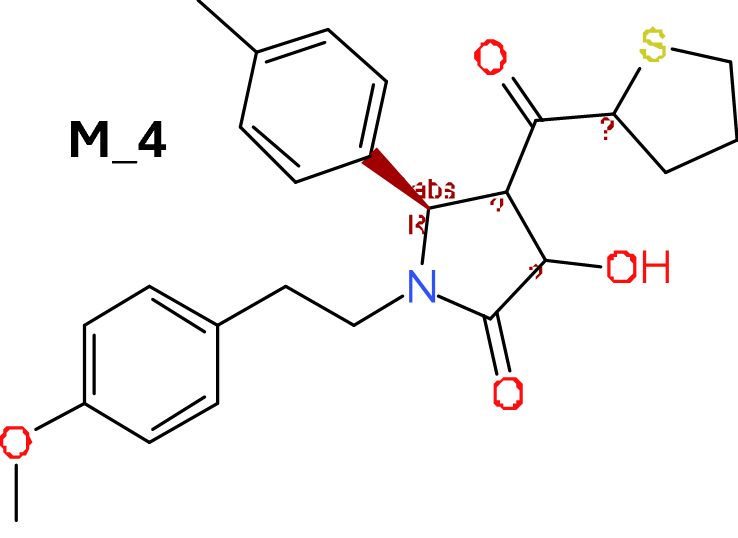
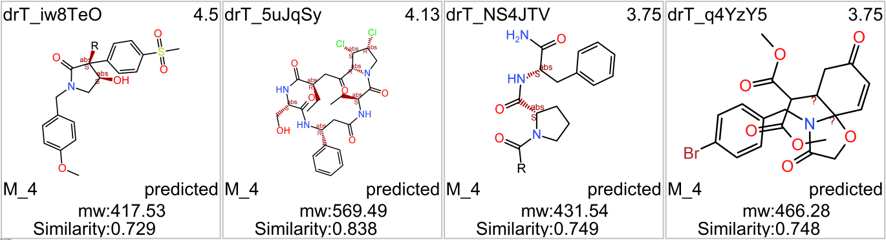
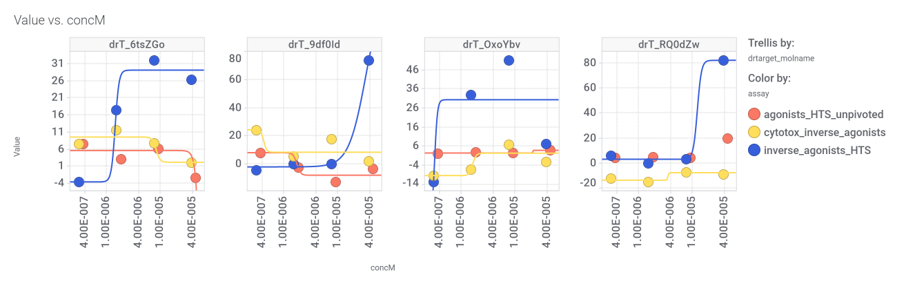
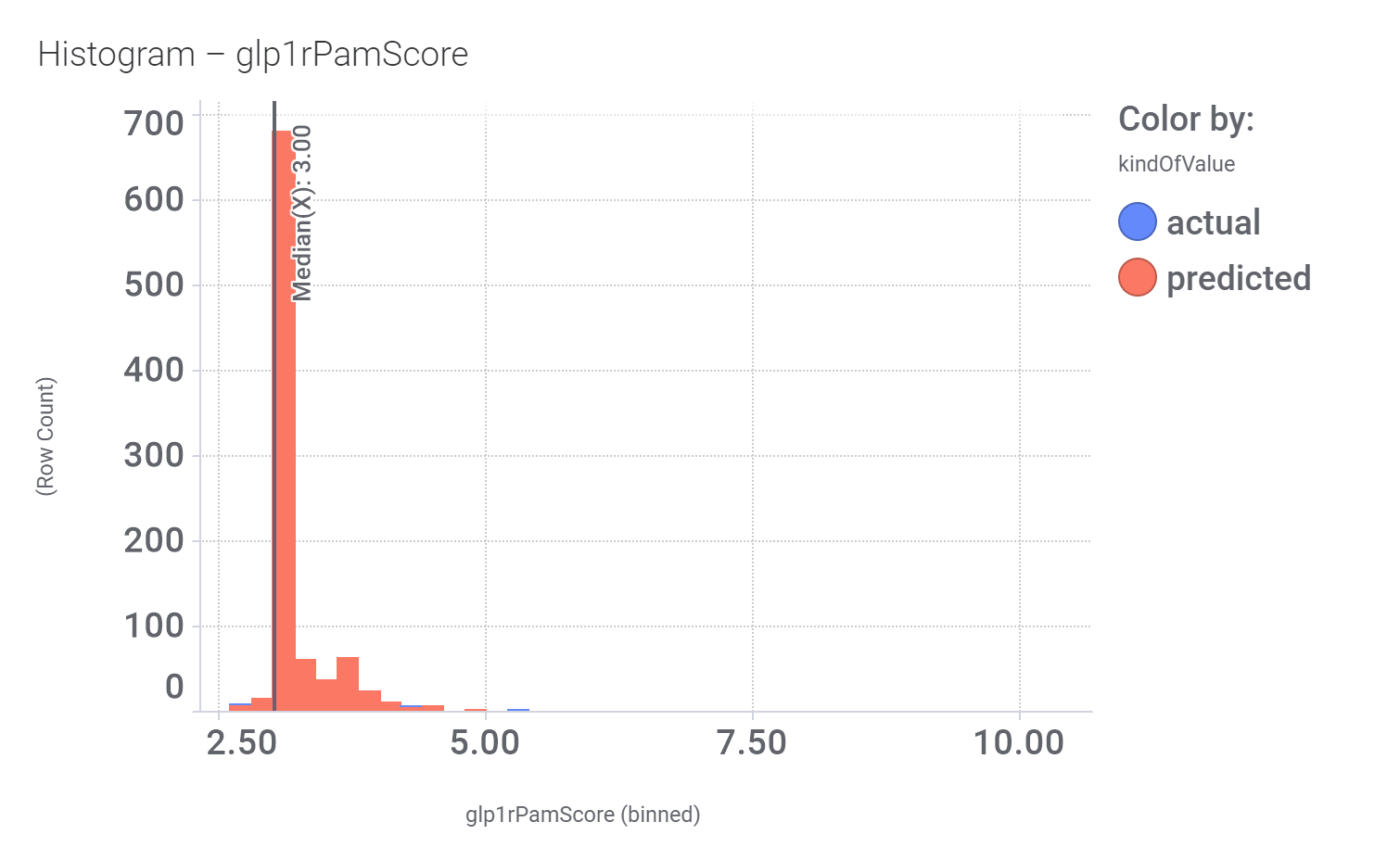
