The built-in databases
To build the databases, the creation of an intermediate layer to transform some of the public components was needed. Bioconductor R package downloads the SQLiteProteinOnly DB, which contains the results for the assays that have been carried out with single proteins. The chemical information of the compounds used, can be downloaded as ASN, and then converted to json with popular python scripts. The disease link information is released by the Open Targets web as json files, that can be easily imported into MongoDB, and then, relevant data passed to SQLite DBs through csv export/import functions. ChemBl DB was directly downloaded through the ChemBl site, and a protein to gene to ensemble converter Uniprot was incorporated to either DB downloading and importing the csv file. This converter is especially important, as not all the tables contain the same target/gene/protein identifiers, and the uniprot converting module helps to link some interesting tables.
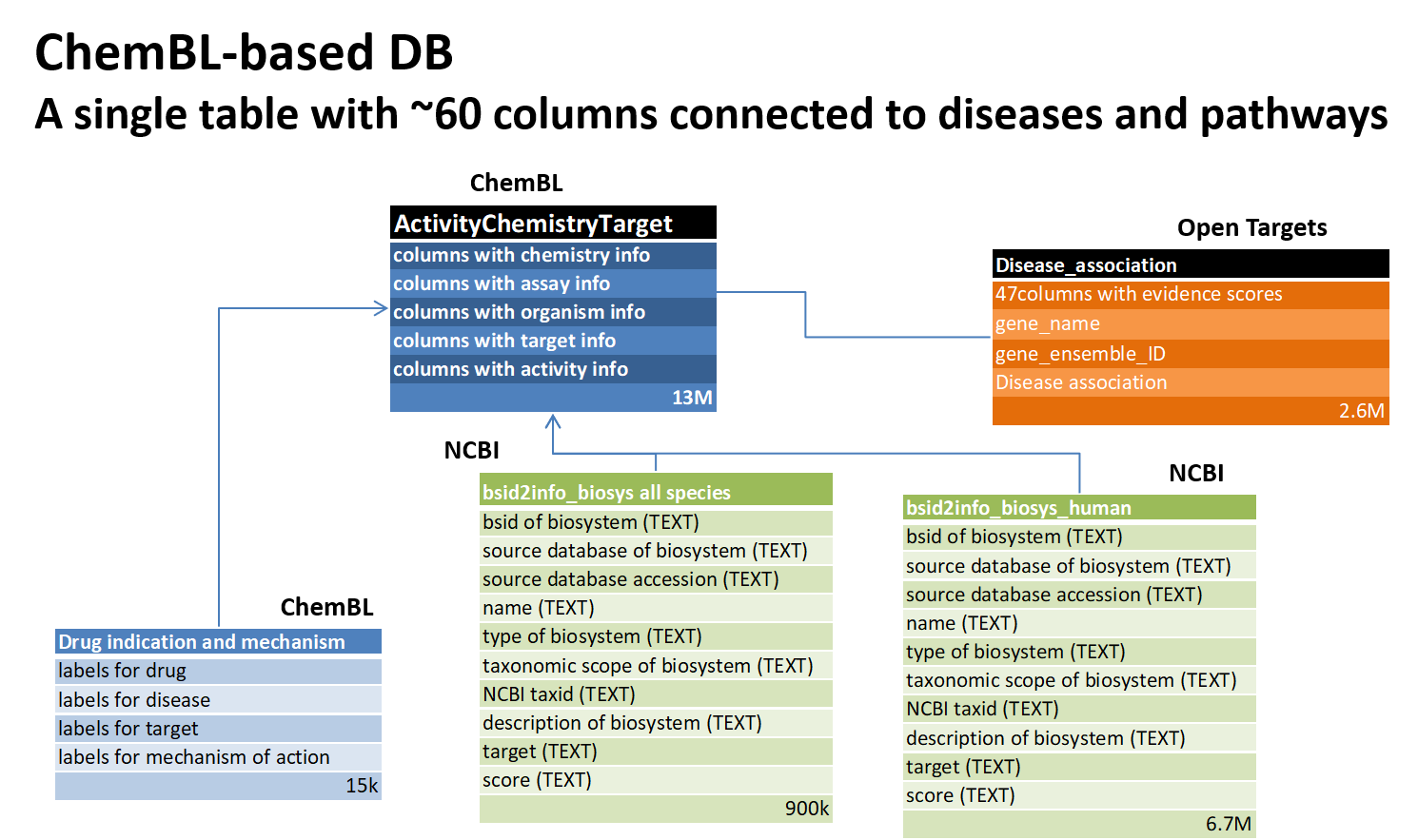
ChemBl-based-database
ChemBl allows direct download of the database in several formats: MySQL, Oracle, Postgree and SQLite. For this work we have chosen the last format because there is already a downloadable SQLite DB for NCBI, and there were several, easy and documented R libraries with good literature references. In order to make ML consultation easier, the variables considered useful for ML procedures were selected from the 76 tables of the ChemBl DB. The Biosystems table from NCBI was edited and transformed to generate a human biosystem table, both containing all biosystems identity labels. Both were then imported into the transformed ChemBl DB. Last but not least, two disease association tables : “Disease association” from open Targets and “drug indication and mechanism” from ChemBl, this was deliberately excluded from the query below as it caused an increase in the final table size by causing replicates, as a disease may be cured by several drugs and a single drug may work on several diseases. See the schema below.
