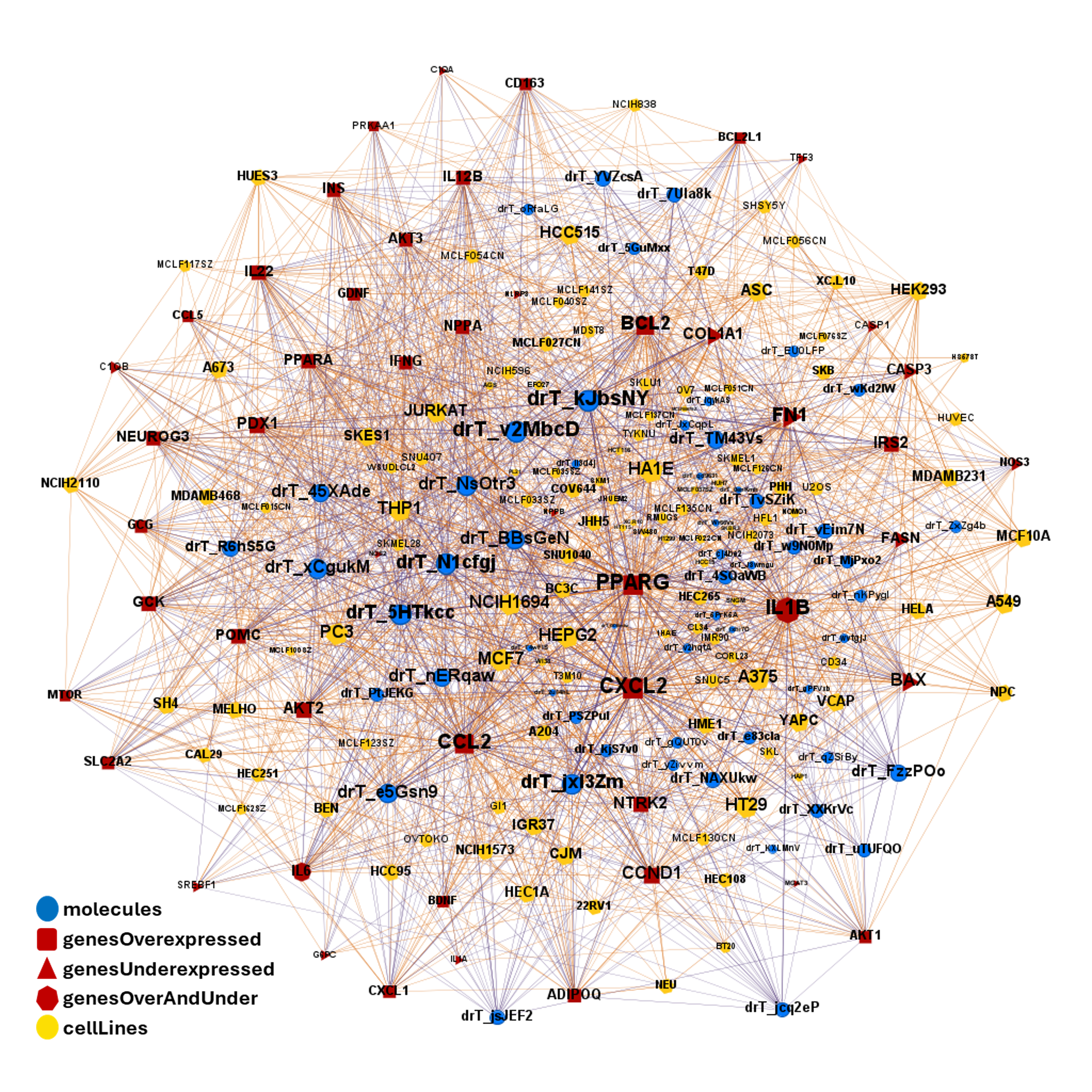
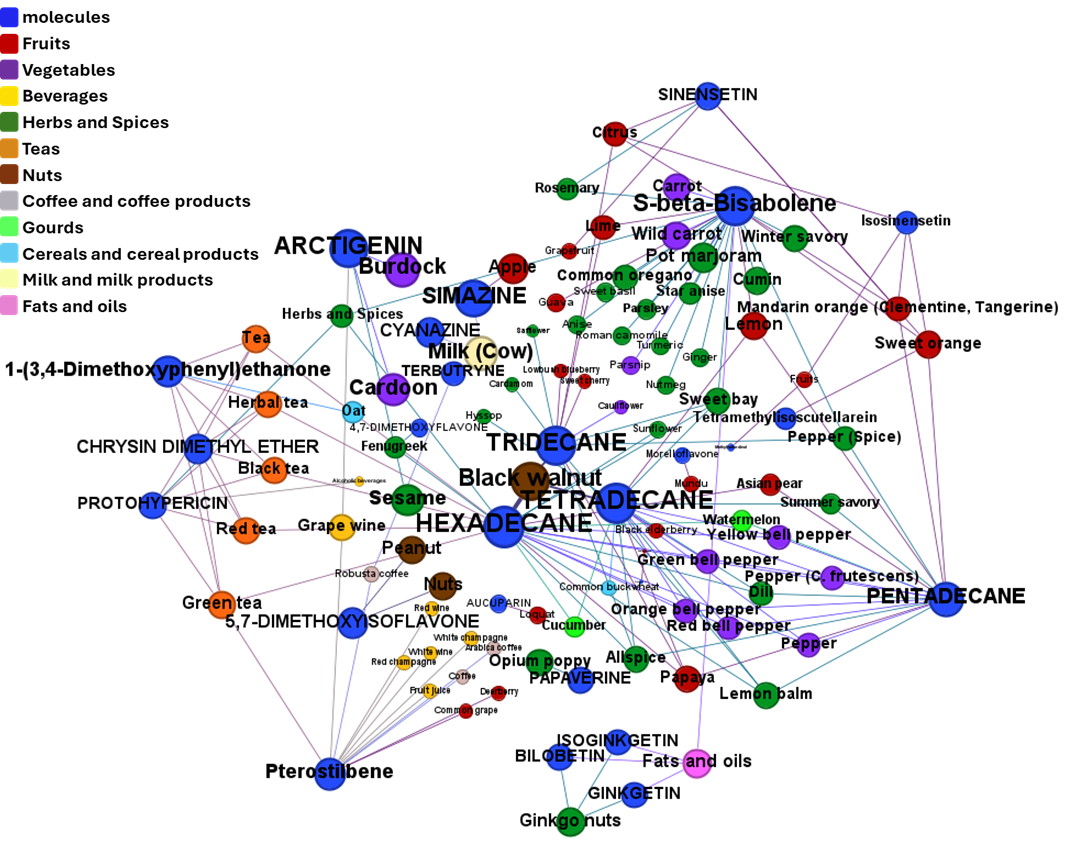
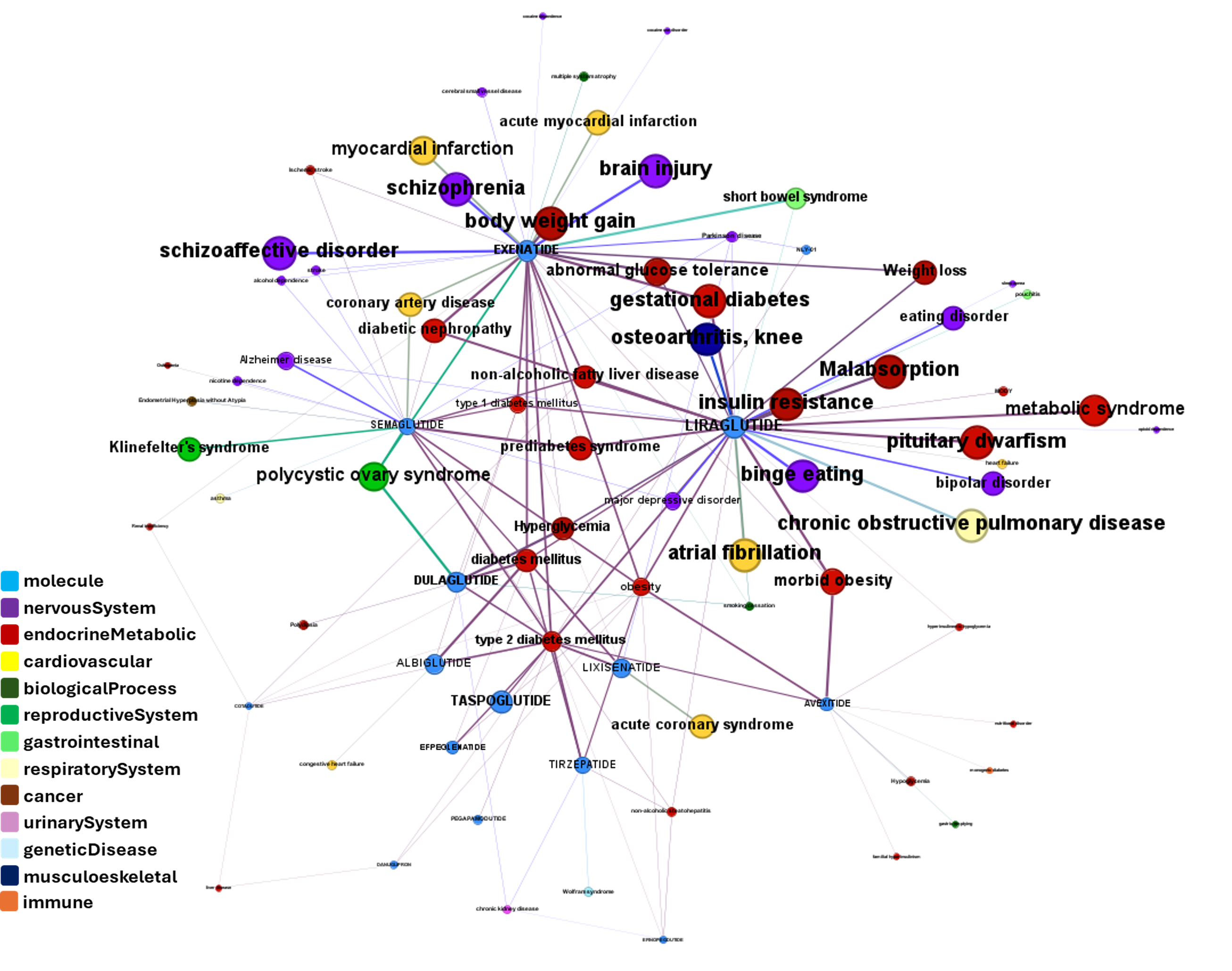