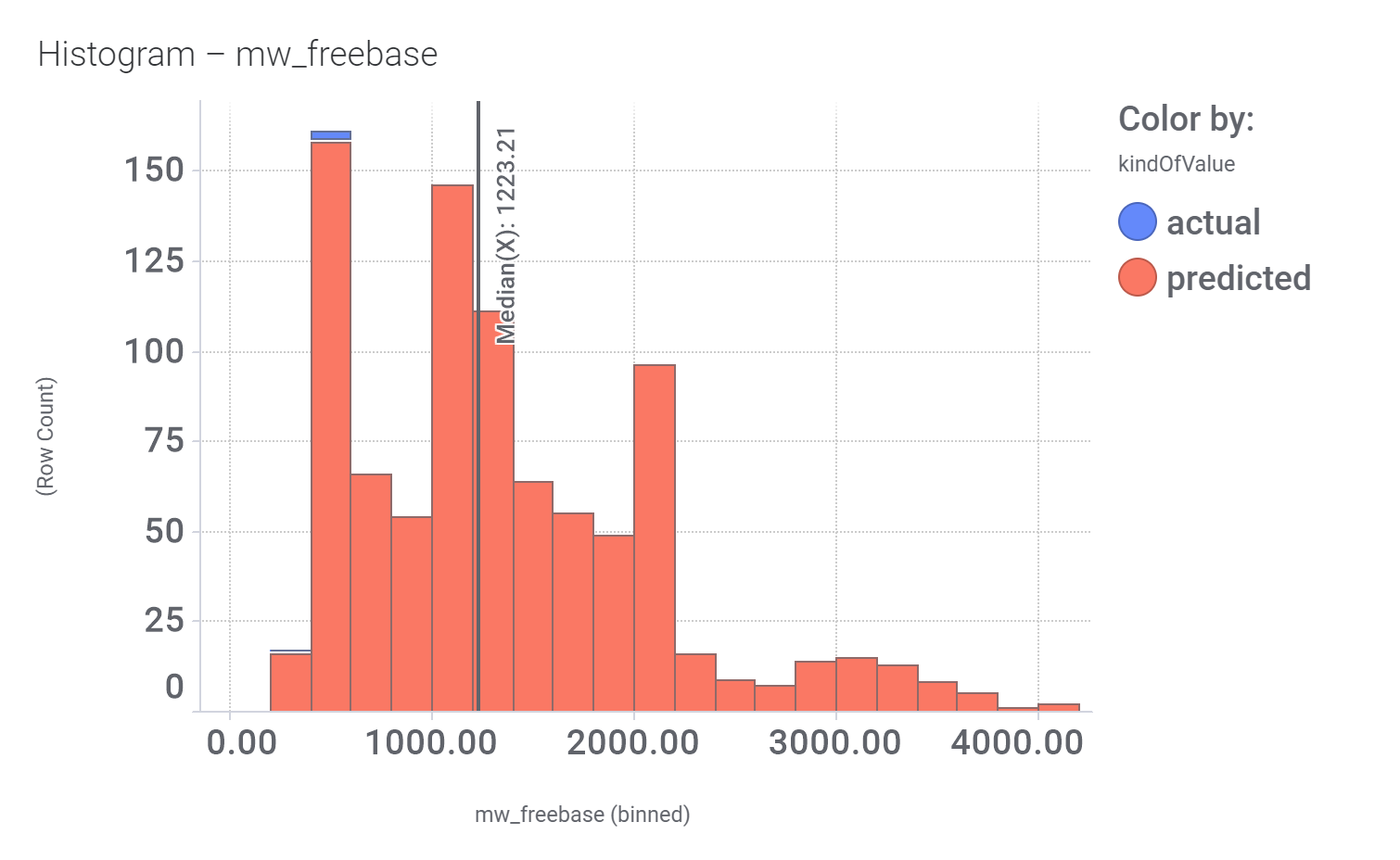
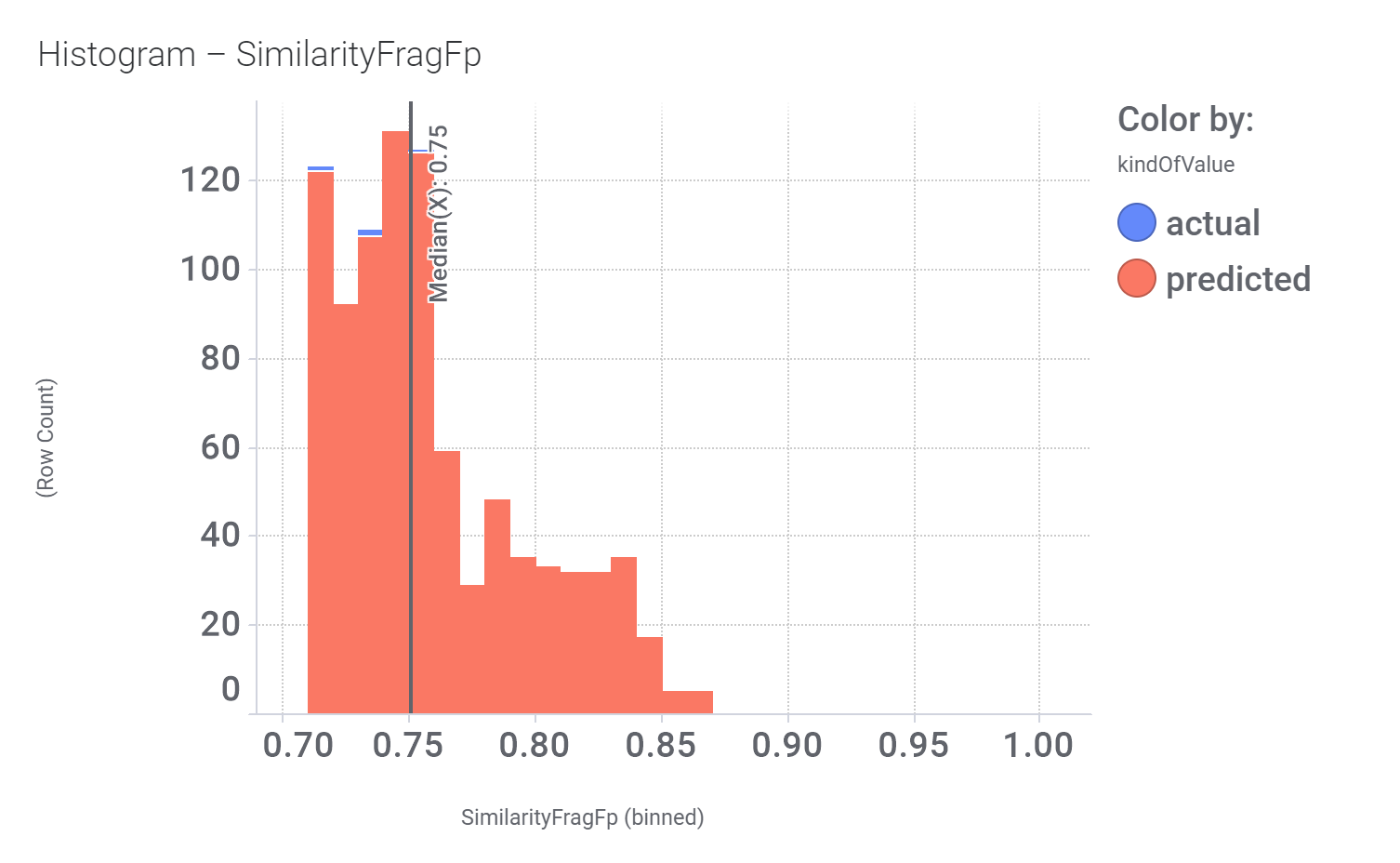
M_4 comes from a structure‐based virtual screening of 5689 compounds from the ZINC database. More than 900 compounds among the GLP1R predicted active molecules show > 70% similarity with M_4.