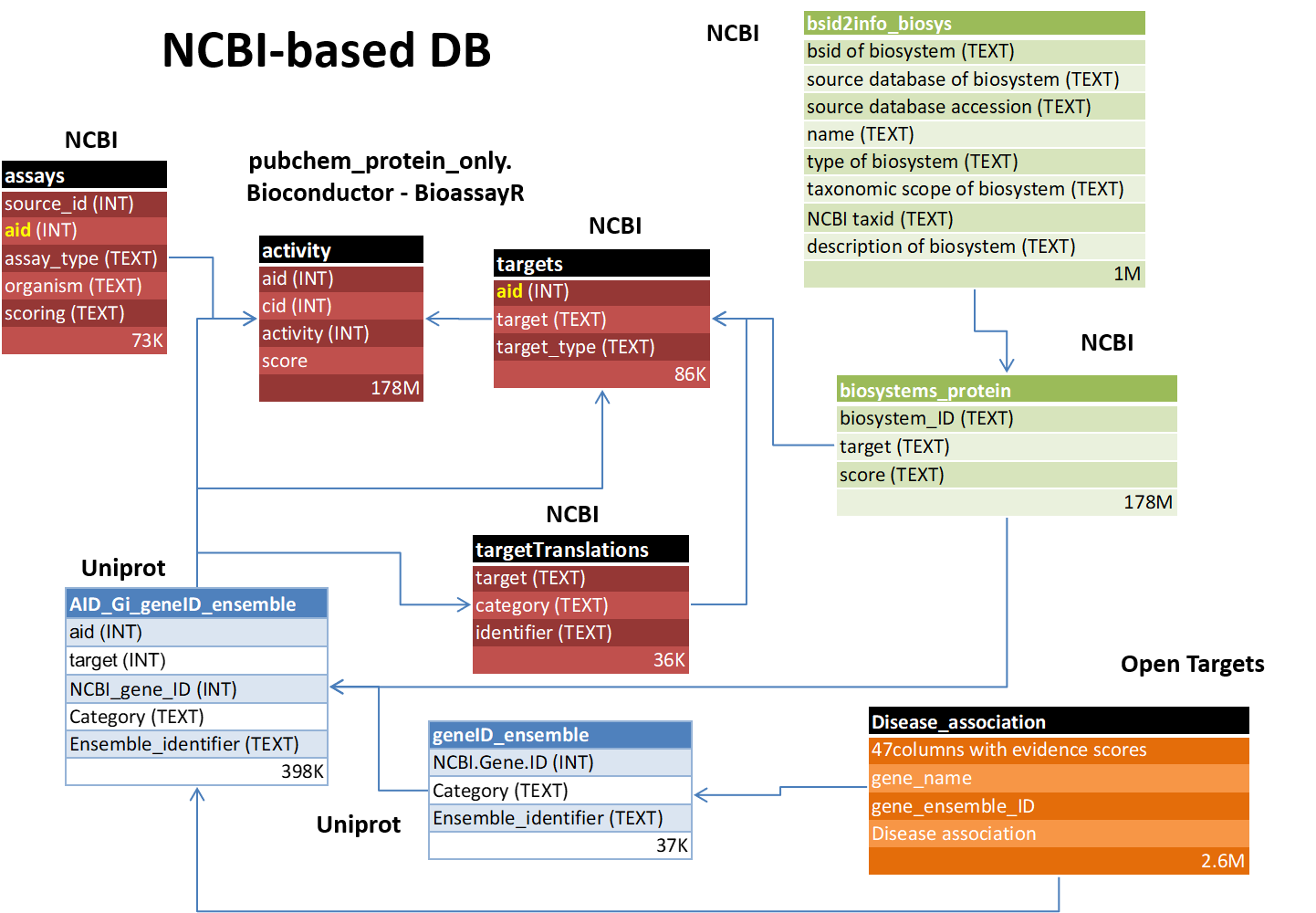
NCBI-based DB
Bioconductor R package downloads the SQLiteProteinOnly DB, which contains the results for the assays that have been carried out with single proteins. The chemical information of the compounds used, can be downloaded as ASN, and then converted to json with popular python scripts. The disease link information is released by the Open Targets web as json files, which can be easily imported into MongoDB, and then, relevant data passed to SQLite DBs through csv export/import functions .