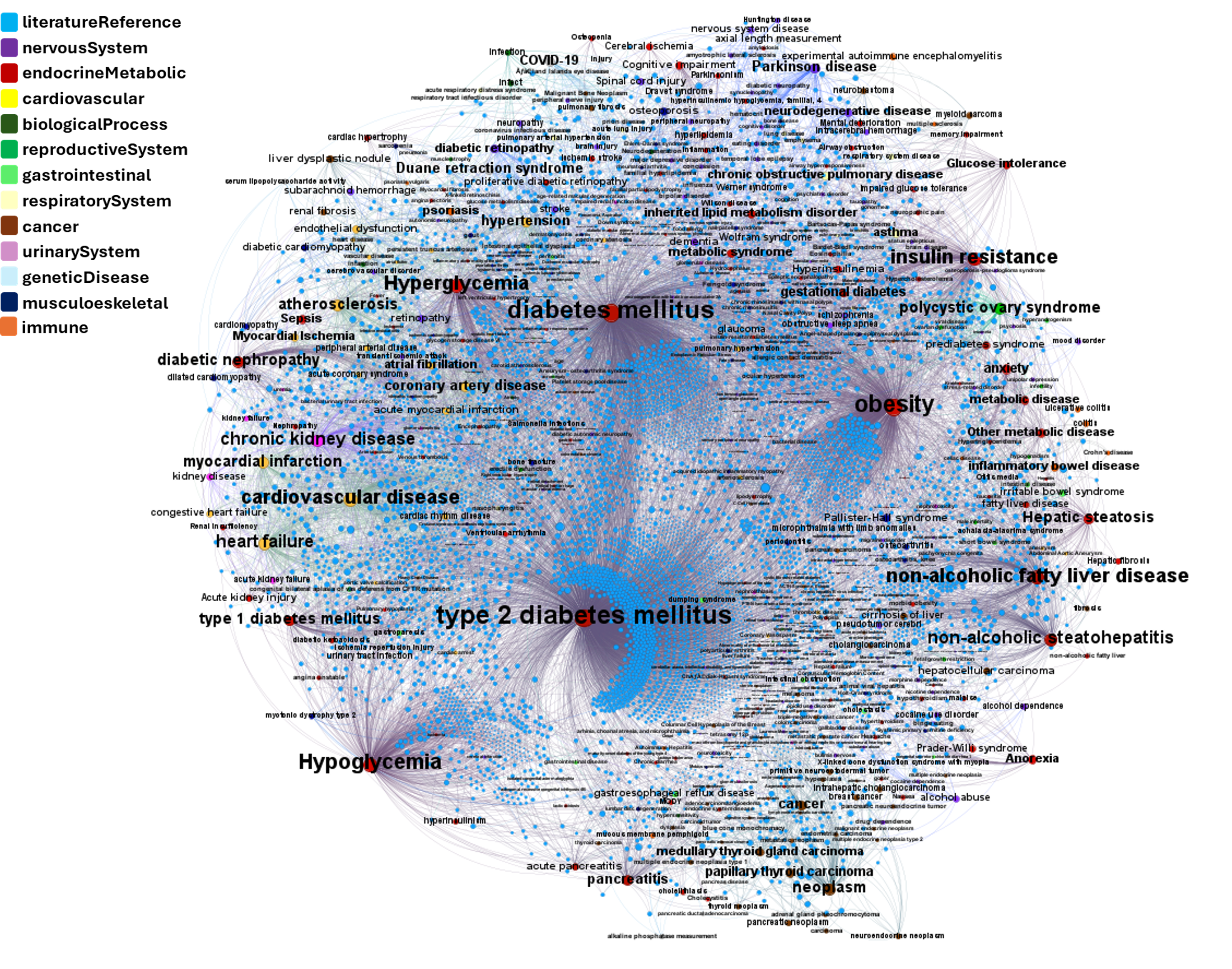
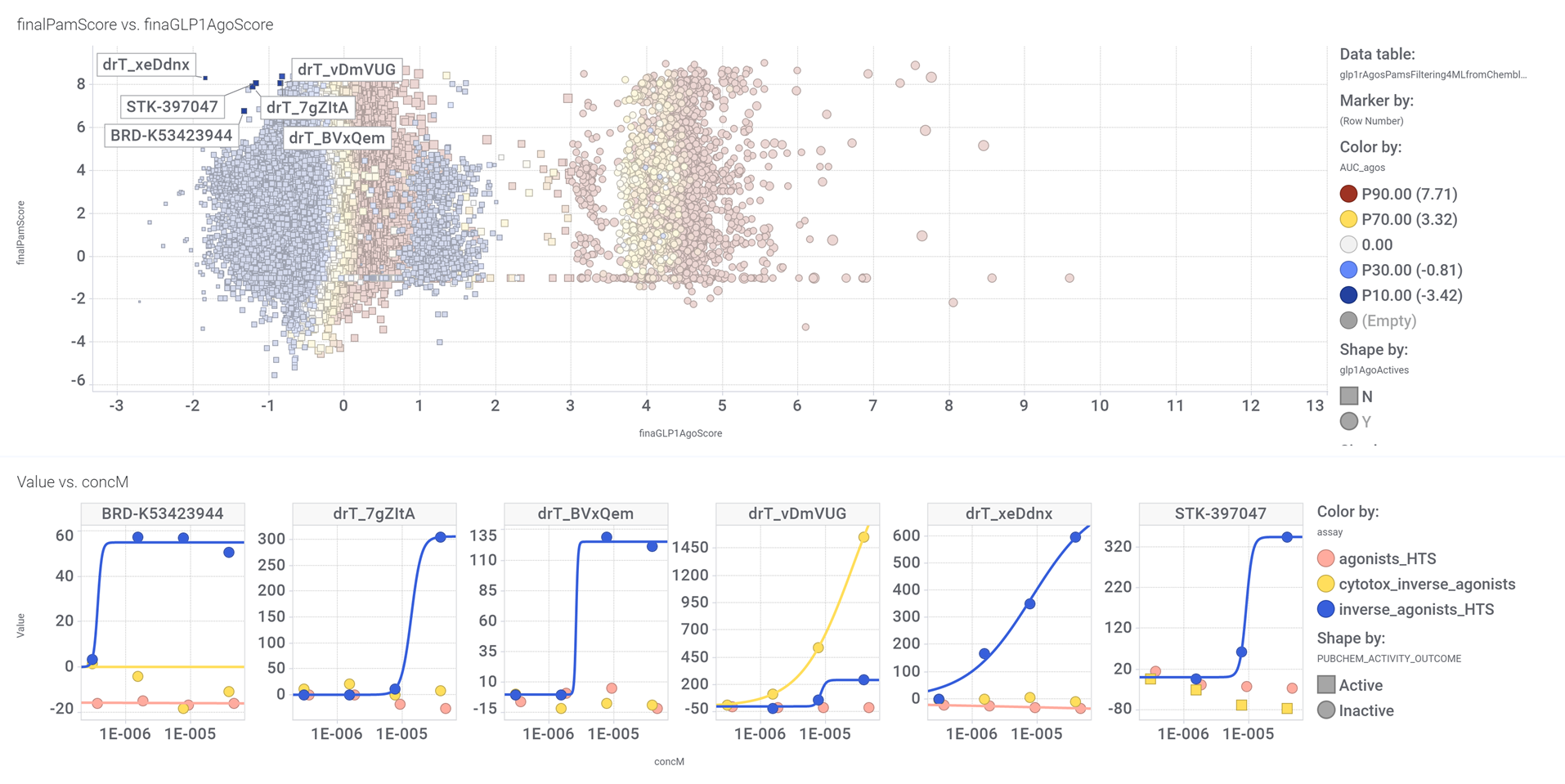
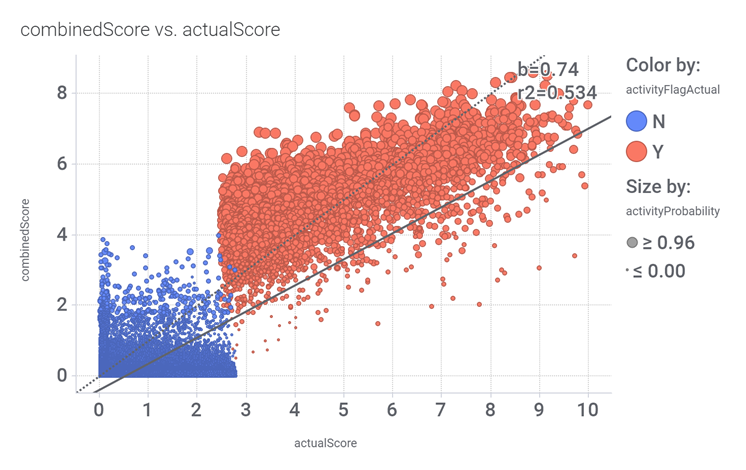