Some exemplars below.
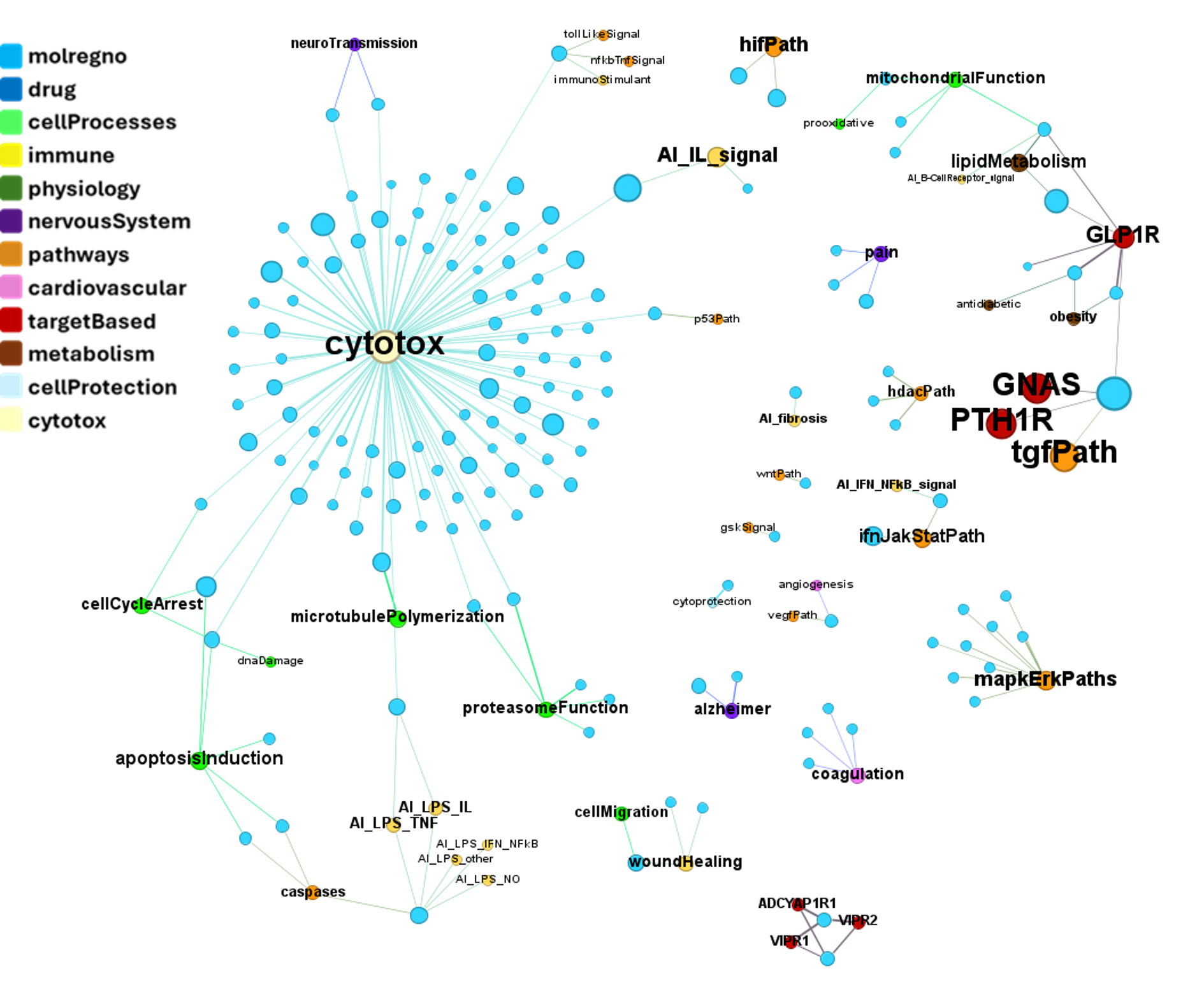
Interactions map with relevant ChEMBL phenotypes.