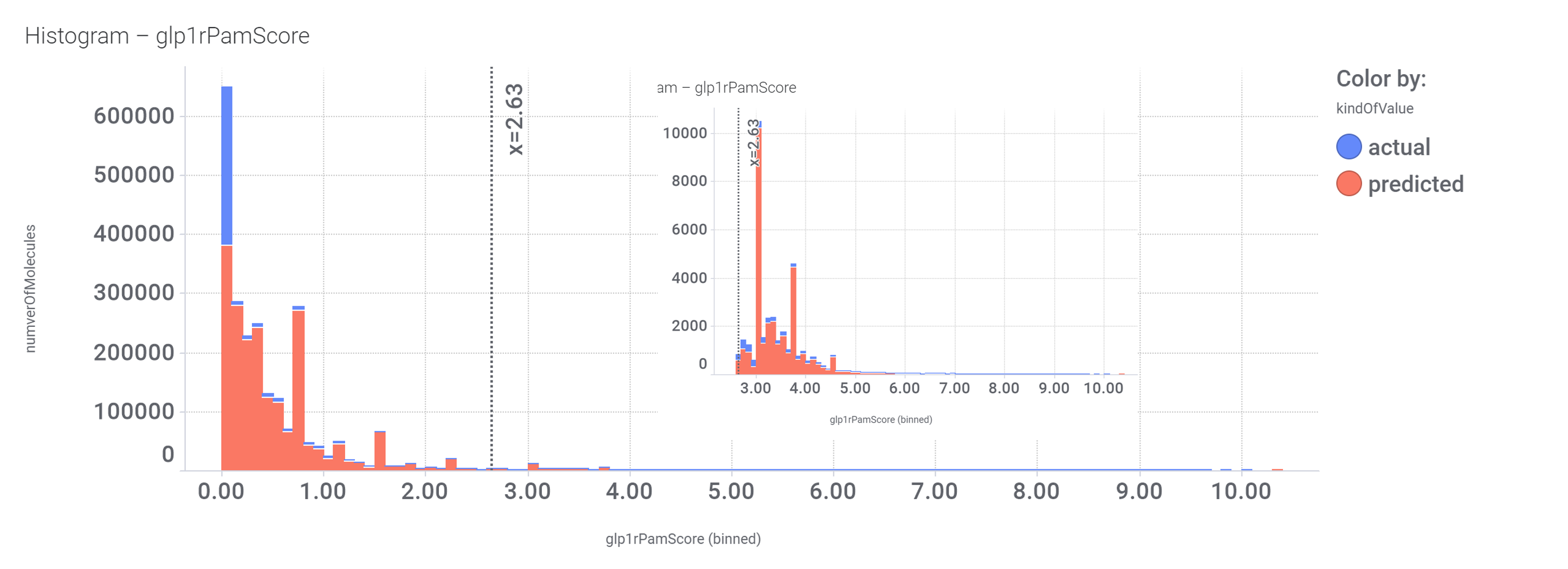
drTarget applies machine learning models on public DBs to perform a virtual screening on 2.3M molecules finding >37k potential positive allosteric modulators with a predicted activation score.
From this virtual screen, drTarget analyzes all activities of selected active PAMs
on relevant ChEMBL phenotypes.