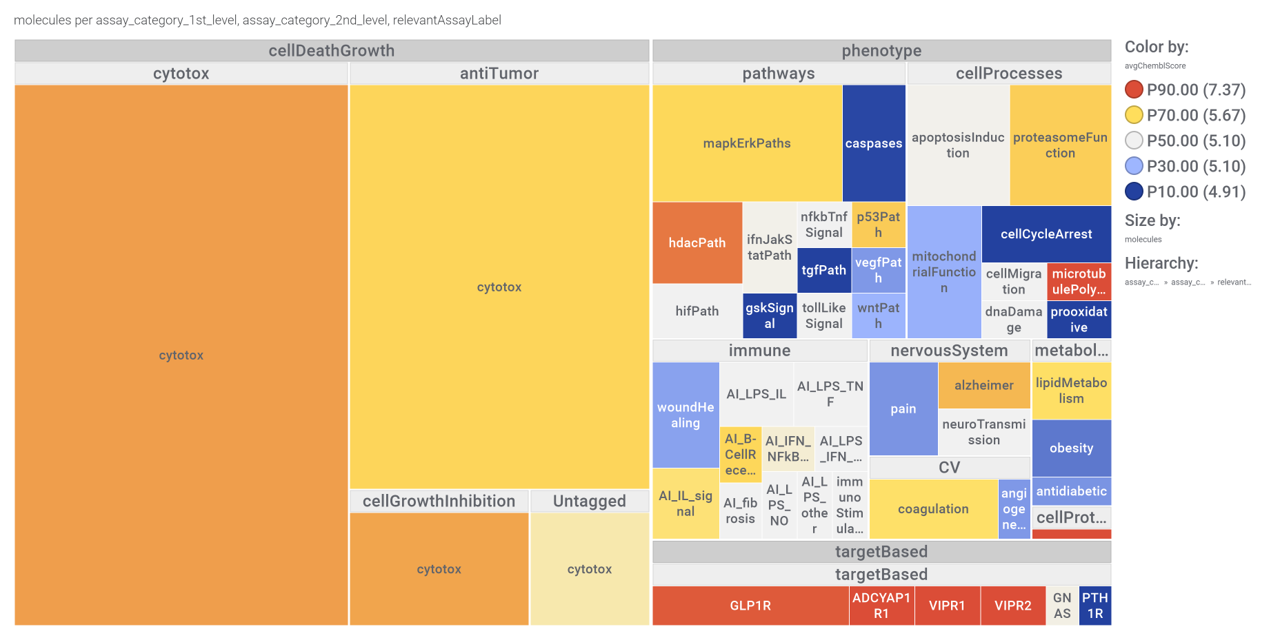
Can also be represented in the shape of a treeMap.
4 of these compounds have