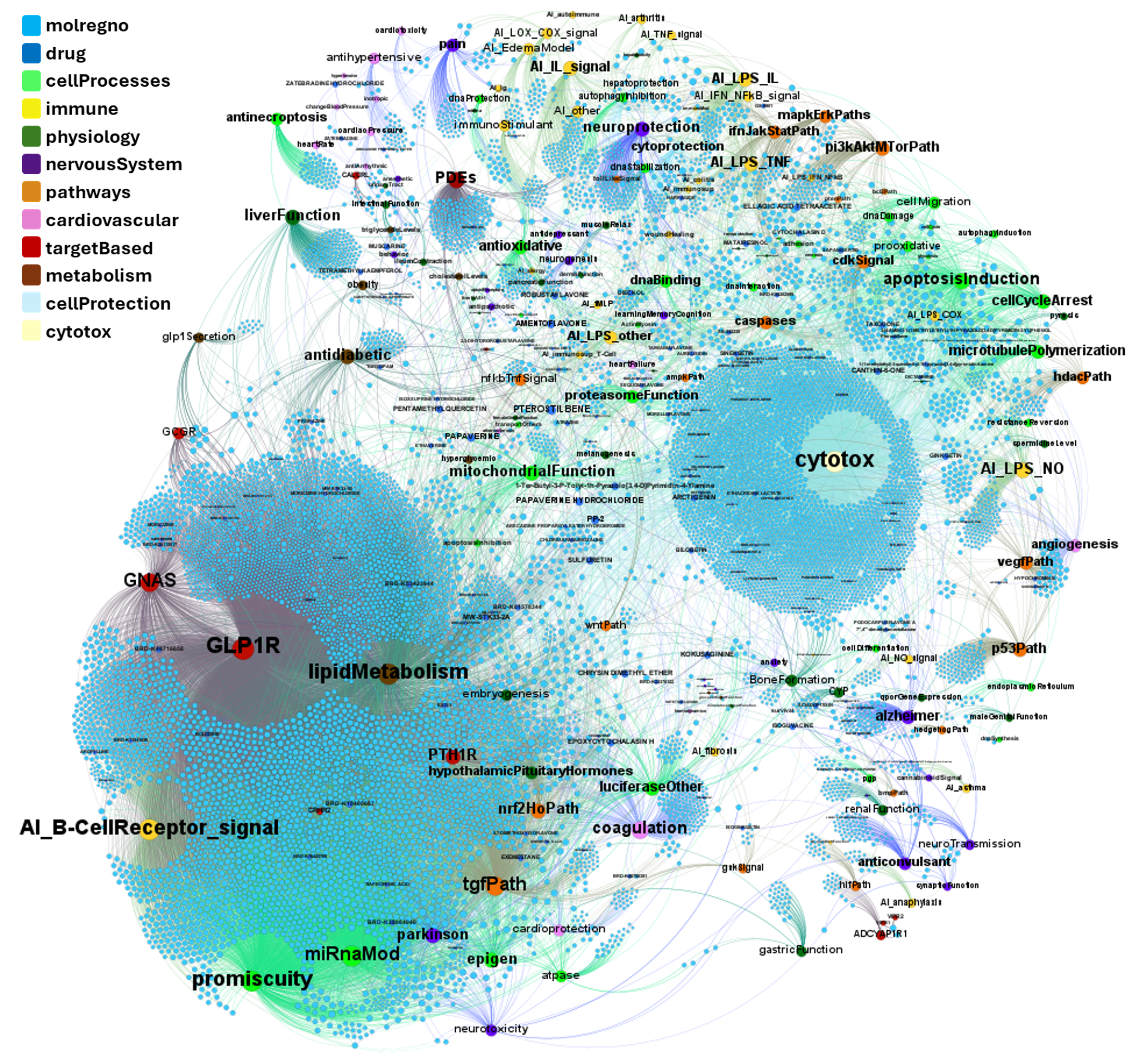
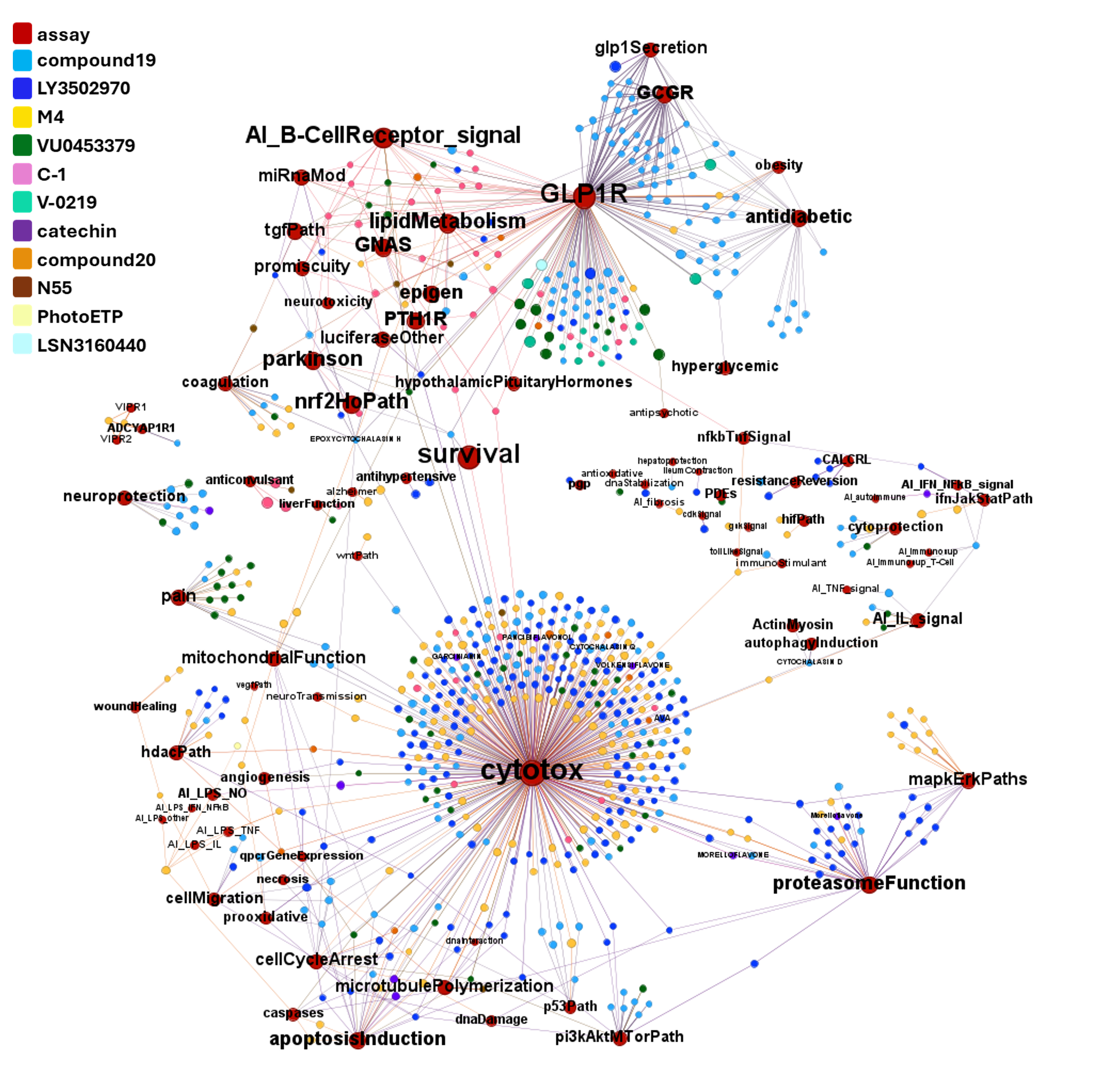
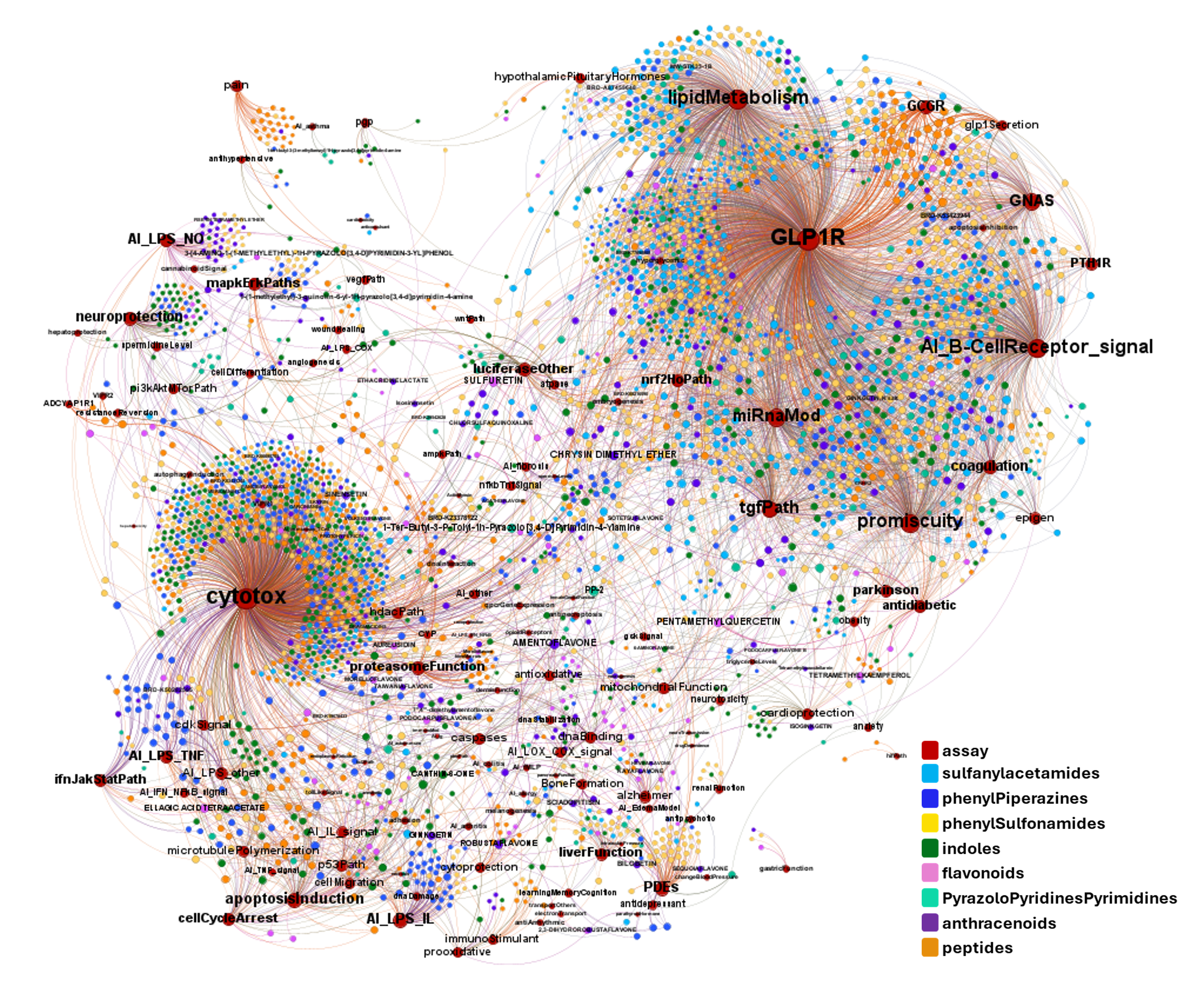
A network graph view here…
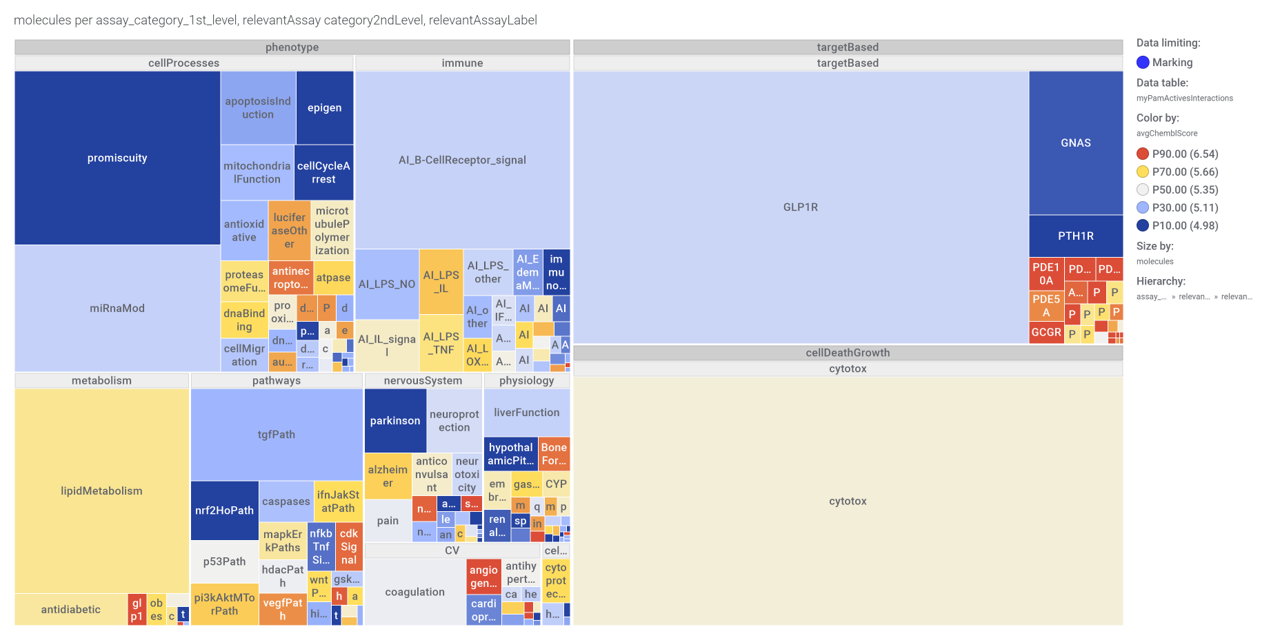
…Or a treeMap
Find below particular examples of these interactions with some analogues of known GLP1R PAMs or some selected molecular series.
Find out effects of GLP1R predicted active compounds on gene expression and their occurrence in foods.
Or explore GLP1R associations with diseases, scored by OpenTargets algorithms.
See also data sources and machine learning models used.
drTarget uses 400k values from a full curve GLP1R inverse agonists screens stored in PChem DB plus all ChEMBL GLP1R positive allosteric modulation assays and literature references describing activities of well characterized GLP1R PAMs.