shown some activity in PChem screens.
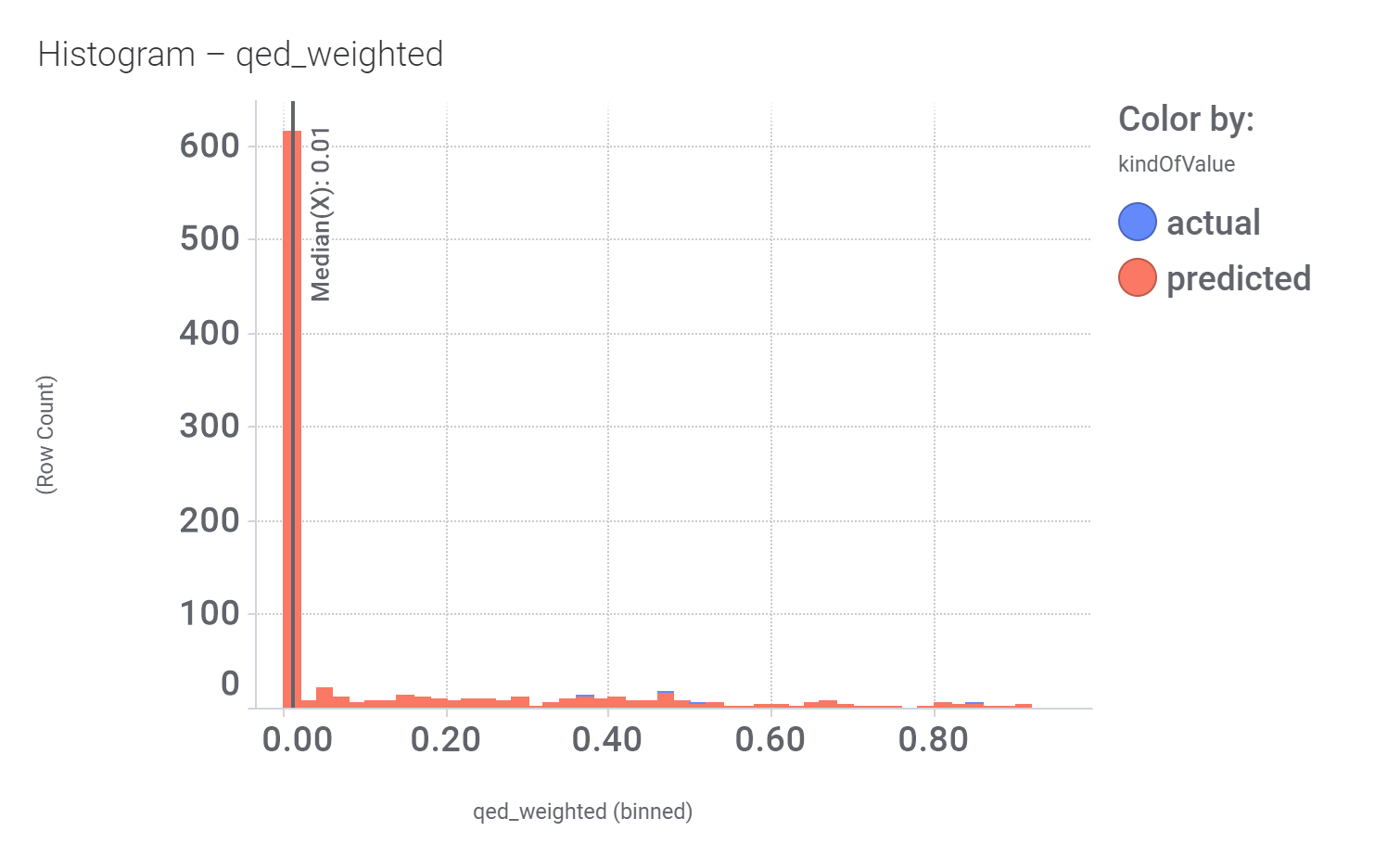
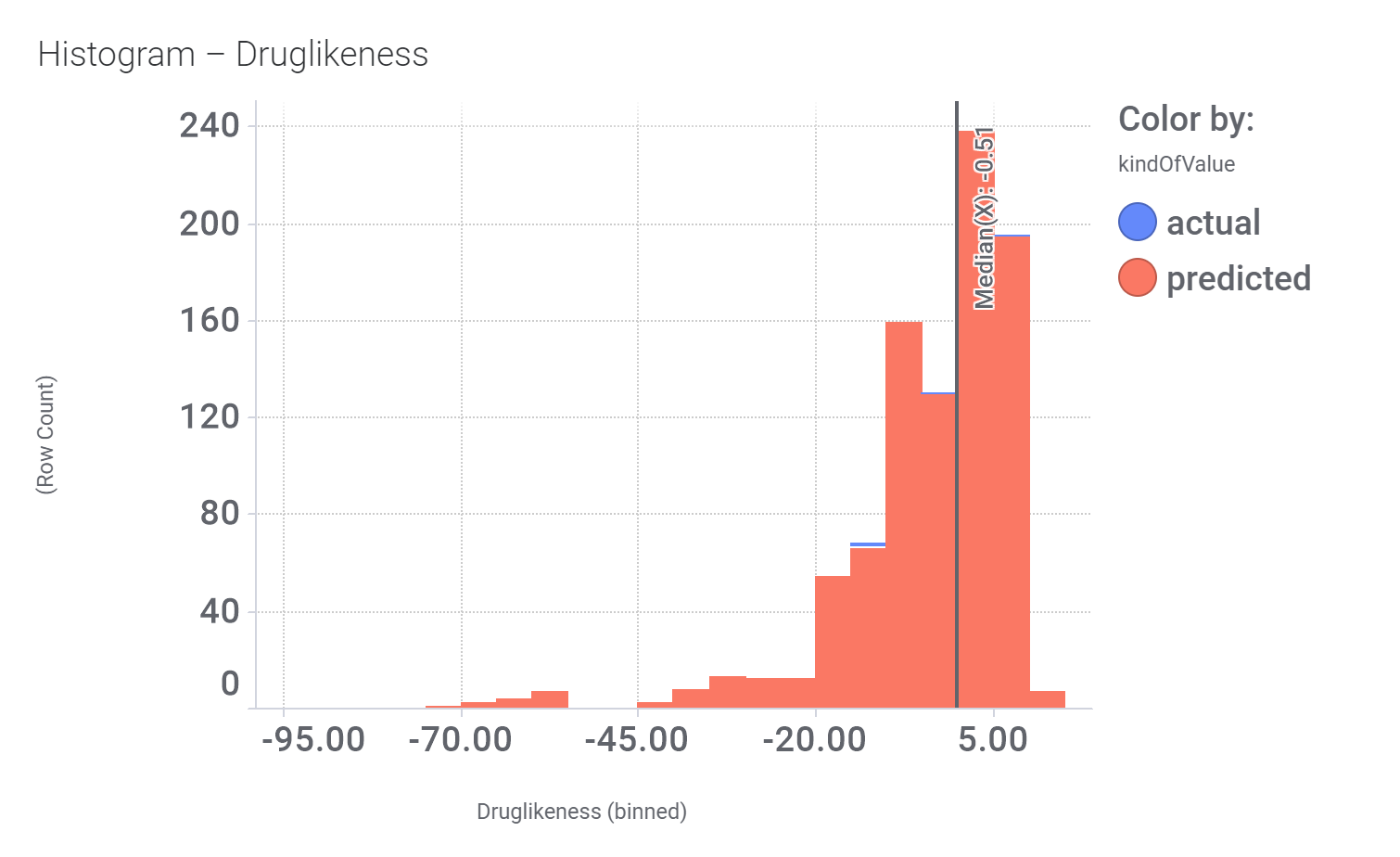
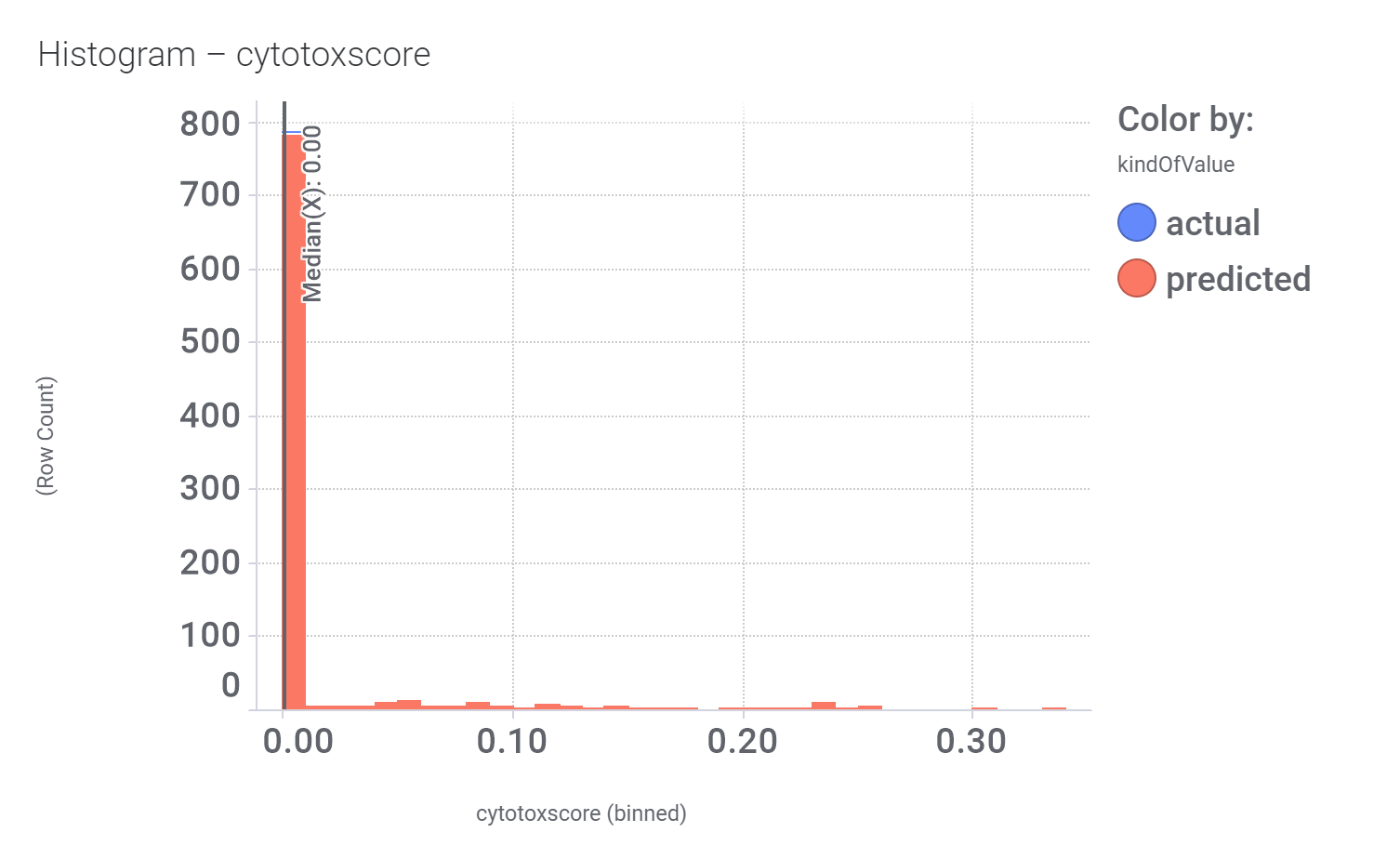
And these are their bio-physicochemical and developability properties.
Druglikeness estimated by QedWeighted
Druglikeness estimated by DataWarrior
References
Br J Pharmacol. 2022 Feb; 179(4): 511–525. doi: 10.1111/bph.15446
Non‐peptide agonists and positive allosteric modulators of glucagon‐like peptide‐1 receptors: Alternative approaches for treatment of Type 2 diabetes
Faisal Malik 1 and Zhijun Li 1
Abstract
Glucagon‐like peptide‐1 (GLP‐1) receptors belong to the pharmaceutically important Class B family of GPCRs and are involved in many biologically significant signalling pathways. Its incretin peptide ligand GLP‐1 analogues are effective treatments for Type 2 diabetes. Although developing non‐peptide low MW drugs targeting GLP‐1 receptors remains elusive, considerable progress has been made in discovering non‐peptide agonists and positive allosteric modulators (PAMs) of GLP‐1 receptors with demonstrated efficacy. Many of these compounds induce biased signalling in GLP‐1 receptor‐mediated functional pathways. High‐quality structures of GLP‐1 receptors in both inactive and active states have been reported, revealing detailed molecular interactions between GLP‐1 receptors and non‐peptide agonists or PAMs. These progresses raise the exciting possibility of developing non‐peptide drugs of GLP‐1 receptors as alternative treatments for Type 2 diabetes. The insight into the interactions between the receptor and the non‐peptide ligand is also useful for developing non‐peptide ligands targeting other Class B GPCRs.
3.2.2. Compound M_4 and C‐1
Following the publication of the structure of two Class B GPCRs, the CRF1 receptor (PDB ID: 4K5Y) and the glucagon receptor (PDB ID: 4L6R), our group constructed the homology models of human GLP‐1 receptors, based on those two crystal structures and carried out structure‐based virtual screening of 5689 compounds from the ZINC database (Redij, Chaudhari, et al., 2019). These compounds have similar physicochemical properties to those of potential low MW agonists of GLP‐1 receptors and were identified through ligand‐based similarity search. Eight top‐ranked compounds from virtual screening were selected and evaluated using the GLP‐1 receptor‐dependent luciferase reporter system. Two compounds were confirmed to activate human GLP‐1 receptors in a dose‐dependent manner and one synergized with GLP‐1 to stimulate GLP‐1 receptor activity, compound 17 (M_4) (Figure 3, [17]). Using in vitro insulin secretion assay in INS‐1832/13 cells, compound M_4 induced glucose‐dependent insulin secretion.
When the cryo‐EM structure of GLP‐1 receptors at the active state became available (Y. Zhang et al., 2017), we carried out another round of structure‐based screening studies using this structure and identified another compound as a PAM for GLP‐1 receptors, compound 18 (C‐1) (Figure 3, [18]) (Redij, Ma, et al., 2019). Using the same GLP‐1 receptor‐dependent luciferase reporter system, compound C‐1 activates human GLP‐1 receptors in a dose‐dependent manner. When combined with GLP‐1, C‐1 improves GLP‐1’s affinity and efficacy to human GLP‐1 receptors. Using in vitro insulin secretion assay in INS‐1832/13 cells, compound C‐1 (9.7 μM), induced insulin secretion at the similar level as that of GLP‐1 (181 nM). Combined with GLP‐1, C‐1 again showed the synergistic effect in stimulating insulin secretion. Despite its modest activity, this compound demonstrates favourable drug‐like properties. For instance, with the molecular weight of 399, this compound represents one of the smallest known PAMs for the GLP‐1 receptor.
ACS Omega. 2019 Jan 31; 4(1): 961–970. doi: 10.1021/acsomega.8b03052. PMCID: PMC6648429. PMID: 31459371
Structural Modeling and in Silico Screening of Potential Small-Molecule Allosteric Agonists of a Glucagon-like Peptide 1 Receptor
Tejashree Redij,†⊥ Rajan Chaudhari,‡⊥# Zhiyu Li,§ Xianxin Hua,∥ and Zhijun Li*†‡
Abstract
The glucagon-like peptide 1 receptor (GLP-1R) belongs to the pharmaceutically important class B family of G-protein-coupled receptors (GPCRs), and its incretin peptide ligand GLP-1 analogs are adopted drugs for the treatment of type 2 diabetes. Despite remarkable antidiabetic effects, GLP-1 peptide-based drugs are limited by the need of injection. On the other hand, developing nonpeptidic small-molecule drugs targeting GLP-1R remains elusive. Here, we first constructed a three-dimensional structure model of the transmembrane (TM) domain of human GLP-1R using homology modeling and conformational sampling techniques. Next, a potential allosteric binding site on the TM domain was predicted computationally. In silico screening of druglike compounds against this predicted allosteric site has identified nine compounds as potential GLP-1R agonists. The independent agonistic activity of two compounds was subsequently confirmed using a cAMP response element-based luciferase reporting system. One compound was also shown to stimulate insulin secretion through in vitro assay. In addition, this compound synergized with GLP-1 to activate human GLP-1R. These results demonstrated that allosteric regulation potentially exists in GLP-1R and can be exploited for developing small-molecule agonists. The success of this work will help pave the way for small-molecule drug discovery targeting other class B GPCRs through allosteric regulations.